Abstract
NetAffx (http://www.affymetrix.com) details and annotates probesets on Affymetrix GeneChip microarrays. These annotations include (i) static information specific to the probeset composition; (ii) sequence annotations extracted from public databases; and (iii) protein sequence-level annotations derived from public domain programs, as well as libraries of hidden Markov models (HMMs) developed at Affymetrix. For each probeset, NetAffx lists the probe sequences, and the consensus sequence interrogated by the probes; for the larger chip sets, interactive maps display this sequence data in genomic context. Sequence annotations include Gene Ontology (GO) terms and depiction of GO graph relationships; predicted protein domains and motifs; orthologous sequences; links to relevant pathways; and links to public databases including UniGene, LocusLink, SWISS-PROT and OMIM.
INTRODUCTION
Affymetrix expression microarrays are widely used in biomedical research. The microarrays consist of sets of DNA probes, each chosen carefully to record expression of specific genes. The set of probes relating to a gene is referred to as a probeset, and the sequence which is best associated with the transcribed region being interrogated by the probeset is referred to as its representative sequence.
At one time, there was little functional information provided with Affymetrix chips: each probeset was assigned an ID and a brief functional description, derived from GenBank. NetAffx is provided to detail probesets, and to describe the probesets in a functional context. The information provided for each probeset falls into two categories: static information and annotations.
The static information for each probeset details the probe sequences, and describes what the probes were designed to interrogate. The sequence annotations refer to the information about the representative sequence for a probeset. These include annotations available in UniGene (1), GenBank, LocusLink (2), model organism databases, SWISS-PROT (3), OMIM, etc.
Where possible, further annotations are provided to put the probesets into a functional framework. These annotations include Gene Ontology (GO) terms (4), GenMAPP (5) pathways and protein sequence analysis. Protein sequences are annotated using public databases including Pfam (6) and BLOCKS (7), and in-house libraries of hidden Markov models (HMMs) trained for recognition of SCOP, EC and GPCR protein families.
NetAffx is built around a searchable SRS interface that allows users to search for probesets matching specified criteria, including annotation terms, and to identify any probesets relevant to a user-specified DNA sequence. In this paper, we detail NetAffx content, organization and access methods.
SYSTEM ARCHITECTURE
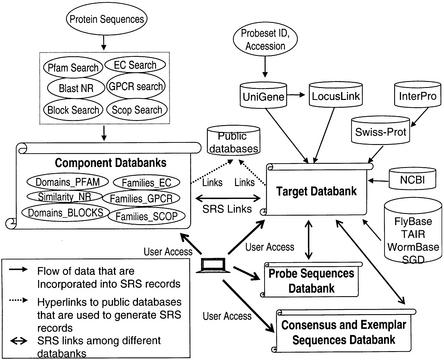
There are two main functions of NetAffx. One is to provide users with detailed descriptions of individual probesets. The second is to allow users to group probesets according to annotation type or category. These functionalities are provided through an SRS (Sequence Retrieval System, LionBio) databank management system and query interface. For each cataloged Affymetrix GeneChip microarray, an anchoring databank called Target summarizes all the annotations for the probesets. For example, the HG-U133 Target Databank is a compilation of probeset annotations and target sequence information for all the probes represented on the human genome HG-U133 A and B arrays. Supporting and providing more details to the Target databanks are Array Consensus and Exemplar Sequence databanks, and Annotation Component Databanks. Annotation Component Databanks hold detailed protein domain and similarity analysis results including alignments. All these supporting databanks are linked to the Target databanks in the SRS system as illustrated in Figure 1.
Figure 1.
Netaffx data flow. Protein sequences are annotated via the GRAPA battery of HMMs and PSI-BLAST models, in addition to Pfam, BLAST and BLOCKS searches. These annotations are consolidated into a unified XML format which is then indexed and loaded into SRS. These separate databanks (Domains_PFAM, Similarity_NR, Domains_BLOCKS, Families_EC, Families_GPCR, Families_SCOP) are summarized in the main Netaffx databank, the Target databank. Other annotations in the Target databank include those extracted from public databases according to the GenBank accession number of the representative sequences, as described in the text. Links exist from the Target databank to databanks of probe sequences and consensus and exemplar sequences. SRS databanks are indicated by the scroll-like icon.
STATIC PROBESET INFORMATION
Static probeset information includes the representative sequence (GenBank sequence accession, textual description, etc.), the UniGene cluster identifier for this gene, the subcluster from which the probeset is derived, and the set of sequence identifiers comprising the subcluster. The representative sequence is chosen during chip design as a sequence which is best associated with the transcribed region being interrogated by the probeset. The UniGene cluster is called the ‘Archival Reference Group’ in NetAffx because the UniGene cluster maintained by NCBI may change or be removed after the array design. The sequence identifiers in the subcluster are called the ‘cluster members’ in NetAffx.
Consider HG-U133 probeset 200697_at. The representative mRNA sequence has the GenBank accession NM_000188.1 The GenBank record indicates that the sequence definition is ‘hexokinase 1’ with gene symbol HK1. The sequence is a member of UniGene cluster Hs.118625, and its transcript identifier is Hs.118625.0. Note that there can be one or more transcripts per UniGene cluster due to different isoforms, polymorphisms, paralogs, or sequencing artifacts.
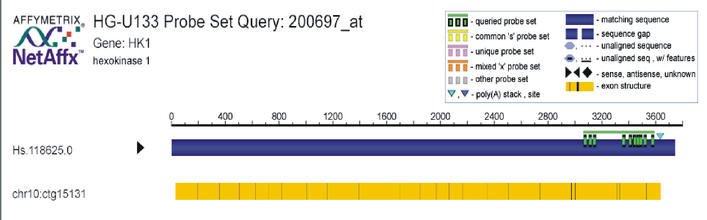
For several newer Affymetrix chips, the static probeset data is depicted graphically. Figure 2 illustrates the data provided for probeset 200697_at. The large blue band represents the exemplar/consensus for the subcluster corresponding to this probeset. The target sequence is shown by the horizontal green bar at the upper right, with the vertical green bars detailing locations of the probe sequences. The yellow bar at the bottom describes the exon structure. The black bars represent splice sites, with the introns collapsed in order to save space. This information is presented as an interactive map. The user can access data for each probeset or transcript by clicking on the corresponding element.
Figure 2.
The interactive probeset query map for probeset 200697_at from the human chip set HG-U133.
SEQUENCE ANNOTATIONS EXTRACTED FROM PUBLIC DATABASES
Annotations derived from public databases include descriptive and functional annotations of the gene sequence from current NCBI releases of the UniGene, LocusLink and Homologene databases (8). Where possible, each probeset is associated with one UniGene entry and one LocusLink entry. The UniGene identifier is determined according to the probeset's representative sequence. If the representative sequence is found in the current UniGene database, then that identifier is associated with the probeset. The UniGene title, gene symbol and cytogenetic bands are also extracted from the UniGene database. LocusLink information such as GO terms are assigned to the probeset via the LocusLink identifier in the UniGene record. In addition, Homologene gives some homolog/ortholog relationships for probesets on other Affymetrix chips. Probesets corresponding to ortholog genes across arrays for different organisms are identified using the ortholog data in the HomoloGene database at NCBI. The latest UniGene identifier corresponding to each probe set is used to assign ortholog probes across different arrays.
Sometimes the representative sequence is not found in the current UniGene release or it is not possible to determine which UniGene identifier corresponds to the representative sequence. This may occur because the representative sequence has been removed from GenBank or excluded from the UniGene build process. It may also occur when the representative sequence refers to an annotated mRNA in a DNA sequence. In those cases, a new UniGene assignment is inferred from other mRNA sequences from the original UniGene cluster believed to represent the same transcript.
Annotations are also derived from the SWISS-PROT database. A probeset's representative sequence is linked to SWISS-PROT accession through GenBank accession and its derived protein gi number. Additional protein domain and pathway annotations are subsequently derived from the InterPro database (available at http://www.ebi.ac.uk/interpro) and GenMAPP (5) (http://www.genmapp.org/), whose protein annotations are based on SWISS-PROT sequences.
Continuing with the probeset 200697_at example, the representative sequence NM_000188.1 is found in the UniGene cluster Hs.118625. Using the UniGene record for this cluster, the probeset is assigned the gene symbol ‘hexokinase 1’ and gene symbol ‘HK1’, and is linked with LocusLink record 3098. From LocusLink, we extract the GO terms ‘glycolysis’ and ‘hexokinase’, and an association with SWISS-PROT record P19367. This in turn links the probeset to a number of pathway maps related to glycolysis and metabolism.
PROTEIN SEQUENCE ANNOTATIONS
NetAffx provides protein annotations derived by sequence homology using the GRAPA method (9) on collections of hidden Markov models (HMMs) representing well-characterized protein families. These collections include: (i) Structural Classification of Proteins (SCOP), with models representing structural families of protein domains; (ii) Enzyme Classification (EC) with models representing all known enzymes, organized by protein structure, enzymatic reaction and substrate identity; and (iii) G protein-coupled receptors (GPCRs) with highly optimized models representing structurally and functionally-related families of this well-characterized class of transmembrane proteins.
NetAffx also provides protein domain annotations obtained by searching Pfam and BLOCKS databases. Top hits from BLAST analysis of protein sequences against the GenBank nonredundant (nr) database are also pre-computed.
Membrane proteins, which span the cell membrane, are often not biochemically characterized. However, they can still be identified as a class because the characteristic amphiphilic helices, which span the cell membrane, can be reliably identified. NetAffx annotates transmembrane portions of protein sequences using TMHMM (10) (http://www.cbs.dtu.dk/services/TMHMM/).
A parallel pipeline to GRAPA, called PSIGRAPA, uses PSI-BLAST (11) to categorize kinases according to the Hanks kinase scheme (12) (http://pkr.sdsc.edu/html/pk_classification/pk_catalytic/pk_hanks_class.html) and the Cytochrome P450 enzymes, as organized by Degtyarenko and Kulikova (13).
Consensus sequences for each probeset were aligned to the GenBank nonredundant protein sequence database using BLASTx (14). Of the probesets for previously unannotated EST-only clusters on the HG-U133 human array set, ∼27% showed significant similarity to a known human protein. The best BLASTx hit is now provided for the following species: Homo sapiens, Rattus norvegicus, Drosophila melanogaster, Arabidopsis thaliana, Mus musculus, Caenorhabditis elegans, Saccharomyces cerevisiae, Pan troglodytes, Macaca mulatta and Danio rerio.
Detailed protein annotations and the alignments are indexed in SRS databanks specific to a search method. For all protein annotations, NetAffx provides hyperlinks to outside sources (BLOCKS, Pfam, PDB, SCOP, GPCR), and pathways [KEGG (15) and GenMAPP (5)].
Consider probeset 200697_at. NetAffx provides BLAST hits to two human hexokinase sequences and several curated orthologs in rat and mouse. Additional annotations link this probeset to the hexokinase sequence family in BLOCKS and Pfam, the hexokinase structural family in SCOP, and the hexokinase D enzyme in EC.
FUTURE DIRECTIONS
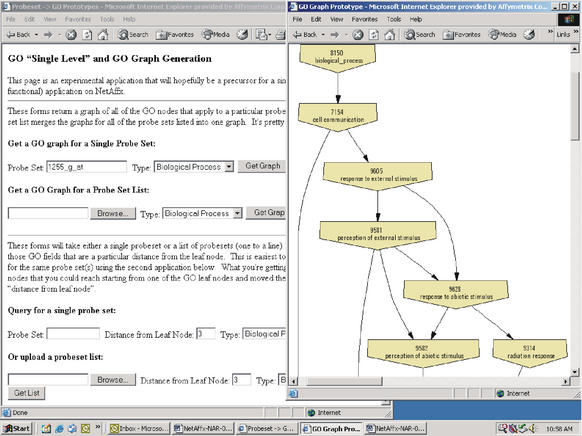
Gene Ontology is widely accepted as the standard for vocabulary describing the biological process, molecular function, and cellular component for genes. In addition to providing the readily available GO terms for annotated genes, we provide graphical, interactive views of the biological process GO sub-graph. These graphs allow the user to visually determine the relationships between a set of probesets based on their locations in the GO graph, thus, aiding in the biological interpretation of a complex set of results. See Figure 3. Furthermore, given the various lengths (or degrees of resolution) of GO paths associated with each probeset, one can examine the GO terms for a particular set of probesets based on level within the graph. By clicking on a specific GO term in the subgraph, a list of probesets with annotations at or downstream to this term will be retrieved. This functionality allows the partitioning of probesets based on the molecular function, biological process or cellular component of genes.
Figure 3.
Prototype interactive GO sub-graph for a set of probesets. One can rapidly generate a sub-graph for the GO terms associated with a single probeset or set of probesets. Part of the Biological Process subgraph is shown for probeset 1255_g_at for the U133 human GeneChip.
A computational approach to finding the relationships within the biological process GO graph is being developed. This method will assign a fingerprint to each probeset, such that high level functionality is flagged as present or not. These functional categories include terms like: cell growth, cell death, cell adhesion, embryogenesis, hematopoeisis, aging, etc. A matrix of probesets and GO functionality may be sorted such that subgroups of functionally-related probesets can be readily identified. Along with the signalling pathways, GO fingerprints will be useful for rapidly determining biological relevance of probesets from gene expression experiments.
Further efforts are being made to curate pathways of all types in conjunction with the GenMAPP group at the Gladstone Institute.
AVAILABILITY
NetAffx is freely available on the web at http://www.affymetrix.com/. Researchers are not required to pay for access to the database, nor do they need to pay to download the data to be used for their own personal research and publications.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to acknowledge our long-term collaborators on signalling pathways, Bruce Conklin, Kam Dahlquist, and Nathan Salomonis of the Gladstone Institute at UCSF.
REFERENCES
- 1.Schuler G.D. (1997) Pieces of the puzzle: expressed sequence tags and the catalog of human genes. J. Mol. Med., 75, 694–698. [DOI] [PubMed] [Google Scholar]
- 2.Pruitt K.D. and Maglott,D.R. (2001) RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res., 29, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O'Donovan C., Martin,M.J., Gattiker,A., Gasteiger,E., Bairoch,A. and Apweiler,R. (2002) High-quality protein knowledge resource: SWISS-PROT and TrEMBL. Brief. Bioinform., 3, 275–284. [DOI] [PubMed] [Google Scholar]
- 4.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S., Eppig,J.T. et al. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahlquist K.D., Salomonis,N., Vranizan,K., Lawlor,S.C. and Conklin,B.R. (2002) GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nature Genet., 31, 19–20. [DOI] [PubMed] [Google Scholar]
- 6.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henikoff J.G., Greene,E.A., Pietrokovski,S. and Henikoff,S. (2000) Increased coverage of protein families with the blocks database servers. Nucleic Acids Res., 28, 228–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler D.L., Church,D.M., Lash,A.E., Leipe,D.D., Madden,T.L., Pontius,J.U., Schuler,G.D., Schriml,L.M., Tatusova,T.A., Wagner,L. et al. (2002) Database resources of the National Center for Biotechnology Information: 2002 update. Nucleic Acids Res., 30, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shigeta R., Siani-Rose,M.A. and Kulp,D. (2001) Currents in Computational Molecular Biology 2001, pp. 247–248.
- 10.Krogh A., Larsson,B., von Heijne,G. and Sonnhammer,E.L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol., 305, 567–580. [DOI] [PubMed] [Google Scholar]
- 11.Altschul S.F. and Koonin,E.V. (1998) Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem. Sci., 23, 444–447. [DOI] [PubMed] [Google Scholar]
- 12.Hanks S. and Quinn,A.M. (1991) Protein kinase catalytic domain sequence database: Identification of conserved features of primary structure and classification of family members. Methods Enzymol., 200, 38–62. [DOI] [PubMed] [Google Scholar]
- 13.Degtyarenko K.N. and Kulikova,T.A. (2001) Evolution of bioinorganic motifs in P450-containing systems. Biochem. Soc. Trans., 29, 139–147. [DOI] [PubMed] [Google Scholar]
- 14.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 15.Wixon J. and Kell,D. (2000) The Kyoto encyclopedia of genes and genomes—KEGG. Yeast, 17, 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]