Abstract
We introduce the PSSH (‘Protein Sequence-to-Structure Homologies’) database derived from HSSP2, an improved version of the HSSP (‘Homology-derived Secondary Structure of Proteins’) database [Dodge et al. (1998) Nucleic Acids Res., 26, 313–315]. Whereas each HSSP entry lists all protein sequences related to a given 3D structure, PSSH is the ‘inverse’, with each entry listing all structures related to a given sequence. In addition, we introduce two other derived databases: HSSPchain, in which each entry lists all sequences related to a given PDB chain, and HSSPalign, in which each entry gives details of one sequence aligned onto one PDB chain. This re-organization makes it easier to navigate from sequence to structure, and to map sequence features onto 3D structures. Currently (September 2002), PSSH provides structural information for over 400 000 protein sequences, covering 48% of SWALL and 61% of SWISS-PROT sequences; HSSPchain provides sequence information for over 25 000 PDB chains, and HSSPalign gives over 14 million sequence-to-structure alignments. The databases can be accessed via SRS 3D, an extension to the SRS system, at http://srs3d.ebi.ac.uk/.
INTRODUCTION
A database of sequence-to-structure alignments is a convenient way to rapidly find 3D structural information for a given protein sequence. For example, the Protein Mutation Database (PMD, 1) uses such a database to map the location of coding SNPs (single nucleotide polymorphisms) onto 3D structure, hence providing insight into the functional consequences of the mutation. Another example, MODBASE (2), provides pre-calculated 3D homology models for all known protein sequences with significant similarity to an experimentally determined 3D structure.
In constructing such databases, a crucial decision is which criteria to use for fold assignment, i.e. which structures should be listed as related to a given sequence, and which not. In general, two naturally evolved sequences more than 30% identical (and with more than 100 residues in the aligned region) have the same fold within 95% confidence limit (3); below this threshold, structural homology can sometimes be inferred using sequence profile and threading methods, however the criteria for safe inference are then less clear. The criteria used directly determine the breadth of coverage, i.e. how many sequences are related to at least one structure. PMD uses a simple, very conservative criterion (>50% sequence identity), hence it has very accurate fold assignment, but narrow coverage; in contrast, MODBASE uses sophisticated criteria that achieve wide coverage and high assignment accuracy (Table 1).
Table 1. Comparison of sequence-to-structure alignment databases.
| Database | Accuracy of sequence alignment | Accuracy of fold assignment | Breadth of coverage (% of SWALL) | Depth of coverage (structures per sequence) |
|---|---|---|---|---|
| PSSH | High | 95% | 48% | 34 |
| MMDB | Low | <95% (?) | ∼48% (?) | ∼34 (?) |
| MODBASE | High | 95% | 40% | 2 |
| PMD | Low | ≫95% | 12% | 1 |
The table compares PSSH with similar databases, based on information provided in respective publications and web-sites. Sequence alignment accuracy is indicated as ‘high’ for profile-based global alignments, and ‘low’ for pair-wise BLAST alignments. Fold assignment accuracy is the probability that the sequence has the same fold as the aligned structure. Breadth of coverage is the percentage of all known sequences that have at least one matching structure in the database. Depth of coverage is the average number of structures per sequence. For MMDB, the fold accuracy, breadth, and depth are not published, so we provide estimates (indicated by question marks). The depth and breadth of coverage is approximately similar to PSSH, however since MMDB is based on pair-wise BLAST, it probably has lower accuracy for both alignment and for fold assignment than PSSH (12). PMD has a stringent criterion for fold assignment, thus it has higher accuracy than the 95% threshold used in PSSH and MODBASE.
Especially in the context of structural genomics, coverage breadth is clearly important in assigning function to novel sequences. However, we believe that most users are interested in proteins of known biological, medical, or industrial importance for which several related 3D structures are already available (with the clear exception of membrane proteins). In these cases, more important is coverage depth, i.e. how many structures are matched to each sequence. Deep coverage allows the user to choose which of the matching structures are the most relevant, e.g. those that match a domain of particular interest, interact with a certain ligand or protein partner, or that have a particular mutation. Both PMD and MODBASE have shallow coverage (Table 1); for MODBASE, this is partly a trade-off due to the computational expense of model building.
The MMDB database, integrated into the Entrez system, provides broad and deep coverage, returning all related 3D structures for a given sequence (4). However as MMDB is based on pair-wise BLAST alignments, the accuracy of alignment and fold assignment is lower than can be achieved with more sophisticated profile-based methods (Table 1). While alignment accuracy is not a major issue above, say, 60% sequence identity, it becomes increasingly crucial for lower levels of identity, both for homology modelling and for mapping sequence features.
Our goal here was to develop a sequence-to-structure database to be integrated into the SRS system (5), primarily to enable mapping of sequence features from diverse databases onto 3D structures. Together with a novel macromolecular graphics system (6), these databases comprise SRS 3D, a new optional module of SRS (7). Our requirements for these databases were: deep coverage to handle the most important use cases (i.e. well studied proteins) and validated estimate of assignment accuracy, so that the significance of assignments can be made obvious to users who may not be experts on molecular modelling.
We chose to base this database on HSSP (‘Homology-derived Secondary Structure of Proteins’), a database of structure-to-sequence alignments, where each entry shows all sequences that are related to a 3D structure (8). HSSP is one of the family of databases created by the Sander group. The other family members are: FSSP (‘Families of Structurally Similar Proteins’), where each entry relates to one PDB entry (9) and gives the structural alignment to all other (non-redundant) PDB entries with significantly similar structures (10); and DSSP (‘Database of Secondary Structure of Proteins’), where each entry relates to one PDB entry and gives the secondary structure of each residue, plus other information such as geometrical features and solvent exposure (11).
HSSP features all of the aspects we consider crucial: accurate alignments (based on sequence family profiles), depth of coverage and a validated assignment accuracy. We implemented the new assignment criteria derived by Rost (12); the resulting database (called here HSSP2) has a better coverage for a given level of accuracy than in the previous HSSP database. The coverage is also significantly better than can be achieved using BLAST or PSI-BLAST alone (12). However, the HSSP and HSSP2 databases are organized around structures, quite the reverse of the typical use case, namely where a user wants to find all related structures for a given sequence. Therefore, we have reorganized the data to produce the PSSH (‘Protein Sequence-to-Structure of Homologies’) database, as described in this paper.
In addition, HSSP has a problem with atom numbering, namely handling PDB insertion codes. Dealing with PDB residue numbering is a central problem with sequence–structure alignment. While the full sequence of a protein contained in a PDB entry is in general given in the PDB SEQRES records, the sequence implicit from the structure coordinate section often contains gaps due to unresolved residues. Therefore, in general, the SEQRES record is the better one to use for sequence searches. Furthermore, ‘insertion codes’ are often used in the numbering of residues, since the natural sequence numbering is changed to a ‘canonical’ numbering of a protein family, making it necessary to account for gaps or insertions in the family alignment. HSSP does not account for insertion codes; thus, for some structures HSSP alignments cannot be mapped directly onto structures.
SYSTEMS AND METHODS
We wrote several Perl scripts and modules that carry out the following steps to create the PSSH database.
Generating PDBequiv
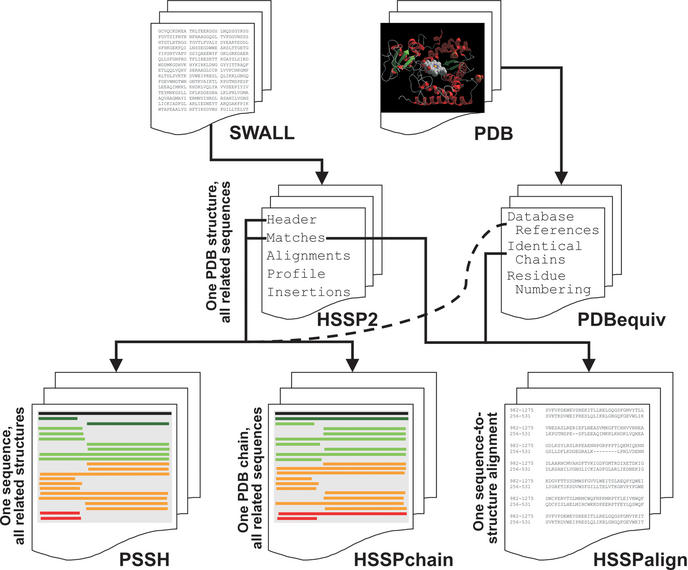
From each chain in a PDB entry, we created an entry in an intermediate database, PDBequiv (Fig. 1), consisting of the following fields: ‘Database References’, the SWISS-PROT accession number from the DBREF record; ‘Identical Chains’, a pointer to other chains in the same PDB entry with more than 99% sequence identity based on SEQRES records; ‘Residue Numbering’, the residue number and insertion code for each residue in the SEQRES record, determined by aligning the sequences in SEQRES to those in the ATOM records.
Figure 1.
Schematic representation of the derivation of the PSSH-related databases. For each PDB entry, all related sequences in SWALL are aligned using MaxHom2 and the alignment details are stored as one entry in HSSP2. For each PDB chain, we store additional information in an intermediate database, PDBequiv. Each HSSP2 entry is then processed to generate the remaining databases: all sequences that align onto one PDB chain are stored as one HSSPchain entry; each individual alignment in HSSP2 is stored as one entry in HSSPalign, with additional information extracted from PDBequiv. As each HSSP2 alignment is read, it is also appended to a separate file named by the sequence accession number, hence accumulating the PSSH database.
Generating HSSP2
HSSP2 was generated from SWALL (13) and PDB (Fig. 1) as introduced by Sander and Schneider (3):
For each structure in the PDB, SWALL is pre-filtered using BLASTP (14) with a very unrestrictive threshold E-value of 1000, so that even very poor matches are retrieved.
All matching sequences are aligned with the structure using MaxHom alignment, based on the Smith–Waterman algorithm (15), and modified as described by Sander and Schneider (3). Similarity is measured using the McLachlan matrix (16).
A similarity-based homology threshold is used to determine the sequences that can safely be assumed (with 95% confidence) to have the same fold as the structure, within the aligned regions.
Sequences that fall inside the threshold are used to generate a profile based on the sequence family.
Steps 2–4 are repeated, this time using the generated profile as a reference.
A final list of aligned sequences is obtained; the final alignment incorporates information about all related sequences.
In step 3, instead of employing the cutoff curve derived by Sander and Schneider, we used the new curve that was recommended by Rost (12) after repeating the analysis based on the vastly increased PDB data available in 1999.
Each HSSP2 entry refers to one PDB structure and has the following sections: ‘Header’ lists the PDB chains used for the sequence matching (for PDB structures with identical chains, only one is used); ‘Proteins’ lists accession numbers of all matching sequences and a brief description of each alignment (i.e. alignment length, sequence identity, and matching residue ranges); ‘Alignments’ lists the alignment details for all matching sequences, ignoring all insertions; ‘Profile’, for each sequence position gives the amino acids profile calculated from all alignments; ‘Insertions’ lists details of alignment insertions in the matching sequences.
Processing HSSP2
HSSP2 was processed, splitting the information into the HSSP-derived databases (Fig. 1). For each HSSP2 entry, for each chain we added an entry to the HSSPchain database consisting of the PDB code, chain identifier, then the accession numbers of all matching sequences to that chain and a brief description of each alignment from the ‘Proteins’ section. For every individual alignment, the ‘Database References’ from PDBequiv were used to assess whether the chain refers to the same protein as the matched sequence; this information was also added to the HSSPchain entry. For every individual alignment, precisely the same information was also appended to separate files, named according to the sequence accession number, hence accumulating the PSSH database. For each individual alignment, the alignment details were extracted from the ‘Alignment’ and ‘Insertions’ sections, combined with the ‘Residue Numbering’ and stored as an individual entry in HSSPalign. A unique key for each match is created by combining the sequence accession number with the PDB code and chain identifier. The alignment is stored in a concise machine-readable format (residue ranges of all ungapped parts of the alignment). In order to be able to use the PDB residue numbering (including insertion codes) in this concise form of the alignment, we refer to the PDBequiv database. Using information about identical chains, the alignment database is completed to contain the alignment between a protein sequence and all matching chains in a PDB structure, e.g. including both chains of a homodimer structure.
Post-processing the databases
The scripts further did the following operations: each PSSH entry was sorted by sequence identity; in all three databases, non-overlapping matches between the same chain and protein sequence were merged to make a single alignment; all database entries were concatenated, with separator characters, into large files (≤2 GB) for easy management.
Database updating
When run in updating mode, the scripts only generate new HSSP2 files if the corresponding PDB files have changed or been added, or if relevant sequences have changed or been added. Likewise, only HSSP2 files that have been changed since the last update are processed. Subsequently, information about unchanged matches is extracted from the old databases and added to the new ones.
RESULTS
PDBequiv
For BLASTing against the PDB, the user can now use the more complete SEQRES records; using PDBequiv, these records are aligned onto the structure residue numbers, including insertion code information.
PSSH
In PSSH, all structural information for a given sequence is in one entry, so a user interested in a particular sequence just needs to find and bookmark the entry. As the PDB is updated, the entry will also be updated. Users are generally more interested in starting queries from a sequence, rather than a structure, hence the arrangement of data in PSSH is more useful than in HSSP. Currently, PSSH contains 414 851 entries, giving structural information for about 48% of all known protein sequences in SWALL, and 61% for all SWISS-PROT entries (including updates). By comparison, less than 2% of SWALL sequences are directly linked to a PDB structure.
HSSPalign
The PSSH entry gives the user all the information needed to choose which of the related structures matches the domain(s) he is interested in. Once the user has selected a PDB entry, the alignment details can be easily found by looking up the corresponding HSSPalign entry (via the unique key). Compared with HSSP, the HSSPalign format is easier to understand and use, and it has residue numbering corrected for PDB insertion codes. Currently, HSSPalign has about 14 million sequence-to-structure alignments.
HSSPchain
Finally, HSSPchain makes it easy for a user who wants to find all sequences related to a given chain of a PDB entry. Currently, HSSPchain has 25 528 entries.
DISCUSSION
Processing the HSSP database, integrating the resulting databases into the database management system SRS and developing a suitable structural viewer has increased the number of sequences that can easily be viewed in structural context more than ten-fold. Replacing the cutoff used in HSSP with the more sensitive cutoff criterion derived by Rost (12), we were able to further increase this number by a factor of two, achieving a good balance of accuracy and coverage compared to similar databases [Table 1 and (12)].
In order to increase sensitivity in the detection of related sequences further, we plan to test using the HSSP profiles for scanning new sequences (instead of PDB sequences only). However, since many structurally related proteins show only a very remote sequence similarity (17), this twilight zone, where distinction between true and false matches is not possible based on sequence alone, promises the highest yield in sequence-to-structure matches. Therefore, we will also evaluate the application of threading methods in order to extend the range of alignments collected in HSSP further into the twilight region.
While Sander and Schneider found the alignments collected in HSSP to be rather reliable, not all the available information is used, since only one structure is regarded at a time. Therefore, we will try to improve the quality of a family alignment by using structural superimposition as guideline where structures of both aligned sequences are available.
As the PSSH and HSSPalign databases collect matches between sequences and their homologous structures and the corresponding alignments, this set of databases provides all the necessary information for calculating homology models. Thus, the databases we have integrated into SRS also facilitate homology modeling, since the user only needs to import an alignment stored in HSSPalign into his favorite modeling program.
However, in order to get a quick overview of the implications of the protein structure on the three-dimensional arrangement of the sequence features it is often sufficient to inspect the template structure, since the backbone deviations in the core of related structures usually are very small.
A variety of biological databases, such as SWISS-PROT, Pfam (18) and OMIM (19), contain a wealth of information about sequence features, e.g. domain boundaries, active site residues, phosphorylated or glycosylated residues, sites of sequence variations, especially those involved in diseases. Visualizing these features on 3D structures gives valuable insight into protein function. Mapping of the sequence features onto a related structure, therefore, proves useful in such diverse fields as proteomics, functional genomics and drug design. Integrating the HSSPalign database into the database integration system SRS (5) facilitates this mapping of sequence features onto three-dimensional structures as described in (7). The graphical overviews provided in the SRS setup of the PSSH database, help the user to identify structures relevant to a protein of interest, especially for multi-domain proteins. Thus in summary, the database setup introduced here enables biochemists to easily keep up to date with structural implications of newly emerging protein data (e.g. from structural genomics).
Acknowledgments
ACKNOWLEDGEMENTS
Thanks to Reinhard Schneider for useful discussions. This work was partly funded by a grant from the German Federal Ministry for Science and Education (`Protein Structure Factory' grant 01 GG 9817).
REFERENCES
- 1.Kawabata T., Ota,M. and Nishikawa,K. (1999) The protein mutant database. Nucleic Acids Res., 27, 355–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pieper U., Eswar,N., Stuart,A.C., Ilyin,V.A. and Sali,A. (2002) MODBASE, a database of annotated comparative protein structure models. Nucleic Acids Res., 30, 255–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sander C. and Schneider,R. (1991) Database of homology-derived structures and the structural meaning of sequence alignment. Proteins, 9, 56–68. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y., Anderson,J.B., Chen,J., Geer,L.Y., He,S., Hurwitz,D.I., Liebert,C.A., Madej,T., Marchler,G.H., Marchler-Bauer,A., Panchenko,A.R., Shoemaker,B.A., Song,J.S., Thiessen,P.A., Yamashita,R.A. and Bryant,S.H. (2002) MMDB: Entrez's 3D-structure database. Nucleic Acids Res., 30, 249–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Etzold T., Ulyanov,A. and Argos,P. (1996) SRS: information retrieval system for molecular biology data banks. In Doolittle,R.F. (ed.), Methods in Enzymology. Academic Press, San Diego, Vol. 266, pp. 114–128. [DOI] [PubMed]
- 6.Fries K. and O'Donoghue,S.I. (2002) Navigating around the building blocks of life. Adv. Imaging, 17, 18–19, 39. [Google Scholar]
- 7.O'Donoghue S.I., Meyer,J.E.W., Schafferhans,A. and Fries,K. (2002) The SRS 3D module: integrating sequence, structure, and annotation data, in preparation. [DOI] [PubMed]
- 8.Dodge C., Schneider,R. and Sander,C. (1998) The HSSP database of protein structure-sequence alignments and family profiles. Nucleic Acids Res., 26, 313–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holm L. and Sander,C. (1994) The FSSP database of structurally aligned protein fold families. Nucleic Acids Res., 22, 3600–3609. [PMC free article] [PubMed] [Google Scholar]
- 11.Kabsch W. and Sander,C. (1983) Dictionary of Protein Secondary Structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers, 22, 2577–2637. [DOI] [PubMed] [Google Scholar]
- 12.Rost B. (1999) Twilight zone of protein sequence alignments. Protein Eng., 12, 85–94. [DOI] [PubMed] [Google Scholar]
- 13.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith T.F. and Waterman,M.S. (1981) Identification of common molecular subsequences. J. Mol. Biol., 147, 195–197. [DOI] [PubMed] [Google Scholar]
- 16.McLachlan A.D. (1971) Tests for comparing related amino acid sequences. J. Mol. Biol., 61, 409–424. [DOI] [PubMed] [Google Scholar]
- 17.Rost B. and O'Donoghue,S.I. (1997) Sisyphus and protein structure prediction. Comput. Appl. Biosci., 13, 345–356. [DOI] [PubMed] [Google Scholar]
- 18.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Online Mendelian Inheritance in Man, OMIMTM (2000) McKusick-Nathans Institute for Genetic Medicine, Johns Hopkins University (Baltimore, MD) and National Center for Biotechnology Information, National Library of Medicine (Bethesda, MD), http://www.ncbi.nlm.nih.gov/omim/.