Abstract
A high resolution map of the human genome previously has been constructed by using the G3 panel of human/hamster radiation hybrid cell lines and >15,000 unique human genetic markers. By determining whether human DNA sequences are present or absent in each of the hybrids, localization of single genes may routinely be achieved at ≈250-kb resolution. In this paper we have tested whether similarly precise localization might be achieved by phenotypic screening of the hybrids to facilitate positional cloning of unknown genes. We assayed the susceptibility of each of the hybrid cell lines to transduction by retroviral vectors bearing different retroviral envelope proteins that recognize receptors present on human but not on hamster cells. The results for each of the retroviral vectors were informative and allowed precise localization of the receptor genes for the RD114 cat endogenous retrovirus, xenotropic murine leukemia virus, and type C feline leukemia virus. After cloning of the receptors for these retroviruses, we found that standard genotypic mapping by PCR gave results that were nearly identical to those from phenotypic mapping. These experiments show that precise gene localization by phenotypic assay of radiation hybrids is practical and was not appreciably impacted by the known instability of such hybrid cells. This technique should be applicable to many other human genes having discernible phenotypes in hamster cells and, with completion of the human genome project, will allow rapid identification of unknown genes on the basis of phenotype.
The Stanford Human Genome Center (SHGC) G3 panel of radiation hybrid cell lines consists of 83 clones of hamster cells that contain multiple independent fragments of human DNA. The panel was generated by fusing irradiated human cells with thymidine kinase-negative hamster cells and selecting clones that express human thymidine kinase (1). At the radiation dose used (10,000 rad), the human DNA is broken into fragments with an average size of 4 megabases (Mb), and each hybrid contains ≈18% of the human genome. By using PCR to screen these hybrids for the presence or absence of >15,000 unique human DNA markers, a high-resolution map of the human genome has been developed. DNA from these hybrids now can be screened for the presence of any new unique sequence and the results can be submitted to a web-based server to determine the precise location of the DNA sequence within ≈250 kb (http://www-shgc.stanford.edu).
It also is possible to screen radiation hybrid cell lines for gene expression and thereby to map genes by phenotype. For example, in 1975, Goss and Harris (2) were the first to use radiation hybrids to determine linkage of four phenotypic markers scattered on the human X chromosome. However, although there is a long history of the use of human/hamster hybrid cell lines to localize and establish linkage maps for human genes based on phenotypic assay, this technique has not been applied to the high-resolution radiation hybrid panels that are now available. Previous studies have provided relatively crude localization data, and it was suspected that the known genetic instability of such hybrids, or lack of expression of genes located on short genomic fragments, might limit the precision of such analyses.
To examine whether genes could be precisely localized based on phenotypic assay of radiation hybrid cell lines, we have attempted to localize several retrovirus receptor genes. Retroviruses can use many different receptors for cell entry (3–5), and we have been interested in the identification and characterization of these cell-surface molecules. Knowledge of these receptors is important for an understanding of the evolution of retroviruses, for treatment of diseases caused by retroviruses, and for gene therapy applications involving retroviral vectors. Several retroviruses can infect human but not hamster cells, making receptor localization by phenotypic screening of human/hamster radiation hybrid cell lines possible. In this study, we have used retroviral vectors packaged into virions bearing different retroviral envelope (Env) proteins (pseudotypes) to screen the hybrid cells for the presence of the cognate virus receptors. By using this approach, we have been able to localize the receptors for the RD114 cat endogenous virus, the xenotropic murine leukemia virus (MuLV), and feline leukemia virus type C (FeLV-C). We subsequently cloned the first two of these receptors whereas others cloned the FeLV-C receptor, and we show that the results of genotypic mapping closely match those from phenotypic mapping.
Materials and Methods
Cell Culture.
HT-1080 human fibrosarcoma cells (American Type Culture Collection CCL-121) were grown in DMEM supplemented with 10% FBS (HyClone). A23 hamster cells and the A23-derived radiation hybrid clones (1) were grown in MEM-α supplemented with 10% FBS. The radiation hybrids were grown for ≤8 weeks before phenotypic analysis.
Retroviral Vectors.
Moloney MuLV-based retroviral vectors encoding human placental alkaline phosphatase and neomycin phosphotransferase (LAPSN, ref. 6) or green fluorescent protein and neomycin phosphotransferase (LNCG, ref. 7) were used to measure transduction rates. Helper-free retroviral vectors pseudotyped with Env proteins from the RD114 cat endogenous retrovirus, xenotropic MuLV, or 10A1 MuLV were produced by using FLYRD18 (8), PX/LAPSN or PX/LNCG (9), or PT67 (10) retrovirus packaging cells, respectively. Vesicular stomatitis virus G protein (VSV-G) pseudotype vectors were produced as described (11). To generate retroviral vectors bearing the FeLV-C Env, a DNA fragment containing the FeLV-C Sarma Env coding region (12) was inserted downstream of the cytomegalovirus immediate-early promoter, in place of the β-galactosidase cDNA, in the expression vector pCMVβ (CLONTECH) to make pCSI-EFSC. Cell lines producing FeLV-C pseudotype vectors were generated by introducing the pCSI-EFSC plasmid and the LAPSN vector into LGPS cells (13) that express the Moloney murine leukemia virus Gag-Pol proteins, and a clone that produced the highest titer of the LAPSN vector (PFSC/LAPSN c4) was identified. For measurement of transduction, cells were exposed to the retroviral vectors in the presence of 4 μg/ml Polybrene (Sigma), the cells were fed the next day, and on day 2 after vector addition the cells were stained for alkaline phosphatase expression as described (14) or were examined for green fluorescent protein expression with a fluorescence microscope. Results are expressed in alkaline phosphatase-positive or green fluorescent protein-positive focus-forming units (FFU) per ml of vector.
Chromosomal Gene Localization.
The chromosomal localizations of the retrovirus receptor genes were determined by PCR analysis of G3 radiation hybrid panel chromosomal DNA samples obtained from Research Genetics (Huntsville, AL). The human RDR gene was detected by using primers R1b15FN (5′-TGGCTGCTGGAGTACATGTG-3′) and R1b15RO (5′-CCCAGTGGGGGCTAGAATTC-3′) to produce a predicted 196-bp product. The human XPR1 gene was detected by using the primers X56F (5′-GAATGGTTGAAACCGGACATTG-3′) and X56R (5′-GCTTCATGAATGAAGGTACTGC-3′) to produce a predicted 144-bp product. The location of the human FLVCR gene was determined previously (15) by using the primers 5′-GCCCCTCTGTTTCAGCATTA-3′ and 5′-CTTGGTCTGTGGGACTGTCA-3′ to produce a predicted 273-bp product. The primers listed above are given in the order of forward and reverse primers with respect to the direction of gene transcription.
Results
The ability of several Env proteins to mediate retroviral vector transduction of human and hamster cells is shown in Table 1. Env proteins from the RD114 cat endogenous virus, xenotropic MuLV, and FeLV-C promoted efficient transduction of HT-1080 human cells but not the A23 hamster cells used to make the G3 panel of radiation hybrid cell lines. In contrast, otherwise identical vectors bearing the 10A1 MuLV Env protein or the surface glycoprotein from vesicular stomatitis virus G protein (VSV-G) could transduce both the human and the hamster cells, showing that the block to transduction of the hamster cells by the retroviral vectors with the RD114, xenotropic MuLV, and FeLV-C pseudotypes is at the level of virus entry mediated by Env. These results provide the basis for phenotypic screening of the radiation hybrids for the presence of the human receptors for the RD114, xenotropic MuLV, and FeLV-C Env proteins.
Table 1.
Transduction of human and hamster cells by retroviral vectors having different Env proteins (pseudotypes)
| Vector pseudotype | Vector transduction of:
|
|
|---|---|---|
| HT-1080 human cells | A23 hamster cells | |
| RD114 | + | − |
| Xenotropic | + | − |
| FeLV-C | + | − |
| 10A1 | + | + |
| VSV-G | + | + |
LAPSN or LNCG vectors with the indicated pseudotypes were added to HT-1080 human and A23 hamster cells. Cells were stained for alkaline phosphatase expression (LAPSN) or were examined for green fluorescent protein expression (LNCG) 2 days after vector exposure, and transduction is indicated as + (≥104 FFU/ml of vector preparation) or − (≤10 FFU/ml).
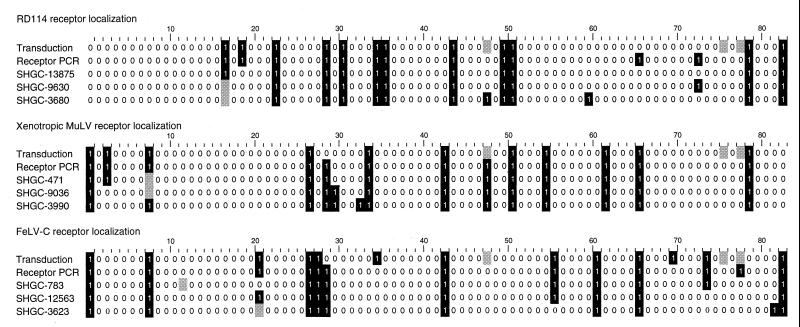
We found that 14–16% of the radiation hybrid cell lines were positive for transduction by retroviral vectors with RD114, xenotropic, or FeLV-C pseudotypes (Fig. 1, first row of each group). These results are consistent with a model involving a single gene encoding each retrovirus receptor and an estimate of the average human DNA content of each hybrid being 18% of the human genome. Evaluation of the results by using the SHGC radiation hybrid web server (http://www-shgc.stanford.edu) revealed that the receptors for RD114, xenotropic MuLV, and FeLV-C were localized at distances of 6, 6, and 21 centirays (cR), or about 140, 140, and 500 kb, from ordered markers at chromosome positions 19q13.3, 1q25.1, and 1q32.1, respectively. One centiray is defined as the distance over which radiation breakage occurs at 1% frequency, and for the radiation dose used to generate the G3 radiation hybrid panel (10,000 rad), 1 cR corresponds to ≈24 kb. Logarithm of odds (lod) scores (log10 of the likelihood ratio) for these linkages were very high, 12.1, 12.1, and 9.1, respectively.
Figure 1.
Results of screening radiation hybrids for the presence of retrovirus receptors by phenotype (Transduction) and genotype (Receptor PCR) as compared with results from the PCR analysis of closely linked markers. The radiation hybrid clones are arrayed horizontally with numbering equivalent to the standard used for hybrid DNA analysis. A “1” on a black background indicates that the radiation hybrid was transduced at a titer of ≥104 FFU/ml in the case of transduction analysis or that a PCR product was detected in the case of DNA analysis. A “0” indicates that the hybrid was transduced at a titer of <10 FFU/ml in the case of transduction analysis or that a PCR product was not detected in the case of DNA analysis. The shaded boxes indicate unavailable or ambiguous data. Note that the radiation hybrid cell lines numbers 48, 76, and 78 were not available for transduction analysis. The third through fifth rows of data for each receptor represent the PCR results for the closest SHGC-ordered markers for each receptor.
These results prompted us to initiate positional cloning efforts to identify the genes encoding the three retrovirus receptors, but in the meantime we and others successfully isolated human cDNAs encoding the RD114 receptor (RDR) (7, 16), the xenotropic MuLV receptor (XPR1) (9, 17, 18), and the FeLV-C receptor (FLVCR) (15, 19) by expression cloning using retroviral cDNA expression libraries. We designed PCR primers to detect the receptor genes in DNA from the radiation hybrids to determine whether the genotypic mapping would give the same results as did the phenotypic mapping. Indeed, the genotypic mapping gave nearly identical chromosomal positions for the three receptors as compared with those determined by phenotypic mapping, as described below. Note that the DNA samples that were used for the PCR analysis were not prepared from the radiation hybrid cell lines that we were growing, but were the standard DNA samples that are supplied to the research community by Research Genetics. Thus, the good correlation between phenotypic and genotypic mapping data indicates the relative stability of the gene expression pattern in these radiation hybrid cell lines.
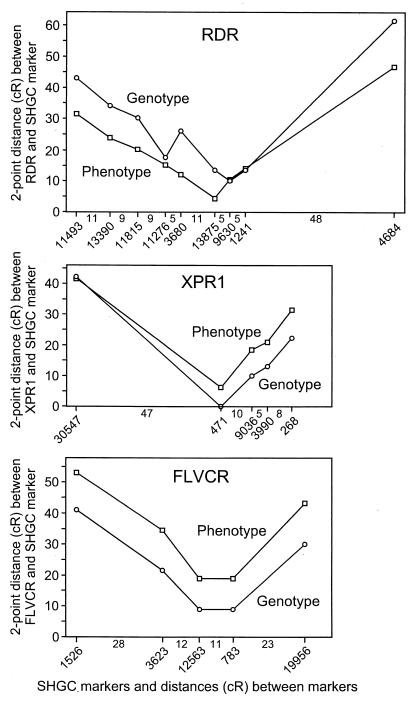
Fig. 1 shows the high similarity among the radiation hybrid bar codes for receptor phenotype, genotype, and several linked markers. To help establish the validity of the phenotypic mapping data, we calculated the two-point distances between the receptor phenotypes or genotypes and the ordered markers on the SHGC map and plotted these results against SHGC map distances determined by statistical analysis of the PCR results from multiple markers over the whole human genome (Fig. 2). An ideal result would be a V-shaped curve with arms at 45° angles from the horizontal, indicating a direct correspondence between the two-point distances and the SHGC map distance. A close to ideal result was observed for the XPR1 receptor phenotypic and genotypic data (Fig. 2). Moreover, both the phenotypic and the genotypic data position XPR1 close to marker SHGC-471. A similar result was obtained for the FLVCR receptor (Fig. 2), but in this case the phenotypic analysis positioned FLVCR farther away from all of the markers than did the genotypic analysis. For the RDR receptor, a V-shaped curve was observed for the phenotypic data, but there is some anomalous behavior for the genotypic data near the presumed location of the gene (Fig. 2), which might be explained by PCR errors. For RDR the phenotypic data appear to give more precise localization for RDR than do the genotypic data.
Figure 2.
Two-point distances between retrovirus receptors and ordered markers (x axis) in comparison to SHGC-ordered marker map distance (y axis). The location and identification numbers of the SHGC-ordered markers are shown below the diagrams with the distances (cR) shown between markers. All distances are drawn to the same scale.
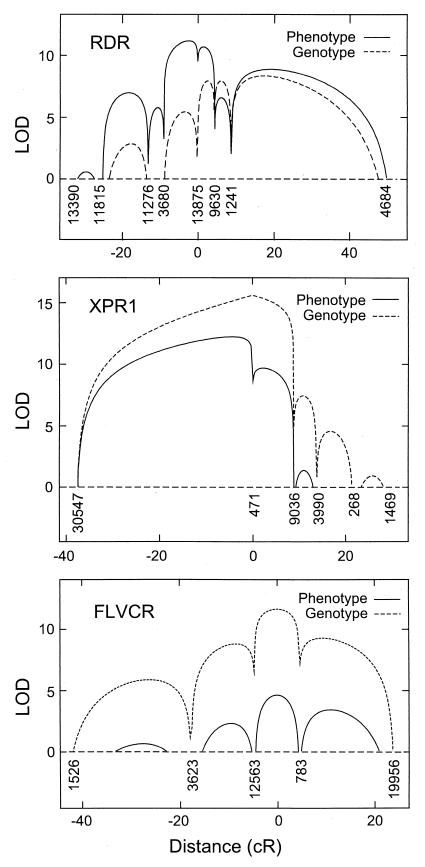
In an attempt to determine the confidence of the phenotypic positional assignments, we calculated the multipoint lod scores as a function of distance on the SHGC ordered maps (Fig. 3). For the RDR phenotypic data, the highest lod scores were obtained in the interval between markers 3,680 and 9,630, with much lower lod scores (>2 lower) at other positions, indicating a high confidence (>95%) that the gene responsible for this phenotype is within this 16 cR (≈380 kb) interval. As with the two-point analysis (Fig. 2), the RDR genotypic data were not as well behaved and provided less accurate positional information. For XPR1, the phenotypic data position the gene between markers 30,547 and 471 with high confidence, but markers in this region are sparse, resulting in less precise localization to a region of about 25 cR (≈600 kb) where the lod scores are within 2 lod of the peak. Markers are more frequent to the right of marker 471, leading to more definitive exclusion of XPR1 from this region. For FLVCR, both the phenotypic and genotypic data position the receptor between markers 12,563 and 783, a distance of 11 cR (≈260 kb). Thus, the phenotypic mapping of these receptors gave reliable and precise chromosomal positions for the receptor genes.
Figure 3.
Multipoint analysis of the likelihood of linkage (lod) between ordered markers and the retrovirus receptors by phenotypic and genotypic analysis. SHGC-ordered marker numbers are shown below the curves. Map distance is plotted on the x axis. lod scores below 0 are not shown. Multipoint lod scores were computed as described (20) with the computer program radmap written by L.K.
Discussion
We were able to precisely map three retrovirus receptor genes by phenotypic analysis of human/hamster radiation hybrid cell lines, however, our attempts to positionally clone these receptors by using this data were superseded by efforts to clone the genes by an approach involving retroviral cDNA expression libraries. In the future, the most exciting application of the phenotypic mapping technique will come with the fast-approaching completion of the human genome sequencing project (21) and the further development of higher resolution radiation hybrid panels. Availability of the complete sequence of the human genome will allow direct identification of candidate genes based on the phenotypic mapping data and will eliminate the time-consuming task of positional cloning after gene localization. The current resolution of the G3 hybrid panel is ≈250 kb, or ≈8 genes, whereas the TNG4 radiation hybrid panel made by using human DNA exposed to 50,000 rad of radiation provides a resolution of ≈60 kb (http://www-shgc.stanford.edu), narrowing the number of candidate genes to ≈2. Phenotypic assays that might be used to screen the radiation hybrids include binding assays for cell surface proteins, the use of antibodies to detect human protein expression, isoenzyme analysis, protein gel analysis, and other techniques that can distinguish phenotypes of human proteins expressed in hamster cells.
An important finding from this study was that the G3 radiation hybrid cells were sufficiently stable in culture to allow informative phenotypic analysis. The hybrid cell lines that we analyzed came from stocks at Stanford University. Similar stocks previously had been sent to Research Genetics to make the large amounts of genomic DNA used for PCR-based mapping of human genetic markers and for the PCR analysis of the retrovirus receptor genes. We grew the hybrids for up to 8 weeks before phenotypic analysis, and the cell lines were expanded independently to make the genomic DNA at Research Genetics. The independent cultivation of the hybrids might explain why PCR analysis sometimes gave a discordant result as compared with phenotypic analysis for a given hybrid, either PCR-positive and phenotype-negative or PCR-negative and phenotype-positive (Fig. 1). Despite these occasional differences, the analysis of multiple hybrids and the relative stability of the hybrids provided good statistical power to localize genes by phenotype.
Other explanations for a PCR-positive and phenotype-negative result include suppression of expression from an intact gene containing the PCR-amplified region, or loss of a critical region of the gene required for expression that did not include the PCR-amplified region. PCR-negative and phenotype-positive results at first seem more difficult to explain, however, most PCR markers for genes, including those used here, are designed against the 3′ end of cDNAs, and thus the PCR-amplified region might be deleted without loss of gene function. For example, of the three retrovirus receptors examined here, the only examples of PCR-negative and phenotype-positive hybrids were observed for FLVCR, and two hybrids show this result. It turns out that the PCR-amplified region in the 3′ end of the gene is separated by a large 6-kb intron from the majority of the FLVCR coding region, thus deletion of the PCR-amplified region without ablation of the gene phenotype is plausible. Lastly, differences in the sensitivities of phenotype and PCR analysis also could lead to occasional discrepancies in phenotype and PCR results.
We were able to localize all three genes that we examined by phenotypic analysis of the radiation hybrid cell lines. However, it is likely that some human genes are not expressed in the hybrid cells, precluding their localization by this method. In addition, very large genes may frequently be fragmented in the hybrids, such that expression occurs in only a small fraction of the hybrids and leads to uninformative results. This problem is predicted to be more pronounced in hybrids made with very high radiation doses such as those in the TNG4 hybrid panel, where the average human genome fragment size is 800 kb, in contrast to the average size of 4 Mb in hybrids from the G3 panel used here. Despite these potential problems, our results show that phenotypic screening of radiation hybrid cells provides a useful method to localize and clone human genes and builds on the considerable effort already devoted to the generation of high-resolution maps using these hybrids. Once the sequence of the human genome is available, genotypic mapping of new human sequences by using radiation hybrids will become obsolete, but the hybrids will still be a valuable resource for phenotypic mapping.
Acknowledgments
We thank Rebecca Gottschalk, Shannon Brady, and Kathleen McKusick for technical assistance, Panos Ioannou and Linda Ashworth for advice, and the Fondation Jean Dausset/Centre d'Étude du Polymorphisme Humain for provision of human genomic clones from chromosomal regions of interest. These studies were supported by Grants HL54881 and HL36444 from the National Heart, Lung, and Blood Institute of the National Institutes of Health. J.E.J.R. was supported by Fellowship DRG081 from the Cancer Research Fund of the Damon Runyon–Walter Winchell Foundation, and J.-L.B. was supported by a fellowship from the Association Française Contre les Myopathies. L.K. is a James S. McDonnell Centennial Fellow.
Abbreviations
- MuLV
murine leukemia virus
- FeLV-C
feline leukemia virus-C
- FFU
focus-forming units
- Env
envelope protein
- SHGC
Stanford Human Genome Center
- cR
centiray
- lod
logarithm of odds
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.130200097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.130200097
References
- 1.Stewart E A, McKusick K B, Aggarwal A, Bajorek E, Brady S, Chu A, Fang N, Hadley D, Harris M, Hussain S, et al. Genome Res. 1997;7:422–433. doi: 10.1101/gr.7.5.422. [DOI] [PubMed] [Google Scholar]
- 2.Goss S J, Harris H. Nature (London) 1975;255:680–684. doi: 10.1038/255680a0. [DOI] [PubMed] [Google Scholar]
- 3.Hunter E. In: Retroviruses. Coffin J M, Hughes S H, Varmus H E, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1997. pp. 71–119. [Google Scholar]
- 4.Weiss R A. In: The Retroviridae. Levy J A, editor. Vol. 2. New York: Plenum; 1993. pp. 1–108. [Google Scholar]
- 5.Miller A D. Proc Natl Acad Sci USA. 1996;93:11407–11413. doi: 10.1073/pnas.93.21.11407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller D G, Edwards R H, Miller A D. Proc Natl Acad Sci USA. 1994;91:78–82. doi: 10.1073/pnas.91.1.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rasko J E J, Battini J L, Gottschalk R J, Mazo I, Miller A D. Proc Natl Acad Sci USA. 1999;96:2129–2134. doi: 10.1073/pnas.96.5.2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cosset F L, Takeuchi Y, Battini J L, Weiss R A, Collins M K. J Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Battini J L, Rasko J E J, Miller A D. Proc Natl Acad Sci USA. 1999;96:1385–1390. doi: 10.1073/pnas.96.4.1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller A D, Chen F. J Virol. 1996;70:5564–5571. doi: 10.1128/jvi.70.8.5564-5571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartz S R, Vodicka M A. Methods. 1997;12:337–342. doi: 10.1006/meth.1997.0487. [DOI] [PubMed] [Google Scholar]
- 12.Riedel N, Hoover E A, Gasper P W, Nicolson M O, Mullins J I. J Virol. 1986;60:242–250. doi: 10.1128/jvi.60.1.242-250.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller A D, Garcia J V, von Suhr N, Lynch C M, Wilson C, Eiden M V. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fields-Berry S C, Halliday A L, Cepko C L. Proc Natl Acad Sci USA. 1992;89:693–697. doi: 10.1073/pnas.89.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Quigley J G, Burns C C, Anderson M M, Lynch E D, Sabo K M, Overbaugh J, Abkowitz J L. Blood. 2000;95:1093–1099. , and erratum (2000) 95, in press. [PubMed] [Google Scholar]
- 16.Tailor C S, Nouri A, Zhao Y, Takeuchi Y, Kabat D. J Virol. 1999;73:4470–4474. doi: 10.1128/jvi.73.5.4470-4474.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tailor C S, Nouri A, Lee C G, Kozak C, Kabat D. Proc Natl Acad Sci USA. 1999;96:927–932. doi: 10.1073/pnas.96.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y L, Guo L, Xu S, Holland C A, Kitamura T, Hunter K, Cunningham J M. Nat Genet. 1999;21:216–219. doi: 10.1038/6005. [DOI] [PubMed] [Google Scholar]
- 19.Tailor C S, Willett B J, Kabat D. J Virol. 1999;73:6500–6505. doi: 10.1128/jvi.73.8.6500-6505.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Slonim D, Kruglyak L, Stein L, Lander E. J Comput Biol. 1997;4:487–504. doi: 10.1089/cmb.1997.4.487. [DOI] [PubMed] [Google Scholar]
- 21.Collins F S, Patrinos A, Jordan E, Chakravarti A, Gesteland R, Walters L. Science. 1998;282:682–689. doi: 10.1126/science.282.5389.682. [DOI] [PubMed] [Google Scholar]