Abstract
The TRANSFAC® database on eukaryotic transcriptional regulation, comprising data on transcription factors, their target genes and regulatory binding sites, has been extended and further developed, both in number of entries and in the scope and structure of the collected data. Structured fields for expression patterns have been introduced for transcription factors from human and mouse, using the CYTOMER® database on anatomical structures and developmental stages. The functionality of Match™, a tool for matrix-based search of transcription factor binding sites, has been enhanced. For instance, the program now comes along with a number of tissue-(or state-)specific profiles and new profiles can be created and modified with Match™ Profiler. The GENE table was extended and gained in importance, containing amongst others links to LocusLink, RefSeq and OMIM now. Further, (direct) links between factor and target gene on one hand and between gene and encoded factor on the other hand were introduced. The TRANSFAC® public release is available at http://www.gene-regulation.com. For yeast an additional release including the latest data was made available separately as TRANSFAC® Saccharomyces Module (TSM) at http://transfac.gbf.de. For CYTOMER® free download versions are available at http://www.biobase.de:8080/index.html.
INTRODUCTION
Gene expression, and in particular transcription, in eukaryotic cells is an important process that is regulated in a complex way, through an intricate system of mutual interactions of transcription factors, whose effects (activation/repression) are mediated via DNA binding sites on their target genes. Within a multicellular organism each cell type or tissue, at a specific developmental stage, has its own characteristic gene expression profile that is defined, at least in part, by the presence of a specific combination of transcription factors.
The TRANSFAC® database, which was developed more than a decade ago to model factor-site interactions (1,2), has been subject to different improvements, modifications and extensions in structure and content over the years (3–9). Some of the latest changes that will be described in the present contribution were done with the intention to lead to a better understanding of tissue-specific expression of genes. Expression patterns were introduced for transcription factors using the CYTOMER® database of anatomical structures and developmental stages as a basis (10,11). Also the functionality of the Match™ tool which is designed for searching potential binding sites for transcription factors in DNA sequences (12) was enhanced through profiles (groups of binding matrices) for transcription factors specific for certain tissues or states.
CONTENT OF TRANSFAC®
TRANSFAC® is maintained internally as a relational database, from which public releases are made available via the web. The release consists of six flat files. At the core of the database is the interaction of transcription factors (FACTOR) with their DNA-binding sites (SITE) through which they regulate their target genes (GENE). Apart from genomic sites, ‘artificial’ sites which are synthesized in the laboratory without any known connection to a gene, e.g., random oligonucleotides, and IUPAC consensus sequences are also stored in the SITE table. Sites must be experimentally proven for their inclusion in the database. Experimental evidence for the interaction with a factor is given in the SITE entry in form of the method that was used (gel shift, footprinting analysis,…) and the cell from which the factor was derived (factor source). The latter contains a link to the respective entry in the CELL table. On the basis of those, method and cell, a quality value is given to describe the ‘confidence’ with which an observed DNA-binding activity could be assigned to a specific factor. From a collection of binding sites for a factor nucleotide weight matrices are derived (MATRIX). These matrices are used by the tool Match™ to find potential binding sites in uncharacterized sequences, while the program Patch™ uses the single site sequences (and consensi given in the IUPAC 15-letter code), which are stored in the SITE table. According to their DNA-binding domain transcription factors are assigned to a certain class (CLASS). In addition to the more ‘planar’ CLASS table a hierarchical factor classification system has been proposed as well some time ago (13) and has been developed further since then. In Table 1 the number of entries in the different tables/flat files are given for the current public release. TRANSFAC® contains data from a wide variety of eukaryotic organisms, ranging from human to yeast.
Table 1. Number of entries in the different tables of the TRANSFAC® database (release 6.0) and the TRANSFAC® Saccharomyces Module (TSM; release 3.0).
| Table | TRANSFAC® Release 6.0 | TSM Release 3.0 |
|---|---|---|
| FACTOR | 4219 | 370 |
| Homo sapiens | 960 | — |
| Mus musculus | 714 | — |
| Drosophila melanogaster | 204 | — |
| Caenorhabditis elegans | 105 | — |
| Arabidopsis thaliana | 230 | — |
| Saccharomyces cerevisiae | 334 | 370 |
| others | 1672 | — |
| SITE | 6627 | 825 |
| Genomic sites | 5064 | 592 |
| Artificial sites | 1308 | 209 |
| Consensus sequences | 255 | 24 |
| MATRIX | 336 | 35 |
| GENE (all entries) | 1755 | 563 |
| Homo sapiens | 449 | — |
| Saccharomyces cerevisiae | 155 | 563 |
| Others | 1151 | — |
| GENE (entries with SITE links) | 1275 | 245 |
| CLASS | 44 | 17a |
| CELL | 1432 | 13 |
aOnly those entries were counted which have a factor from Saccharomyces cerevisiae assigned.
THE TRANSFAC® SACCHAROMYCES MODULE (TSM)
The early completion of the whole genome sequence in 1996 gave yeast a headstart in the now rapidly developing field of genome-wide expression analysis (14). In order to make sense of the vast amount of yeast-related data and to extract conclusions and hypotheses that are biologically meaningful, sophisticated systems of knowledge representation are needed. An ongoing effort to provide the scientific community with an integrated data collection and knowledge resource is the Comprehensive Yeast Genome Database (CYGD). It is a joint endeavour of several European yeast laboratories and comprises a number of specialized databases (15).
As part of the CYGD project, the TRANSFAC® database was massively updated with yeast data (16) and is now being integrated into the CYGD framework. In parallel to being integrated into CYGD, the TRANSFAC® yeast data were made publicly accessible as the Saccharomyces Module TSM (Table 1).
APPLICATION OF CYTOMER® FOR TRANSCRIPTION FACTOR EXPRESSION PATTERNS
CYTOMER® is a database on physiological systems, developmental stages, anatomical structures and substructures, and their constituting cell-types for particular organisms (10,11). We have now completed CYTOMER® for human and Caenorhabditis elegans, work is in progress for mouse. The relational structure of CYTOMER® comprises five tables, four of them are catalogs of organs, cells, developmental stages and physiological systems. The ORGAN table is itself hierarchically organized and represents an ontology of anatomical structures and substructures as they occur at the particular developmental stage. For human, an organ tree is constructed for the adult organism as well as for characterized embryonic stages (in the current version: Carnegie stages 1 to 17). The central table of CYTOMER® is HUB, which is a list that links entries of the five other tables. Each entry in this table corresponds to the particular cell type within a particular organ or suborgan and physiological system at the given developmental stage. Thus, the HUB table represents anatomical/histological knowledge about which cells occur in which organs and at what stages of development. Being complemented by descriptions and definitions, CYTOMER® provides a comprehensive ontology on human's anatomy and ontogenesis.
The CYTOMER® database has been applied to map expression patterns of genes. Presently, we provide descriptions of expression of human and mouse genes encoding transcription factors collected in the TRANSFAC® database. Descriptions of factor expression patterns are released as a part of the TRANSFAC® FACTOR table. Presently, in the public release expression patterns of the following families of transcription factors are characterized: GATA-factors, nuclear receptors (e.g., androgen and estrogen receptor) and a number of homeobox factors. Entries of the CYTOMER® HUB table have been linked with human and mouse transcription factor entries in the TRANSFAC® FACTOR table of the relational database. This structure allows us to present exact information about temporal and spatial characteristics of gene expression. In addition, the method used for the experimental detection of mRNA or protein expression is given (Table 2). Expression levels are provided in a semiquantitative way by assigning one of seven levels from ‘none’ to ‘very high’.
Table 2. Methods used for the experimental detection of mRNA or protein expression.
| Method | Detected molecule |
|---|---|
| Northern blot total RNA | m-RNA (poly A) |
| RNA (undefined) | |
| RNA-in situ hybridization (not further specified) | RNA (undefined) |
| RNA-in situ hybridization (radioactive) | RNA (undefined) |
| RNA-in situ hybridization (non-radioactive) | RNA (undefined) |
| RT–PCR total RNA | m-RNA (poly A) |
| RNA (undefined) | |
| Immunohistochemistry/immunocytochemistry | protein |
| Western blot | protein |
| Dot blot (RNA) total RNA | m-RNA (poly A) |
| RNA (undefined) | |
| Dot blot (protein) | protein |
| RNAse protection assay total RNA | m-RNA (poly A) |
| RNA (undefined) |
Describing transcription factor expression patterns through the link between the CYTOMER® and TRANSFAC® databases has several advantages over the previously existing description in free text fields (CP=cell-specific-positive for those expression sources where a certain factor has been shown to be expressed in, and CN=cell-specific-negative for those expression sources where evidence for the absence of a certain factor has been published). Gene expression patterns are described now in a computer-readable format, giving the possibility to perform better queries and searches of expression patterns. Experimental methods and references are linked now to expression patterns. CYTOMER® provides a comprehensive overview on all spatial and temporal expression patterns.
ENHANCEMENTS OF INTRA- AND INTER-LINKING (A CENTRAL ROLE FOR THE GENE TABLE)
The GENE table is one of the central tables of the TRANSFAC® database. It is not only jointly used by several of our own databases, TRANSPATH® (17), PathDB® (8,9), S/MARt DB™ (18), and TRANSCompel® (19). Recently, the GENE table has been extended to one of the major link sources to external databases, including BRENDA (20), LocusLink, OMIM and RefSeq (21).
The GENE table serves to list the transcription factor binding sites within a gene regulatory region, and thus showing them in a context. Alongside these sites the factors binding to them are shown as well now. (Also in the FACTOR table the regulated genes are listed now aside the binding sites, providing direct links from factors to target genes). In addition to these factor-gene links based on protein-DNA binding, in those cases where the gene encodes a transcription factor, links from gene to the encoded factor have been introduced and vice versa. In this structure, a particular transcription factor, as a gene product, is always linked to one gene. Along with this, the same gene entry could be linked to several transcription factors in those cases when a gene encodes for several products as a result of alternative start of transcription, splicing, start of translation, or polyadenylation. For instance, the human gene hnf-4a encodes for at least four different splice variants that are transcription factors with different functional properties due to the differences in particular protein domains (gene id HS$HNF4A, factors ids T00373, T02421, T02425, T02428). For many transcription factors, it is known that the gene encoding a particular factor is itself regulated by this factor, either positively or negatively. These autoregulatory feedback loops are presented now in the GENE table, for example for the human and mouse genes encoding transcription factors c-Jun, c-Fos, c-Myc, c-Myb, E2F1, CRE-BP1, C/EBP-α, RAR-β, RAR-γ, SRY.
In cases, where proteins are encoded which are part of the signal transduction network of the cell, links from GENE to the MOLECULE table in the TRANSPATH® database (17) were added. Together with the links from MOLECULE (TRANSPATH®) to FACTOR (TRANSFAC®) these links are intended as steps towards an integration of the gene regulation data of TRANSFAC® into the overall regulatory network of the cell.
Beside this, the GENE table contains additional fields for synonyms and for chromosomal localization now, and references about transcriptional regulation of a gene are listed as well.
MATCH™: ENHANCEMENT BY TISSUE- AND STATE-SPECIFIC PROFILES
TRANSFAC® 6.0 is accompanied by the new public version of Match™ (12). This tool performs searches for putative transcription factor binding sites in DNA sequences based on weight matrices. Match™ uses the library of weight matrices collected in the MATRIX table of the TRANSFAC® database. We have developed a WWW interface and a graphical representation of the program output.
The algorithm of the Match™ uses two values to score putative hits: the matrix similarity score and the core similarity score resembling herein the previously published MatInspector algorithm (22). The core similarity weights the quality of a match between the sequence under study and the core sequence of a matrix which consists of the five most conserved consecutive positions in a matrix. The matrix similarity score is a weight for the quality of a match between the sequence and the whole matrix. Both scores range from 0 to 1 where 1 denotes the exact match.
The new version of Match™ provides several specific profiles as well as a tool, the Match™ Profiler, for creation and modification of profiles by the user. A profile is a set of matrices and their cut-offs designed for function-driven searches within regulatory regions of genes whose function is partially known. Currently, we provide immune cell-, muscle-, liver- and cell cycle-specific profiles. The liver-specific profile, for instance, contains matrices for liver-enriched factors of HNF-1, -3, -4, C/EBP and SREBP families. Matrices for widely expressed transcription factors, both inducible (GR, NF-κB, STAT, AP-1, CREB) and constitutive (Sp1, TBP, NF-1, YY1, USF), are included in this profile as well. These widely expressed factors are known to bind DNA sites and regulate transcription of genes in liver, in many cases by cooperation with liver-specific factors. Examples of liver-specific gene regulation confirming involvement of both liver-enriched and ubiquitous factors, are collected in the databases TRANSFAC® and TransCOMPEL®. The liver-specific profile can be applied for the regulatory regions of genes that are known to be expressed in liver, but function and mechanisms of this regulation are not known in detail.
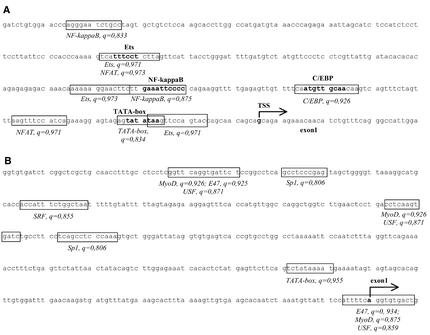
Examples of profile application are shown in Figure 1. The immune-specific profile (with modified cut-offs) was applied to the promoter region of the human IL-12 p40 subunit gene. In this gene, four binding sites are known: Ets, NF-κB, C/EBP and TATA-box (23). NF-κB and C/EBP cooperatively regulate the IL-12 p40 promoter (24). All known sites as well as additional potential binding sites are found by Match™ with the immune-specific profile (Fig. 1A). Another example addresses a gene with unknown function. It is just known that its mRNA is expressed in skeletal muscles. In this case, we have applied the muscle-specific profile (with modified cut-offs) and found a number of potential sites in the close proximity to the beginning of the first exon as it is annotated in RefSeq (Fig. 1B).
Figure 1.
Application of specific profiles provided by the Match™ program. Potential binding sites found by Match™ are boxed, name of the transcription factor and score for the match are given under the sequence. (A) The immune-specific profile (with modified cut-offs) is applied to find potential binding sites within the promoter sequence of the human IL-12 p40 gene (EMBL accession no. AY008847, positions 2101 to 2460). Known binding sites for transcription factors are shown in bold, the name of the transcription factor is given above the sequence. The transcription start site (TSS) is indicated by an arrow. (B) The muscle-specific profile (with modified cut-offs) is used to find potential binding sites in the 5′ region of the hypothetical gene, LocusLink ID LOC88523. This gene encodes a protein with unknown function, a corresponding EST is shown to be expressed in skeletal muscles. The sequence has been retrieved from RefSeq, the start of the first exon is shown according to RefSeq annotation.
AVAILABILITY
The public releases of TRANSFAC® and of our other databases, PathoDB®, S/MARt DB™, and TRANSCompel®, as well as the public versions of the programs Match™ and Patch™ are all freely available to users from non-profit organizations at http://www.gene-regulation.com/. The TSM is freely available as a standalone resource at http://transfac.gbf.de/ (under ‘Databases’). For Homo sapiens and C. elegans free download versions of the CYTOMER® database are available at http://www.biobase.de:8080/index.html.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank all present and former members of BIOBASE GmbH and the AG Bioinformatics at the German Research Centre for Biotechnology (GBF) for contributing to this work in various ways. This work is supported in part by a grant of the European Commission (contract no. QLRI-CT-1999-01333) and two grants of the German Ministry of Education and Research (BMBF, grant no. 0312432 and 031U210B).
REFERENCES
- 1.Wingender E. (1988) Compilation of transcription regulating proteins. Nucleic Acids Res., 16, 1879–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wingender E., Heinemeyer,T. and Lincoln,D. (1991) In Collins,J. and Driesel,A.J. (eds), Genome Analysis—From Sequence to Function; BioTechForu—Advances in Molecular Genetics. Hüthig Buch Verlag, Heidelberg, vol. 4, pp. 95–108.
- 3.Knüppel R., Dietze,P., Lehnberg,W., Frech,K. and Wingender,E. (1994) TRANSFAC® retrieval program: a network model database of eukaryotic transcription regulating sequences and proteins. J. Comput. Biol., 1, 191–198. [DOI] [PubMed] [Google Scholar]
- 4.Wingender E., Dietze,P., Karas,H. and Knüppel,R. (1996) TRANSFAC®: A database on transcription factors and their DNA binding sites. Nucleic Acids Res., 24, 238–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wingender E., Kel,A.E., Kel,O.V., Karas,H., Heinemeyer,T., Dietze,P., Knüppel,R., Romaschenko,A.G. and Kolchanov,N.A. (1997) TRANSFAC®, TRRD and COMPEL: towards a federated database system on transcriptional regulation. Nucleic Acids Res., 25, 265–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heinemeyer T., Wingender,E., Reuter,I., Hermjakob,H., Kel,A.E., Kel,O.V., Ignatieva,E.V., Ananko,E.A., Podkolodnaya,O.A., Kolpakov,F.A., Podkolodny,N.L. and Kolchanov,N.A. (1998) Databases on transcriptional regulation: TRANSFAC®, TRRD, and COMPEL. Nucleic Acids Res., 26, 362–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heinemeyer T., Chen,X., Karas,H., Kel,A.E., Kel,O.V., Liebich,I., Meinhardt,T., Reuter,I., Schacherer,F. and Wingender,E. (1999) Expanding of the TRANSFAC® database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Res., 27, 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wingender E., Chen,X., Hehl,R., Karas,H., Liebich,I., Matys,V., Meinhardt,T., Prüß,M., Reuter,I. and Schacherer,F. (2000) TRANSFAC®: an integrated system for gene expression regulation. Nucleic Acids Res., 28, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wingender E., Chen,X., Fricke,E., Geffers,R., Hehl,R., Liebich,I., Krull,M., Matys,V., Michael,H., Ohnhäuser,R., Prüß,M., Schacherer,F., Thiele,S. and Urbach,S. (2001) The TRANSFAC® system on gene expression regulation. Nucleic Acids Res., 29, 281–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen X., Dress,A., Karas,H., Reuter,I. and Wingender,E. (1999) A database framework for mapping expression patterns. In Proceedings of the German Conference on Bioinformatics GCB'99. Hannover, Germany, pp. 174–178.
- 11.Fricke E., Land,S., Rotert,S., Karas,D. and Wingender,E. (2001) Cytomer: A database on gene expression sources. Proceedings of the German Conference on Bioinformatics GCB'01. Braunschweig, Germany, pp. 149–151.
- 12.Gößling E., Kel-Margoulis,O.V., Kel,A.E. and Wingender,E. (2001) MATCH™—a tool for searching transcription factor binding sites in DNA sequences. Application for the analysis of human chromosomes. Proceedings of the German Conference on Bioinformatics GCB'01. Braunschweig, Germany, pp. 158–161.
- 13.Wingender E. (1997) Classification scheme of eukaryotic transcription factors. Mol. Biol., 31, 483–497. [PubMed] [Google Scholar]
- 14.Akache B., Wu,K. and Turcotte,B. (2001) Phenotypic analysis of genes encoding yeast zinc cluster proteins. Nucleic Acids Res., 29, 2181–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mewes H.W., Frishman,D., Güldener,U., Mannhaupt,G., Mayer,K., Mokrejs,M., Morgenstern,B., Münsterkötter,M., Rudd,S. and Weil,B. (2002) MIPS: a database for genomes and protein sequences. Nucleic Acids Res., 30, 31–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michael H., Thiele,S. and Wingender,E. (2001) TRANSFAC®_YEAST. Proceedings of the German Conference on Bioinformatics GCB'01. Braunschweig, Germany, p. 208.
- 17.Schacherer F., Choi,C., Götze,U., Krull,M., Pistor,S. and Wingender,E. (2001) The TRANSPATH® signal transduction database: a knowledge base on signal transduction networks. Bioinformatics, 17, 1053–1057. [DOI] [PubMed] [Google Scholar]
- 18.Liebich I., Bode,J., Frisch,M. and Wingender,E. (2002) S/MART DB: a database on scaffold/matrix attached regions. Nucleic Acids Res., 30, 372–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kel-Margoulis O., Kel,A.E., Reuter,I., Deineko,I.V. and Wingender,E. (2002) TransCOMPEL®—a database on composite regulatory elements in eukaryotic genes. Nucleic Acids Res., 30, 332–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schomburg I., Chang,A. and Schomburg,D. (2002) BRENDA, enzyme data and metabolic information. Nucleic Acids Res., 30, 47–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wheeler D.L., Church,D.M., Lash,A.E., Leipe,D.D., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Tatusova,T.A., Wagner,L. and Rapp,B.A. (2002) Database resources of the National Center for Biotechnology Information: 2002 update. Nucleic Acids Res., 30, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quandt K., Frech,K., Karas,H., Wingender,E. and Werner,T. (1995) MatInd and MatInspector—New fast and sensitive tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res., 23, 4878–4884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becker C., Wirtz,S., Ma,X., Blessing,M., Galle,P.R. and Neurath,M.F. (2001) Regulation of IL-12 p40 promoter activity in primary human monocytes: roles of NF-kappaB, CCAAT/enhancer-binding protein beta and PU.1 and identification of a novel repressor element (GA-12) that responds to IL-4 and prostaglandin E(2). J. Immunol., 167, 2608–2618. [DOI] [PubMed] [Google Scholar]
- 24.Plevy S.E., Gemberling,J.H., Hsu,S., Dorner,A.J. and Smale,S.T. (1997) Multiple control elements mediate activation of the murine and human interleukin 12 p40 promoters: evidence of functional synergy between C/EBP and Rel proteins. Mol. Cell. Biol., 17, 4572–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]