Abstract
The PANTHER database was designed for high-throughput analysis of protein sequences. One of the key features is a simplified ontology of protein function, which allows browsing of the database by biological functions. Biologist curators have associated the ontology terms with groups of protein sequences rather than individual sequences. Statistical models (Hidden Markov Models, or HMMs) are built from each of these groups. The advantage of this approach is that new sequences can be automatically classified as they become available. To ensure accurate functional classification, HMMs are constructed not only for families, but also for functionally distinct subfamilies. Multiple sequence alignments and phylogenetic trees, including curator-assigned information, are available for each family. The current version of the PANTHER database includes training sequences from all organisms in the GenBank non-redundant protein database, and the HMMs have been used to classify gene products across the entire genomes of human, and Drosophila melanogaster. PANTHER is publicly available on the web at http://panther.celera.com.
INTRODUCTION
The PANTHER database was designed for high-throughput functional analysis of large sets of protein sequences (1). It has been used to annotate the human genome (2) as well as the Drosophila genome (3). Like databases such as Pfam (4) and SMART (5), PANTHER uses a library of Hidden Markov Models (HMMs) to annotate sequences with information from homologous sequences. However, unlike these databases, the goal of PANTHER is not to annotate individual domains, but the overall biological function(s) of the molecule. Also unlike these other databases, because many protein families have branches that have diverged in function during evolution, the PANTHER library contains HMMs not only for families, but also for functionally distinct subfamilies. In these cases, subfamily annotation allows a much more precise definition of nomenclature and biological function.
PANTHER is composed of two main components: the PANTHER library (PANTHER/LIB) and the PANTHER index (PANTHER/X). PANTHER/LIB is a collection of ‘books’, each representing a protein family as a multiple sequence alignment, an HMM and a family tree. Functional divergence within the family is represented by first dividing the tree into subtrees (subfamilies) based on shared function, and then constructing a distinct HMM for each subfamily. PANTHER/X is an abbreviated ontology for summarizing and navigating molecular (biochemical) functions and biological processes (such as pathways, cellular roles or even physiological functions). Families and subfamilies are defined and named by biologist curators, who then associate each group of sequences with terms in the PANTHER/X ontology.
Protein query sequences can then be scored against the functionally-labelled family and subfamily HMMs. Query sequences are classified with the name and functional assignments of the best-scoring HMM, with the HMM score providing an estimate of the confidence level of the classification. Like other HMM-based approaches, PANTHER classification scales well for genome projects: the curated functional assignment is performed up-front on sets of training sequences that span many organisms, and can then be transferred to other organisms using the labelled HMMs. As a result, the PANTHER database classifies a significantly larger fraction of human genes than does LocusLink (Table 1).
Table 1. The percentage of human genes (approximated by LocusLink entries) having functional ontology classifications from PANTHER and from LocusLink GO associations.
| LocusLink GO | PANTHER/X | |
|---|---|---|
| Molecular function (NP) | 42% | 52% |
| Molecular function (XP) | 0% | 19% |
| Biological process (NP) | 41% | 46% |
| Biological process (XP) | 0% | 17% |
Percentages of genes classified are shown for two sets of LocusLink entries: NP (with a curated RefSeq protein, accession beginning with NP, total: 13 780), and XP (with only a provisional RefSeq entry, accession beginning with XP, total: 38 506). The total number of LocusLink entries that hit a PANTHER HMM is 9276 (67%) for NP, and 9141 (24%) for XP.
PANTHER has been available to Celera Discovery System (CDS) (7) subscribers for almost two years, and is now publicly available to academic users at http://panther.celera.com. The public version uses the GenBank non-redundant protein database to define sets of training sequences for HMMs. These HMMs are used to classify human gene products from LocusLink, and Drosophila melanogaster gene products from FlyBase (http://www.fruitfly.org/sequence/release3download.shtml). The CDS version includes training proteins from the sets curated at Celera, with additional HMM scoring of Celera-curated human and mouse gene products.
BROWSING GENES BY FUNCTION
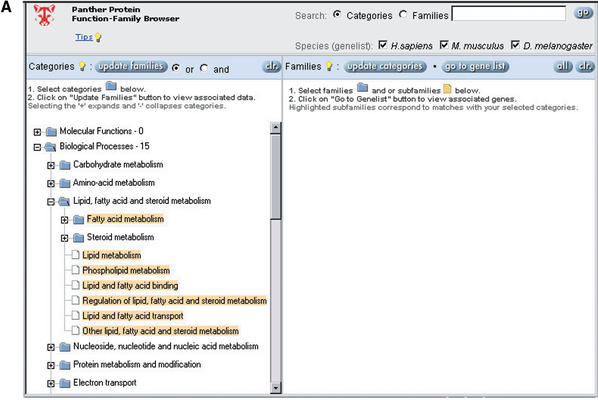
A key feature of PANTHER is that it can be browsed by protein functions, facilitating access to biologists. Browsing of controlled vocabulary terms can be much simpler than trying to construct effective queries in databases that have free text annotations. The primary entry point into PANTHER is the PANTHER Prowler, which uses the file-folder analogy to navigate PANTHER/X molecular functions and biological processes (Fig. 1). The PANTHER/X ontology is essentially hierarchical, though, more accurately, it is a directed acyclic graph as child categories occasionally appear under more than one parent if it is biologically justified. For example, the biological process DNA replication is a child of two categories: (1) nucleoside, nucleotide and nucleic acid metabolism, and (2) cell cycle. PANTHER/X contains many of the same higher-level categories as the more comprehensive Gene Ontology (GO) (8), and has been mapped to GO (3), but is arranged quite differently in order to facilitate navigation and large-scale analysis of protein sets. PANTHER/X also contains a number of vertebrate-specific categories that do not appear in the current release of GO, such as additional developmental and immune system categories.
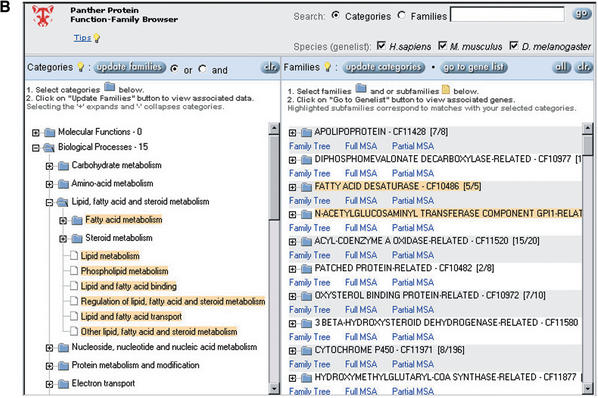
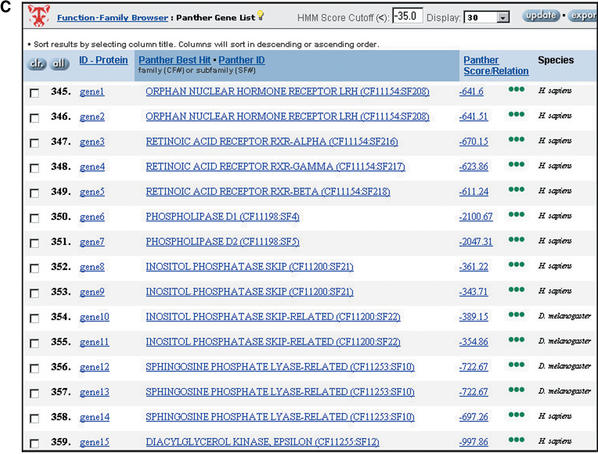
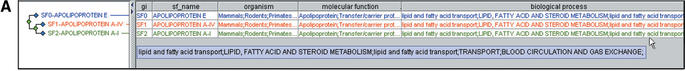
Figure 1.
Browsing the PANTHER database by biological functions. (A) Selection of biological processes under lipid, fatty acid and steroid metabolism (note that categories can be independently selected/deselected, so, for example, steroid metabolism has been deselected). (B) Retrieval of protein families and subfamilies assigned by curators to the selected functional categories. (C) Retrieval of a list of human genes encoding proteins that match the selected family and subfamily HMMs.
After a set of functions is selected, the Prowler retrieves the list of protein families and/or subfamilies that have been previously assigned, by biologist curators, to those functions. A user can make further selections in the family/subfamily list, and then generate a list of proteins or genes that scored significantly against the HMMs for the selected families and subfamilies. In the current version, gene lists are available for LocusLink human genes, and FlyBase Drosophila genes. The LocusLink and FlyBase sequences used to create these gene lists are updated on a monthly basis. Gene lists can be sorted and easily exported in tab-delimited format.
In addition to browsing, PANTHER can be accessed by text searching of curator-assigned family and subfamily names, or of the GenBank identifiers or definition lines of training sequences. Training sequences for the classification can also be searched by BLASTP (9).
SUPPORTING DATA: PHYLOGENETIC TREES, MULTIPLE SEQUENCE ALIGNMENTS AND SEQUENCE ANNOTATION
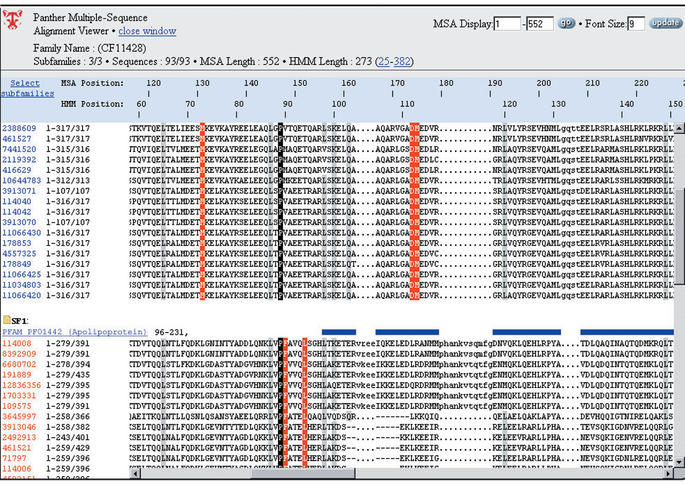
For each PANTHER family, data are available to support the curated classifications. The multiple sequence alignments used to generate the phylogenetic trees can be downloaded and viewed in a web browser. One of the features of the MSA viewer is that it highlights not only family-conserved columns (amino acids conserved across the entire family), but also subfamily-conserved columns (amino acids conserved within a subfamily but not found in other subfamilies). Curator-defined subfamilies have distinct annotations and often distinct functions, so these subfamily-conserved columns provide hypotheses about which residues may mediate functional divergence or specificity (Fig. 2).
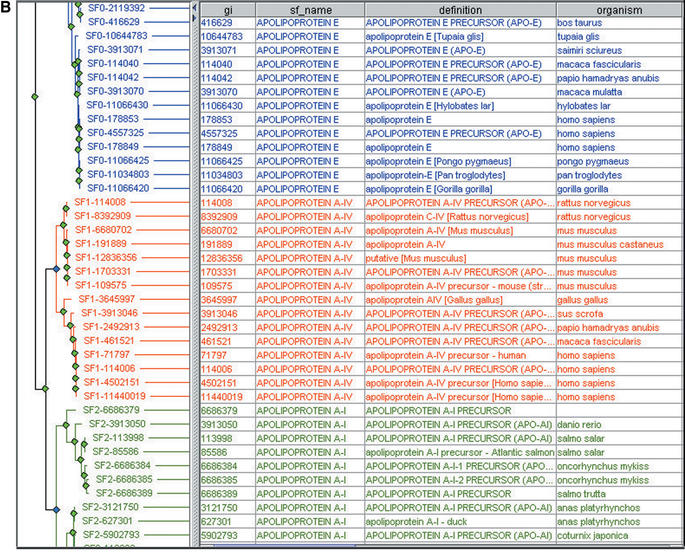
Figure 2.
The PANTHER multiple sequence alignment view, highlighting globally conserved positions (black and gray), and subfamily-specific conservation patterns that may indicate residues important for functional specificity (red). Pfam domains are shown as blue bars, one for each subfamily.
The phylogenetic trees, including the curator-defined subfamily divisions, can be viewed as GIF images. Subfamily nodes can be expanded to view sequence-level annotations from GenBank and SWISS-PROT (10), to verify curator definitions (Fig. 3). We also provide forms to make it easy for users of PANTHER to help correct names and ontology associations, and keep them up-to-date.
Figure 3.
The PANTHER tree-attribute view for verifying curation. (A) The ‘collapsed view’, showing the curator-defined subfamilies and ontology associations. (B) The ‘expanded view’, showing all of the constituent sequences and their annotations.
ACCURATE ASSIGNMENT OF FUNCTION USING HMMS FROM CURATED PROTEIN FAMILIES AND SUBFAMILIES
PANTHER/X functional ontology associations for gene products have been shown to be very accurate (3), primarily due to the emphasis on biologist curation, and to the tree-based homology inference method.
Curators define subfamilies in the context of aphylogenetic tree
Much of the curation of the PANTHER library is performed in the context of a phylogenetic tree (1). Trees are constructed for each family to represent the sequence-level relationships. A biologist curator then reviews the tree, dividing it into subtrees (subfamilies) such that all the sequences in a given subfamily can be given the same name and functional assignments. Names are free-text (following a set of defined guidelines available on the website), while the functional assignments use controlled PANTHER/X ontology terms. The family and subfamily groupings provide sets of training sequences for building HMMs.
The design of PANTHER, and the curation effort in particular, has been biased toward functional annotation and ontology classification. Most of the curation effort is devoted to assigning functions in the context of a phylogenetic tree representation, using functional information from SWISS-PROT and GenBank records, as well as more detailed information, if necessary, in OMIM (http://www.ncbi.nlm.nih.gov/omim/) and PubMed abstracts. A PANTHER family is defined to be as diverse as possible (increasing the number of sequences from which functional inferences can be made) while keeping it tight enough that the resulting tree is accurate. In the current version of PANTHER, we do not hand-curate the alignments or trees, or even demand that families be mutually exclusive; instead, curators judge them on how well they perform functional annotation. The tree-building algorithm is based on a distance metric derived from HMM scoring, so if proteins with the same function are located in the same subtree, the resulting subfamily HMMs will be predictive of function.
Competition between family and subfamily-level HMMs allows appropriate homology-based inference
The family and subfamily HMMs are then used to score sequences that were not in the training set. One of the advantages of PANTHER is the ability to assign specific functions, without overgeneralization. A sequence database search commonly assigns function based on the best hit. The advantage is that this assignment can be very specific, such as a GPCR having serotonin as a ligand. The disadvantage is that it is difficult to know when the query is too distant from the hit, and that the inference of serotonin binding is therefore incorrect. A family database search, on the other hand, will generally be correct in associating a sequence with a family, but cannot capture the specificity of function in divergent families. For example, there are members of the aldo-keto reductase family that function as ion channel subunits. PANTHER combines the advantages of both methods, by including both family and subfamily models in the HMM library. If the best hit is a subfamily HMM, and the HMM score is above the accepted threshold, then a specific annotation can be made, while a family HMM best hit often allows a less specific annotation. Following the example above, a family-level best hit will result in the annotation aldo-keto reductase 2 family member and no curated ontology terms, while a subfamily hit results in the annotation potassium voltage-gated channel, beta subunit ( family 6, subfamily A), and the ontology associations voltage-gated potassium channel (molecular function) and cation transport (biological process).
In the current release of PANTHER, all significant HMM scores are stored for each FlyBase Drosophila protein, and LocusLink human protein. The classification of each gene product is based on the best HMM score. For non-experts, whenever an HMM score is reported, it is accompanied by a ‘relation’ icon that indicates the relative certainty of the classification. As the scores become less significant, the probability becomes higher that the classification is in error. Even using a permissive score cutoff of −35 (‘distantly related’, i.e. the lowest degree of certainty), the total error rate for Drosophila molecular function classifications was shown to be less than 2% (3).
Because PANTHER/LIB comprises over 40 000 HMMs, it is not yet practical to provide a general web interface for HMM scoring of user-defined sequences. However, PANTHER/LIB HMM scoring can be made available as an additional service, or for collaborations.
PANTHER HMM annotations can differ from domain-based HMM annotation
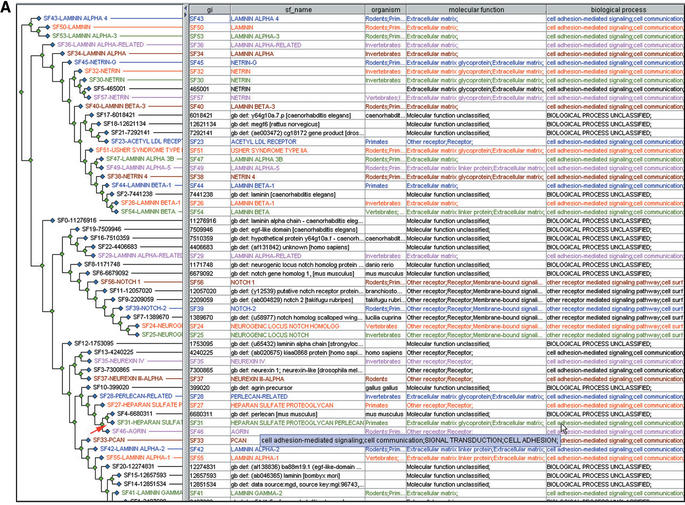
Databases such as Pfam and SMART have used the HMM formalism to provide an extremely useful tool for identifying conserved functional and structural domains in a protein sequence. PANTHER uses HMMs somewhat differently, with the goal of annotating the overall biological function of a protein. Like Pfam and SMART, the PANTHER family-level HMMs often have a functional annotation based on a single domain. PANTHER subfamily-level HMMs (and many family-level HMMs as well), however, can be more informative than the simple sum of the individual domain annotations. For example, the protein encoded by the human gene HSPG2 contains many different domains, including the LDL receptor A domain, epidermal growth factor repeat-like domains, immunoglobulin-like domains and both laminin B and laminin G domains. Each of these domains is found in different combinations across a variety of proteins having divergent functions. The only one of these domains that can be assigned a consistent function is the laminin-type EGF domain, which has been assigned by Interpro to the Gene Ontology (molecular function) term structural molecule. By contrast, the highest scoring PANTHER HMM is the subfamily heparan sulfate proteoglycan perlecan (CF10574:SF31), which is assigned to the PANTHER/X ontology terms (molecular function) extracellular matrix glycoprotein, and (biological processes) cell adhesion and cell adhesion-mediated signalling. This is a specific subfamily of the broader PANTHER family laminin-related (CF10574), which, like the Pfam laminin B and G domains, is not assigned to any functional terms (Fig. 4A).
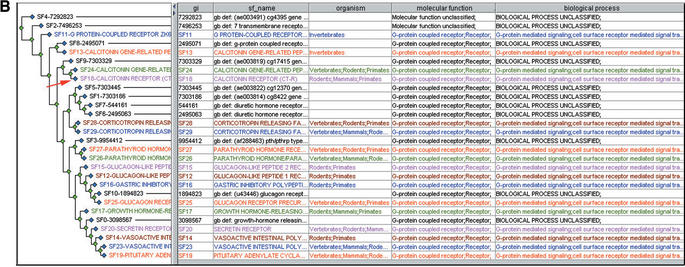
Figure 4.
Examples of PANTHER subfamilies capturing functional divergence. (A) Laminin-related proteins have divergent domain structures (which correlates with divergence within the shared laminin domain), while (B) Secretin-related GPCRs have divergent sequences within a common domain. Both cases can generally be modelled using subfamily HMMs.
Even for single-domain proteins the PANTHER subfamily HMMs often allow for more specific functional inferences than is possible from more general HMMs such as Pfam and SMART. For example, the CALCR gene product hits the Pfam HMM for the secretin-like seven transmembrane receptor family, which is assigned to the GO molecular function G protein-coupled receptor. The highest-scoring PANTHER HMM is the subfamily calcitonin receptor (CF12011:SF18), which is assigned to G protein-coupled receptor, as well as to the biological processes skeletal development and other neuronal activities. The more specific assignments are correct for this subfamily but not for all members in the larger family (Fig. 4B).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Kimmen Sjolander, Gangadharan Subramanian, Mark Yandell, Anthony Kerlavage, Richard Mural and Michael Ashburner for helpful discussions. We thank Matteo di Tommaso, James Jordan, Brian Karlak and Bruce Moxon for critical software engineering assistance. We also thank the many biologists who helped to curate the PANTHER library.
REFERENCES
- 1.Thomas P.D., Campbell,M.J., Kejariwal,A., Mi,H., Karlak,B., Daverman,R., Diemer,K. and Muruganujan,A. PANTHER: a library of protein families and subfamilies indexed by function, submitted. [DOI] [PMC free article] [PubMed]
- 2.Venter J.C., Adams,M.D., Myers,E.W., Li,P.W., Mural,R.J. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- 3.Mi H., Vandergriff,J., Campbell,M., Narechania,A., Lewis,S., Thomas,P.D. and Ashburner,M. Assessment of genome-wide protein function classification for Drosophila melanogaster, submitted. [DOI] [PMC free article] [PubMed]
- 4.Sonnhammer E.L., Eddy,S.R. and Durbin,R. (1997) Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins, 28, 405–420. [DOI] [PubMed] [Google Scholar]
- 5.Schultz J., Milpetz,F., Bork,P. and Ponting,C.P. (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl Acad. Sci. USA, 95, 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pruitt K.D., Katz,K.S., Sicotte,H. and Maglott,D.R. (2000) Introducing RefSeq and LocusLink: curated human genome resources at the NCBI. Trends Genet., 16, 44–47. [DOI] [PubMed] [Google Scholar]
- 7.Kerlavage A., Bonazzi,V., di Tommaso,M., Lawrence,C., Li,P., Mayberry,F., Mural,R., Nodell,M., Yandell,M., Zhang,J. and Thomas,P.D. (2002) The Celera Discovery System. Nucleic Acids Res., 30, 129–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T., Harris,M.A., Hill,D.P., Issel-Tarver,L., Kasarskis,A., Lewis,S., Matese,J.C., Richardson,J.E., Ringwald,M., Rubin,G.M. and Sherlock,G. (2000) Gene ontology: tool for the unification of biology. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 10.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]