Abstract
The HIV reverse transcriptase and protease sequence database is an on-line relational database that catalogues evolutionary and drug-related sequence variation in the human immunodeficiency virus (HIV) reverse transcriptase (RT) and protease enzymes, the molecular targets of antiretroviral therapy (http://hivdb.stanford.edu). The database contains a compilation of nearly all published HIV RT and protease sequences, including submissions to GenBank, sequences published in journal articles and sequences of HIV isolates from persons participating in clinical trials. Sequences are linked to data about the source of the sequence, the antiretroviral drug treatment history of the person from whom the sequence was obtained and the results of in vitro drug susceptibility testing. Sequence data on two new molecular targets of HIV drug therapy—gp41 (cell fusion) and integrase—will be added to the database in 2003.
INTRODUCTION
Antiretroviral drug resistance is a major obstacle to the successful treatment of human immunodeficiency virus type 1 (HIV-1) infection. A large number of retrospective and prospective studies have demonstrated that the presence of drug resistance before starting a treatment regimen is an independent predictor of success of that regimen (1). As a result, several expert panels have recommended that HIV reverse transcriptase (RT) and protease sequencing be done to help physicians select antiretroviral drugs for their patients and genotypic resistance testing has been part of routine clinical care for the past several years (2).
The HIV RT and protease sequence database (HIVRT&PrDB) is intended to assist scientists designing new HIV-1 drugs, clinical investigators studying HIV-1 drug resistance and clinicians using genotypic HIV-1 drug resistance tests (3). The database links sequence changes in the molecular targets of HIV-1 therapy to other forms of data including treatment history and phenotypic (drug susceptibility) data. Data on the virological response (plasma HIV-1 RNA levels) to a new treatment regimen have been added and will soon be accessible over the web.
The HIVRT&PrDB is a relational database with 19 normalized (nonredundant) core tables, 10 look-up tables and about 20 derived tables. The database is implemented using MySQL on a Linux platform. There are several major hierarchical relationships linking key entities in the database: (i) patient treatment history (list of drug regimens and their start and stop dates); (ii) patient isolate (clinical) sequence drug susceptibility result; (iii) isolate (laboratory) drug susceptibility result; and (iv) patient plasma HIV-1 RNA level. Sequences are stored in a virtual alignment with the subtype B consensus sequence; thus amino acid sequences are also represented as lists of differences from the consensus sequence.
The HIVRT&PrDB contains data from more than 420 published papers. Sequences are available on HIV-1 isolates from more than 7000 individuals and from about 500 laboratory isolates containing mutations generated by virus passage or site-directed mutagenesis. About 20 000 drug susceptibility results from tests performed on more than 2000 virus isolates are available. Figures 1 and 2 contain composite alignments showing 193 protease and 395 RT mutations present at a frequency of >0.1% in HIV-1 isolates from treated and untreated persons. Figure 3 shows a summary of the drug susceptibility results available on each of the 16 approved antiretroviral drugs.
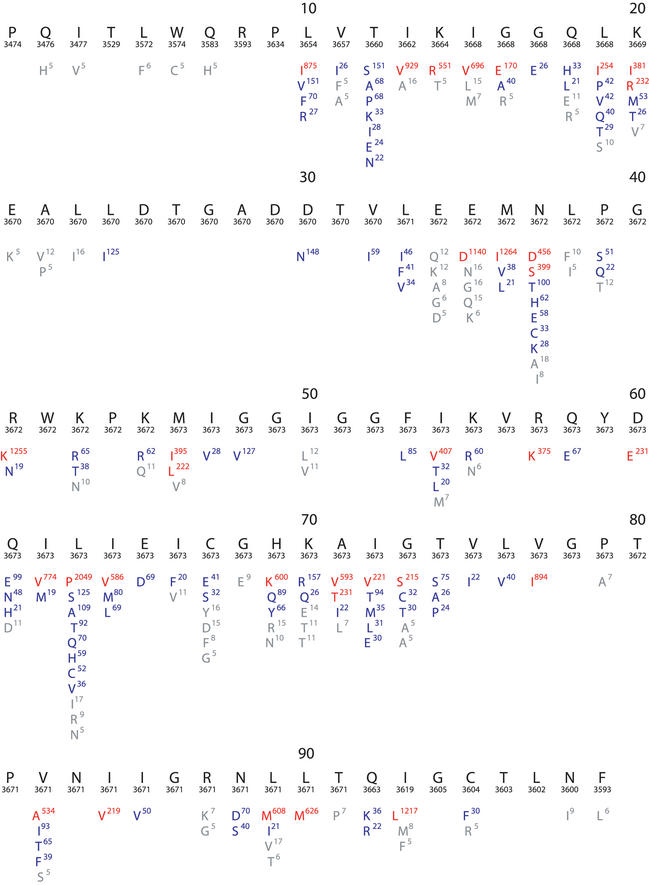
Figure 1.
Composite sequence alignment of HIV-1 protease, positions 1–99. This figure resulted from a query that retrieved all HIV-1 sequences in the database including those belonging to different subtypes and those obtained from treated and untreated individuals. Beneath the numbered consensus sequence is the number of isolates in the database for which sequence information at the position is available. The remaining lines in each row show the frequency of variation at each position in the database. Amino acids shown in red have a mutation rate ≥5%; those in blue have a mutation rate between 1 and 5%; and those in grey of 0.1–1%.
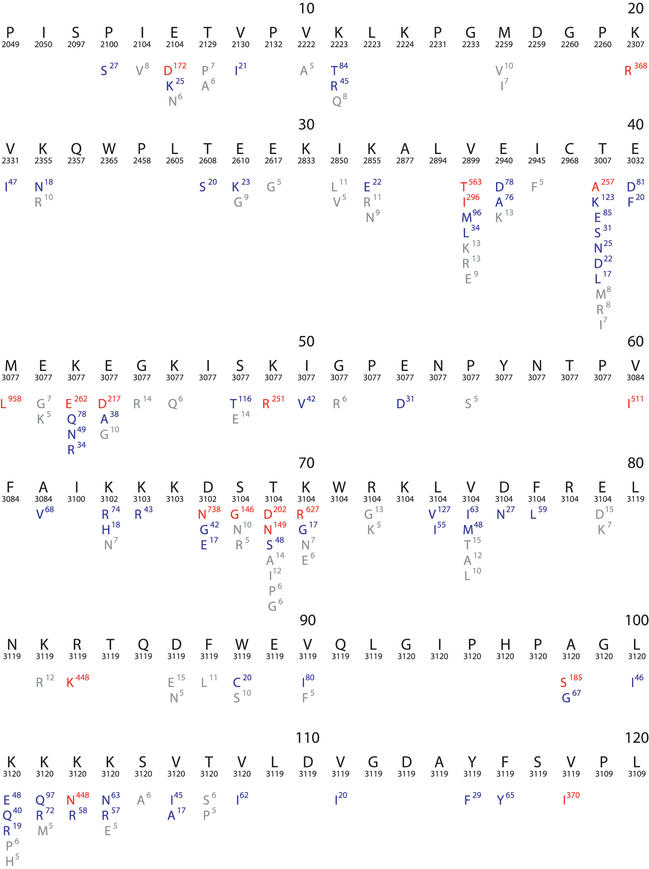
Figure 2.
Composite sequence alignment of HIV-1 RT, positions 1–240. Although the RT enzyme has 560 positions, nearly all drug-resistance mutations are found between positions 40–240. This figure resulted from a query that retrieved all HIV-1 sequences in the database including those belonging to different subtypes and those obtained from treated and untreated individuals. Beneath the numbered consensus sequence is the number of isolates in the database for which sequence information at the position is available. The remaining lines in each row show the frequency of variation at each position in the database. Amino acids shown in red have a mutation rate ≥5%; those in blue have a mutation rate between 1 and 5%; and those in grey of 0.1–1%.
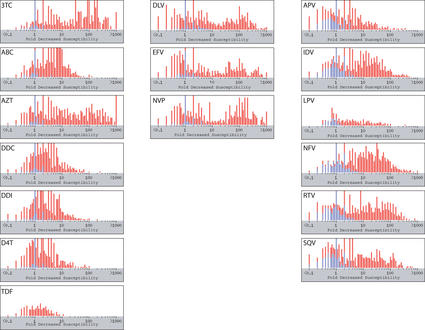
Figure 3.
Phenotypic drug susceptibility data on about 2000 HIV-1 isolates. Drug susceptibility to each of the 16 FDA-approved drugs are shown. The first column contains the nucleoside/nucleotide RT inhibitors: 3TC (lamivudine), ABC (abacavir), AZT (zidovudine), DDC (zalcitibine), DDI (didanosine), D4T (stavudine) and TDF (tenofovir). The second column contains the nonnucleoside RT inhibitors: DLV (delavirdine), EFV (efavirenz) and NVP (nevirapine). The third column contains the protease inhibitors: APV (amprenavir), IDV (indinavir), LPV (lopinavir), NFV (nelfinavir), RTV (ritonavir) and SQV (saquinavir). Each point represents the fold decrease in susceptibility of a single virus isolate compared with the susceptibility of a wildtype control isolate (X-axis). Blue points represent results on tests with no known drug-resistance mutations. Red points represent results on tests of isolates with at least one drug-resistance mutation. The maximum number of tests shown (Y-axis) that yield the same result is 40. A large proportion of these results were obtained using one of three well-characterized recombinant virus drug susceptibility assays (11–13).
The database allows users to retrieve sets of sequences meeting specific criteria. Commonly submitted queries include: (i) the retrieval of sequences of HIV-1 isolates from patients receiving a specific drug treatment, (ii) the retrieval of sequences of HIV-1 isolates containing mutations at specific protease or RT positions, (iii) the retrieval of drug susceptibility data on HIV-1 isolates containing specific mutations or combinations of mutations, and (iv) a summary of data in any particular reference.
Each query initially returns data in the form of a table and each record in the returned table contains 8 or more columns of data. The data returned include: (i) hyperlinks to the MEDLINE abstract and GenBank record, (ii) a list of mutations in the sequence, (iii) a classification of the sequence by patient and time point, (iv) drug treatment history, and (v) additional data depending upon the query (e.g. drug susceptibility results, phylogenetic data, technical data about virus isolation and sequencing). Together with this table, users are given the option of downloading or viewing the raw sequence data in a variety of formats.
SEQUENCE INTERPRETATION PROGRAMS
The database website contains three sequence interpretation programs. The first program, HIVseq, accepts user-submitted RT and protease sequences, compares them to a reference sequence and uses the differences (mutations) as query parameters for interrogating the database (4). HIVseq allows users to examine new sequences in the context of previously published sequences, providing two main advantages. First, unusual sequence results can be detected and immediately rechecked. Second, unexpected associations between sequences or isolates can be discovered when the program retrieves data on isolates sharing one or more mutations with the new sequence.
The second program, a drug resistance interpretation program (HIVdb), accepts user-submitted protease and RT sequences and returns inferred levels of resistance to the 16 FDA-approved antiretroviral drugs. Each drug resistance mutation is assigned a drug penalty score; the total score for a drug is derived by adding the scores associated with each mutation. Using the total drug score, the program reports one of the following levels of inferred drug resistance: susceptible, potential low-level resistance, low-level resistance, intermediate resistance and high-level resistance.
The third program (HIValg), allows researchers to compare the output of different publicly available drug-resistance algorithms on the same sequence or set of sequences. The algorithms used by this program are encoded using a programming platform or Algorithm Specification Interface (ASI) developed to facilitate the comparison of HIV genotypic resistance algorithms. ASI consists of an XML format for specifying an algorithm and a compiler that transforms the XML into executable code.
NEW ADDITIONS PLANNED FOR THE HIVRT&PrDB
Two additions to the database are planned: (i) gp41 sequences and data on resistance to fusion inhibitors. The first fusion inhibitor, enfuvirtide (T-20) has been shown to have potent antiretroviral activity in clinical trials (5,6) and is likely to be approved in 2003. A wide range of mutations in gp41 contributing to T-20 resistance, most occurring between residues 36–45, have been reported, but mutations outside of this region also appear to contribute to drug resistance (7,8); (ii) integrase sequences. A new class of compounds that inhibit HIV-1 integrase have been shown to be active in vitro and in a SHIV rhesus macaque model of infection (9,10).
REFERENCES
- 1.Shafer R.W. (2002) Genotypic testing for human immunodeficiency virus type 1 drug resistance. Clin. Microbiol. Rev., 15, 247–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirsch M.S., Brun-Vezinet,F., D'Aquila,R.T., Hammer,S.M., Johnson,V.A., Kuritzkes,D.R., Loveday,C., Mellors,J.W., Clotet,B., Conway,B. et al. (2000) Antiretroviral drug resistance testing in adult HIV-1 infection: recommendations of an International AIDS Society-USA Panel. JAMA, 283, 2417–2426. [DOI] [PubMed] [Google Scholar]
- 3.Kantor R., Machekano,R., Gonzales,M.J., Dupnik,B.S., Schapiro,J.M. and Shafer,R.W. (2001) Human immunodeficiency virus reverse transcriptase and protease sequence database: An expanded model integrating natural language text and sequence analysis. Nucleic Acids Res., 29, 296–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shafer R.W., Jung,D.R. and Betts,B.J. (2000) Human immunodeficiency virus type 1 reverse transcriptase and protease mutation search engine for queries. Nature Med., 6, 1290–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kilby J.M., Hopkins,S., Venetta,T.M., DiMassimo,B., Cloud,G.A., Lee,J.Y., Alldredge,L., Hunter,E., Lambert,D., Bolognesi,D. et al. (1998) Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nature Med., 4, 1302–1307. [DOI] [PubMed] [Google Scholar]
- 6.Kilby J.M., Lalezari,J.P., Eron,J.J., Carlson,M., Cohen,C., Arduino,R.C., Goodgame,J.C., Gallant,J.E., Volberding,P., Murphy,R.L. et al. (2002) The Safety, Plasma Pharmacokinetic, and Antiviral Activity of Subcutaneous Enfuvirtide (T-20), a Peptide Inhibitor of gp41-Medicated Virus Fusion, in HIV-Infected Adults. AIDS Res. Hum. Retroviruses, 18, 685–693. [DOI] [PubMed] [Google Scholar]
- 7.Wei X., Decker,J.M., Liu,H., Zhang,Z., Arani,R.B., Kilby,J.M., Saag,M.S., Wu,X., Shaw,G.M. and Kappes,J.C. (2002) Emergence of resistant hyman immunodeciency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother., 46, 1896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sista P.R., Melby,T., Greenberg,M., Davison,D., Jin,L., Mosier,S., Mink,M., Nelson,E., Fang,L., Cammack,N. et al. (2002) Characterization of baseline and treatment-emergent resistance to T-20 (enfuvirtide) observed in phase II clinical trials: substitutions in gp41 amino acids 36--45 and enfuvirtide susceptibility of virus isolates. Antivir. Ther., 7, S16–S17. [Google Scholar]
- 9.Grobler J.A., Stillmock,K., Hu,B., Witmer,M., Felock,P., Espeseth,A.S., Wolfe,A., Egbertson,M., Bourgeois,M., Melamed,J. et al. (2002) Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotransferase enzymes. Proc. Natl Acad. Sci. USA, 99, 6661–6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hazuda D.J. and HIV-1 Integrase Inhibitor Discovery Team (2002) A novel HIV-1 integrase inhibitor mediates sustained suppression of viral replication and CD4 depletion in a SHIV rhesus macaque model of infection. Antivir. Ther., 7, S3. [Google Scholar]
- 11.Hertogs K., de Bethune,M.P., Miller,V., Ivens,T., Schel,P., Van Cauwenberge,A., Van Den Eynde,C., Van Gerwen,V., Azijn,H., Van Houtte,M. et al. (1998) A rapid method for simultaneous detection of phenotypic resistance to inhibitors of protease and reverse transcriptase in recombinant human immunodeficiency veirus type 1 isolates from patients treated with antiretroviral drugs. Antimicrob. Agents Chemother., 42, 269–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petropoulos C.J., Parkin,N.T., Limoli,K.L., Lie,Y.S., Wrin,T., Huang,W., Tian,H., Smith,D., Winslow,G.A., Capon,D.J. et al. (2000) A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob. Agents Chemother., 44, 920–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beerenwinkel N., Schmidt,B., Walter,H., Kaiser,R., Lengauer,T., Hoffmann,D., Korn,K. and Selbig,J. (2002) Diversity and complexity of HIV-1 drug resistance: a bioinformatics approach to predicting phenotype from genotype. Proc. Natl Acad. Sci. USA, 99, 8271–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]