Abstract
PEP is a database of Predictions for Entire Proteomes. The database contains summaries of analyses of protein sequences from a range of organisms representing all three major kingdoms of life: eukaryotes, prokaryotes and archaea. All proteins publicly available for organisms were aligned against SWISS-PROT, TrEMBL and PDB. Additionally, the following annotations are provided: secondary structure, transmembrane helices, coiled coils, regions of low complexity, signal peptides, PROSITE motifs, nuclear localization signals and classes of cellular function. Proteins that contain long regions without regular secondary structure are also identified. We have produced a related database of structural domain-like fragments derived from PEP and clusters based on homology between all fragments. The PEP database, fragments and clusters are distributed freely as a set of flat files and have been integrated into SRS. The PEP group of databases can be accessed from: http://cubic.bioc.columbia.edu/pep.
INTRODUCTION
Large-scale genome sequencing has provided us with the building blocks of living organisms. However, to obtain new insights into physiological and biochemical processes, it is essential to analyse and catalogue the structural and functional features of each individual protein in the genome. We refer to all these proteins as the proteome of an organism. With bioinformatics tools becoming more and more accurate, it is now possible to systematically generate various reliable structural and functional annotations for entire proteomes and make the information easily accessible in different ways. Such predictions for entire proteomes suggest conclusions in context of comparative genomics (1–4) and provide crucial information in the context of structural genomics (4).
DATABASE DESCRIPTION
Design
Predictions for Entire Proteomes (PEP) has been created as a generic bioinformatics resource. The objective of predicting features for all constituent peptides of proteomes has been to allow users to data mine proteomes globally, or to retrieve sequences of particular interest and to review predictions on individual sequences. PEP entries constitute the sequences of proteins as given by the Open Reading Frames (ORFs) from sequencing projects. We have dissected the ORFs into putative structural domains or fragments. The fragments in turn have been clustered based upon sequence similarity. The North East Structural Genomics (NESG) consortium (5) is using the fragments and clusters for target selection purposes (http://www.nesg.org).
Content
The PEP database is a summary of analyses for publicly available proteomes (2). All PEP entries were aligned against proteins taken from SWISS-PROT (6), TrEMBL (6) and PDB (7). ORFs were taken from FlyBase (8), WormBase (9) and databases at the NCBI. Protein sequences from each proteome were: (i) aligned against the SWISS-PROT, TrEMBL and PDB using pairwise BLAST (10), PSI-BLAST (11) and the dynamic programming method MaxHom (12); (ii) assigned secondary structure and other sequence based predictions; and (iii) assigned predicted cellular function according to EUCLID (13). The structural and functional features we analysed included:
coiled-coil regions predicted by COILS (14)
percentage relative solvent accessibility predicted by PROFacc (15,16)
transmembrane helices assigned by PHDhtm (16)
low sequence complexity regions according to SEG (17)
long stretches of non-regular secondary structure (NORS) (3)
presence and location of signal peptide cleavage sites identified by SignalP (18)
PROSITE motifs (19)
cellular functional classes assigned by EUCLID (13)
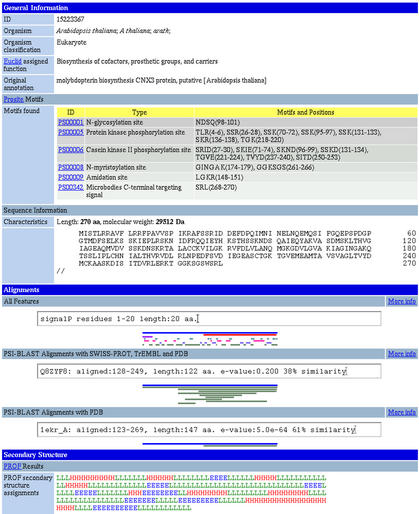
An example of a PEP entry is shown in Figure 1.
Figure 1.
Screen-dump of a PEP entry. Some general information (organism, sequence length and molecular weight) about the PEP sequence is provided and cellular function as predicted by Euclid. The three graphics are interactive when viewed on the web, and the text above each changes according to the region of the sequence being examined. The first graphic, labelled ‘All Features’, shows structural and functional features of the sequence and their positions in different colours. In this example, the 270 amino acid sequence is predicted to have a signal peptide 20 residues in length. Also, a long region from residues 123–268 was shown to have homology with a PDB entry. Additionally, helix, beta-sheet and loop regions are indicated. The second and third graphics show the results of PSI-BLAST alignments of the PEP sequence against SWISS-PROT, TrEMBL and PDB databases.
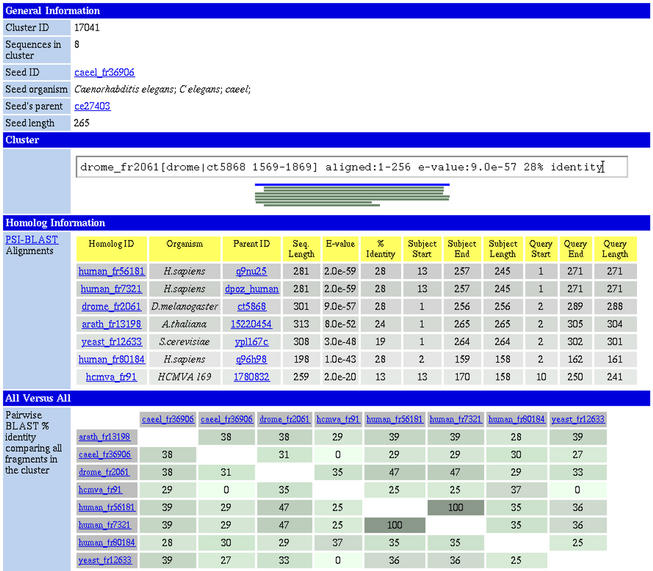
The structural domain-like fragments have been analysed for the same features i.e. database homologies, sequence based features and cellular function. The fragment results are available as a database named CHOP. These fragments have been clustered using PSI-BLAST with an ‘all versus all’ sequence similarity comparison to find distinct protein families. The clusters are also available as a database (Fig. 2).
Figure 2.
Clustering a structural family. PEP contains clusters of proteins sharing a common structural region corresponding to putative structural domains. Given are the alignments of member sequences against the seed of the cluster produced by PSI-BLAST and results of a pairwise BLAST ‘all versus all’ comparisons of all the proteins in the cluster.
Table 1 shows proteomes we have analysed to date. We will analyse more and add the results to PEP in the future. Currently, we are using a 28 node (58 processor) Dell cluster to perform our predictions.
Table 1. Excerpt from list of organisms annotated in PEP.
| Classification | Organism | Number of proteins analysed |
|---|---|---|
| Eukaryotes | Arabidopsis thaliana | 25 542 |
| Caenorhabditis elegans | 20 251 | |
| Drosophila melanogaster | 14 304 | |
| Homo sapiens | 37 271 | |
| Saccharomyces cerevisiae | 6356 | |
| Prokaryotes | Aquifex aeolicus | 1522 |
| Borrelia burgdorferi | 850 | |
| Campylobacter jejuni | 1633 | |
| Chlamydia trachomatis | 894 | |
| Escherichia coli | 4281 | |
| Helicobacter pylori | 1564 | |
| Mycoplasma genitalium | 470 | |
| Mycoplasma pneumoniae | 688 | |
| Neisseria meningitidis | 2065 | |
| Rickettsia conorii | 1374 | |
| Ureaplasma urealyticum | 611 | |
| Archaea | Achaeoglobus fulgidus | 2407 |
| Aeropyrum pernix K1 | 2694 | |
| Halobacterium sp. (strain NRC-1) | 2058 | |
| Pyrococcus horikoshii | 2064 | |
| Sulfolobus solfataricus | 2977 | |
| Virus | Human cytomegalovirus (strain AD169) | 202 |
Availability and interface
The three databases (ORFs, fragments and clusters) are available as flat files and have been integrated into SRS (22). We distribute the full results of the analyses also, although they are quite large in size (gigabytes). The PEP databases can be accessed through the Columbia University Bioinformatics Center (CUBIC) web site: http://cubic.bioc.columbia.edu/pep.
PEP can be searched on many fields (over 40), some examples of which are ‘Euclid assigned function’, ‘number of coiled coil regions’, ‘length of non-regular secondary structure regions’, ‘number of alpha-helices’, ‘number of transmembrane helices’ and ‘length of signal peptide’. The proteomes can also be searched using a range of bioinformatics tools with their own sequences. The flat files can also be downloaded for local investigation.
Acknowledgments
ACKNOWLEDGEMENTS
Thanks to Dariusz Przybylski, Rajesh Nair and Kazimierz Wrzeszczynski (Columbia University) for providing preliminary information and programs. Thanks to the SRS team for their software. The work was supported by the grants 1-P50-GM62413-01 and RO1-GM63029-01 from the National Institutes of Health (NIH). Last, not least, thanks to all those who deposit their experimental data in public databases and to those who maintain these databases.
REFERENCES
- 1.Rost B. (2002) Did evolution leap to create the protein universe? Curr. Opin. Struct. Biol., 12, 409–416. [DOI] [PubMed] [Google Scholar]
- 2.Liu J. and Rost,B. (2001) Comparing function and structure between entire proteomes. Protein Sci., 10, 1970–1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu J., Tan,H. and Rost,B. (2002) Loopy proteins appear conserved in evolution. J. Mol. Biol., 322, 53–64. [DOI] [PubMed] [Google Scholar]
- 4.Liu J. and Rost,B. (2002) Target space for structural genomics revisited. Bioinformatics, 18, 922–933. [DOI] [PubMed] [Google Scholar]
- 5.Montelione G.T. (2001) Structural genomics: an approach to the protein folding problem. Proc. Natl Acad. Sci. USA, 98, 13488–13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TFEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The Flybase Consortium (2002) The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res., 30, 106–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stein L., Sternberg,P., Durbin,R., Thierry-Mieg,J. and Spieth,J. (2001) WormBase: network access to the genome and biology of Caenorhabditis elegans. Nucleic Acids Res., 29, 82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul S.F. and Gish,W. (1996) Local alignment statistics. Methods Enzymol., 266, 460–480. [DOI] [PubMed] [Google Scholar]
- 11.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sander C. and Schneider,R. (1991) Database of homology-derived protein structures and the structural meaning of sequence alignment. Proteins, 9, 56–68. [DOI] [PubMed] [Google Scholar]
- 13.Tamames J., Ouzounis,C., Casari,G., Sander,C. and Valencia,A. (1998) EUCLID: automatic classification of proteins in functional classes by their database annotations. Bioinformatics, 14, 542–543. [DOI] [PubMed] [Google Scholar]
- 14.Lupas A. (1996) Prediction and analysis of coiled-coil structures. Methods Enzymol., 266, 513–525. [DOI] [PubMed] [Google Scholar]
- 15.Rost B. (2001) Review: protein secondary structure prediction continues to rise. J. Struct. Biol., 134, 204–218. [DOI] [PubMed] [Google Scholar]
- 16.Rost B. (1996) PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol., 266, 525–539. [DOI] [PubMed] [Google Scholar]
- 17.Wootton J.C. and Federhen,S. (1996) Analysis of compositionally biased regions in sequence databases. Methods Enzymol., 266, 554–571. [DOI] [PubMed] [Google Scholar]
- 18.Nielsen H., Engelbrecht,J., Brunak,S. and von Heijne,G. (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng., 10, 1–6. [DOI] [PubMed] [Google Scholar]
- 19.Falquet L., Pagni,M., Bucher,P., Hulo,N., Sigrist,C.J., Hofmann,K. and Bairoch,A. (2002) The PROSITE database, its status in 2002. Nucleic Acids Res., 30, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cokol M., Nair,R. and Rost,B. (2000) Finding nuclear localization signals. EMBO Rep., 1, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nair R., Carter,P. and Rost,B. (2003) NLSdb: database of nuclear localization signals. Nucleic Acids Res., 31, 342–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Etzold T. and Argos,P. (1993) Transforming a set of biological flat file libraries to a fast access network. Comput. Appl. Biosci., 9, 49–57. [DOI] [PubMed] [Google Scholar]