Abstract
GlycoSuiteDB is an annotated and curated relational database of glycan structures reported in the literature. It contains information on the glycan type, core type, linkages and anomeric configurations, mass, composition and the analytical methods used by the researchers to determine the glycan structure. Native and recombinant sources are detailed, including species, tissue and/or cell type, cell line, strain, life stage, disease, and if known the protein to which the glycan structures are attached. There are links to SWISS-PROT/TrEMBL and PubMed where applicable. Recent developments include the implementation of searching by 2D structure and substructure, disease and reference. The database is updated twice a year, and now contains over 7650 entries. Access to GlycoSuiteDB is available at http://www.glycosuite.com.
INTRODUCTION
Glycoproteins are widely distributed in nature. They have been found in species from bacteria, viruses and fungi to fish, birds and humans. Their biological functions are many and varied, and there are many instances where the glycan structures have been shown to have significant importance. For example, glycosylation of Asn-319 on rabies virus glycoprotein is essential for the secretion of soluble rabies virus glycoprotein (1). Changes in levels and types of glycosylation are also associated with disease. It has been shown, for example, that detecting changes in glycan structure may be used as a diagnostic for aggressive breast cancer (2).
There exists a wide diversity of glycan structures. This comes from the variation in the type, number and position of individual sugar residues, the degree of branching, and the level of acetylation, methylation, sialylation, phosphorylation and sulfation. This emphasizes the need for a consistent curated catalogue of known glycan structures against which to compare newly discovered structures.
GlycoSuiteDB (3) currently contains glycan structures derived from glycoproteins of many different biological sources, and from free oligosaccharides isolated from biologically important fluids such as milk, saliva and urine. The database has been constructed to allow researchers to search for precedence and thus have more confidence in making assumptions of glycan structure. GlycoSuiteDB assists researchers to see what is already known about glycan structures attached to proteins.
GlycoSuiteDB is available on the web (www.glycosuite.com) and there has been considerable focus on data standardization, which means that it is easily searchable and accurate (4). Queries may be performed using monosaccharide composition, glycan 2D structure and substructure, glycan mass, taxonomy, tissue or cell type, glycoprotein, disease, reference or a combination of these query types. GlycoSuiteDB is extensively linked with the SWISS-PROT protein database (5), the ExPASy tool GlycoMod (6) and PubMed. Further links to other online databases, such as the NCBI taxonomy (7) and the Online Mendelian Inheritance in Man (OMIM) (8) databases, are planned.
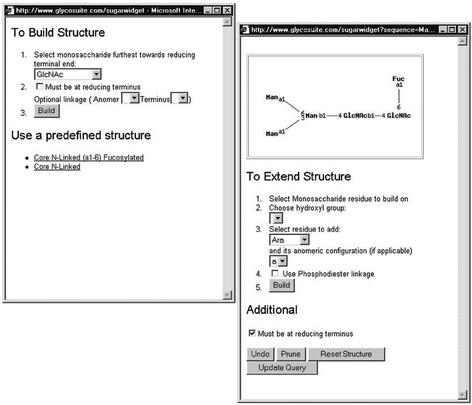
NEW QUERY BY COMPLETE OR PARTIAL 2D STRUCTURE
An exciting new feature has been added to GlycoSuiteDB. We have now enabled searching by complete or partial 2D structure. We have developed a novel search method that ensures that all matching structures are found and we have added a new user-friendly interface for drawing 2D structures or partial structures.
The user may either:
Draw a new structure by choosing a monosaccharide residue from the drop-down list to appear furthest towards (or at) the reducing terminus of the query structure (see Fig. 1). The anomeric configuration and the hydroxyl group on the adjacent residue to which this monosaccharide is attached may also be entered. Note: it is possible to specify if this residue must be located at the reducing terminus (i.e. is the monosaccharide residue directly linked to the protein backbone) or internally with a glycan structure. Or,
Select a predefined structure.
Figure 1.
Query by structure—user interface for building and extending a structure. This structure is then used to directly query the GlycoSuite database.
To extend the structure (see Fig. 1) the user:
Clicks on the monosaccharide residue to build on (it will turn red);
Selects the hydroxyl group to link to;
Chooses a monosaccharide or substituent in the drop-down list;
Selects its anomeric configuration where appropriate; and
Clicks ‘Build’. The user may tick the checkbox for phosphodiester linkage if required.
These steps are repeated as desired. When the query structure is complete the user simply clicks ‘Update Query’. The structure will then be visible in the main GlycoSuiteDB query page and the query can be executed.
Added features: ‘Undo’ will delete the last addition; ‘Prune’ will delete the selected residue and anything attached to the non-reducing terminus (left-hand side) of it; ‘Reset Structure’ will clear the structure and return to the beginning.
This new query type enables the user to rapidly and correctly identify glycan structures containing significant branching and specific epitopes, such as Lewis X or blood group A, that may be of biological importance. This new query type can also be combined with any of the existing query types in an advanced query.
GlycoSuiteDB DATA CONTENT
GlycoSuiteDB is updated at least twice per year. Release 4.0, August 14, 2002, contained more than 7650 entries, extracted from over 740 scientific research articles. Currently, the database contains most O-linked glycans published since 1950, and most N-linked glycans in the literature from the years 1990–2002.
There are 2757 unique glycan structures in the database, 1630 of which are completely characterized (871 N-linked, 689 O-linked and 70 other). There are more than 800 different monosaccharide compositions represented in GlycoSuiteDB.
There are currently 969 individual biological sources represented, from ∼200 different tissue or cell types. Glycan structures isolated from more than 160 species are presented in GlycoSuiteDB.
Glycan structures from ∼500 individual proteins are given and more than 3500 entries have links to protein sequences in SWISS-PROT.
REFERENCES
- 1.Wojczyk B.S., Stwora-Wojczyk,M., Shakin-Eshelman,S., Wunner,W.H. and Spitalnik,S.L. (1998) The role of site-specific N-glycosylation in secretion of soluble forms of rabies virus glycoprotein. Glycobiology, 8, 121–130. [DOI] [PubMed] [Google Scholar]
- 2.Dwek M.V., Ross,H.A. and Leathem,A.J. (2001) Proteome and glycosylation mapping identifies post-translational modifications associated with aggressive breast cancer. Proteomics, 1, 756–762. [DOI] [PubMed] [Google Scholar]
- 3.Cooper C.A., Harrison,M.J., Wilkins,M.R. and Packer,N.H. (2001) GlycoSuiteDB: a new curated relational database of glycoprotein glycan structures and their biological sources. Nucleic Acids Res., 29, 332–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cooper C.A., Harrison,M.J., Webster,J.M., Wilkins,M.R. and Packer,N.H. (2002) Data standardisation in GlycoSuiteDB. Pac. Symp. Biocomput. 2002, 297–309. [DOI] [PubMed] [Google Scholar]
- 5.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper C.A., Gasteiger,E. and Packer,N.H. (2001) GlycoMod—A software tool for determining glycosylation compositions from mass spectrometric data. Proteomics, 1, 340–349. [DOI] [PubMed] [Google Scholar]
- 7.Wheeler D.L., Chappey,C., Lash,A.E., Leipe,D.D., Madden,T.L., Schuler,G.D., Tatusova,T.A. and Rapp,B.A. (2000) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res., 28, 10–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKusick V.A. (1998) Mendelian Inheritance in Man. Catalogs of Human Genes and Genetic Disorders, 12th edn. The Johns Hopkins University Press, Baltimore, MD.