Abstract
TIGRFAMs is a collection of manually curated protein families consisting of hidden Markov models (HMMs), multiple sequence alignments, commentary, Gene Ontology (GO) assignments, literature references and pointers to related TIGRFAMs, Pfam and InterPro models. These models are designed to support both automated and manually curated annotation of genomes. TIGRFAMs contains models of full-length proteins and shorter regions at the levels of superfamilies, subfamilies and equivalogs, where equivalogs are sets of homologous proteins conserved with respect to function since their last common ancestor. The scope of each model is set by raising or lowering cutoff scores and choosing members of the seed alignment to group proteins sharing specific function (equivalog) or more general properties. The overall goal is to provide information with maximum utility for the annotation process. TIGRFAMs is thus complementary to Pfam, whose models typically achieve broad coverage across distant homologs but end at the boundaries of conserved structural domains. The database currently contains over 1600 protein families. TIGRFAMs is available for searching or downloading at www.tigr.org/TIGRFAMs.
INTRODUCTION
TIGRFAMs is a manually curated database of protein families described by hidden Markov models (HMMs) and attached information. It is available by FTP and through the World Wide Web. The salient feature of TIGRFAMs is the tuning of the breadth of each protein family to serve the needs of genome annotation. This is achieved through judicious selection of both cutoff scores and members of the seed alignment for each model. Factors examined during model construction include sequence similarity, evidence of function taken directly from the scientific literature, phylogenetics inferred from carefully constructed sequence alignments, species-specific metabolic context and neighboring genes.
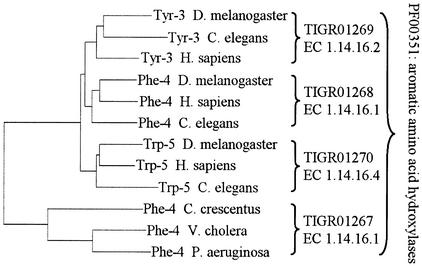
Figure 1 illustrates the tailoring of model range to support annotation. Four non-overlapping models were built from a larger family of aromatic amino acid hydroxylases. A neighbor-joining tree is shown rooted between eukaryotic, monomeric forms and tetrameric bacterial forms. The monomeric forms, although quite closely related to each other, are separated on the basis of both function and phylogenetics into three families, each representing a distinct biochemical activity.
Figure 1.
Neighbor-joining phylogenetic tree of aromatic amino acid hydroxylases. The nodes of a neighbor-joining tree based on aligned sequences are labeled to show assigned function. The tree is shown rooted at the left such that bacterial phenylalanine-4-hydroxylases (Phe-4) represented by TIGR01267, a tetrameric form, comprise the outgroup. Three other HMMs represent monomeric eukaryotic forms of aromatic amino acid hydroxylases (Tyr-3: tyrosine-3-monoxygenase, Trp-5: tryptophan-5-monoxygenase). The four equivalog models are children of the Pfam model PF00351. Note that the three closely related sets of eukaryotic proteins could have been represented by an additional subfamily HMM.
We have previously (1) defined the term ‘equivalog’ to describe the relationship of proteins conserved in function since their last common ancestor. This term stands in contrast to ortholog, the proper term for proteins related purely by speciation since their last common ancestor (2). Orthologs by definition cannot have undergone horizontal gene transfer events, although such events are ubiquitous (3,4). Orthologs are not necessarily conserved in function. Although the term ortholog is used commonly in the literature to imply conserved function, this ambiguous and imprecise usage may lead easily to misinterpretation. We suggest that separating the terms ortholog and equivalog will help clarify discussions of protein sequence homology.
More than half the models in TIGRFAMs are of type equivalog (as are the four TIGRFAMs in Fig. 1). Each TIGRFAMs equivalog model confers a strong prediction of the specific protein function named by the model to any protein that scores above its trusted cutoff. For example, model TIGR00936 is adenosylhomocysteinase, EC 3.3.1.1, with gene symbol ahcY in bacteria. The trusted cutoff score of 600 bits and the noise cutoff of −150 set thresholds for automated and manual annotation. The trusted cutoff sets the bar above which recognition by the HMM may trigger automated assignment of protein name, EC number, gene symbol, GO-IDs (5), use in metabolic reconstruction or phylogenetic profiling, etc. The noise cutoff filters out sequences that clearly belong to different families. Between noise and trusted is a gray zone requiring manual inspection. For this family, the gray zone is populated with a dubious second adenosylhomocysteinase from Archaeoglobus fulgidus and fragmentary sequences from a number of sources.
Over 350 TIGRFAMs models are of type ‘subfamily.’ ‘Superfamily’ is a homology type representing the complete set of proteins having homology over essentially their whole length. Members may vary greatly in function. A subfamily represents a distinct clade within a superfamily. We assign the term subfamily to classify any family that is not necessarily conserved with respect to function but is also not necessarily a complete superfamily.
The breadth of each subfamily model in TIGRFAMs is tuned, where possible, to support annotation. Within a large family of homologous transporters, for example, one phylogenetic clade may contain various heavy metal cation transporters. By reflecting the nature of the clade on the whole rather than the name of a one sample member, a TIGRFAMs subfamily HMM can provide an informative naming suggestion for any new member of the subfamily encountered during genome sequencing and annotation.
A subfamily model may perform two equally important functions in annotation. First, it can represent what is shared among a group of proteins that vary somewhat in function and thus extend the reach of annotation with moderately specific information. Second, it can mark families of proteins in which the danger of overinterpreting homologs as equivalogs can lead to misannotation. Comments within the model can explain the pitfalls of overinterpreting particular protein matches, of following certain legacy annotations, or of overgeneralizing from cursory analysis of pairwise matches.
A listing and description of the various types of models in TIGRFAMs, including equivalog, subfamily and domain can be found at http://www.tigr.org/TIGRFAMs/Explanations.shtml.
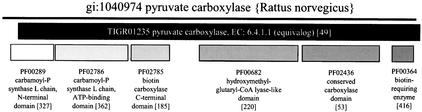
TIGRFAMs is designed to be complementary to Pfam, an invaluable resource for finding homology domains with high sensitivity. Both HMM databases use the same, freely available HMMER package (6) of HMM software. A review comparing Pfam, TIGRFAMs, and SMART has recently been published (7). A graphic illustration of one contrast between TIGRFAMs and Pfam is seen in Figure 2. Six separate domain HMMs from Pfam describe the architecture of the rat pyruvate decarboxylase, but none directly answers the questions ‘What should this protein be called?’ and ‘What does this protein do?’ each has a broad scope, describing regions shared by proteins with various functions. In contrast, a single equivalog model provides annotation for the protein on the whole.
Figure 2.
HMM hit regions for pyruvate carboxylase. The thin line represents the polypeptide sequence. Bars represent hit regions for various HMMs. Numbers in square brackets show the current size of each family. The number for each domain is larger than the number for the equivalog model because each domain is distributed more broadly than solely among pyruvate carboxylases.
The current release of TIGRFAMs, version 2.1, contains 1622 families, of which 837 are classified as equivalogs. An additional 198 are proposed equivalog families whose function is not yet known (‘hypothetical equivalog’). Coverage in newly sequenced bacterial genomes is estimated at 20% for all TIGRFAMs models and 10% for equivalog models, including roughly half of enzymes annotated with complete EC numbers.
TIGRFAMs strives for broad coverage of microbial proteins, but the starting point for model construction is often interest-driven. Areas of special focus have included transporters and DNA repair proteins from both prokaryotes and eukaryotes, bacterial housekeeping proteins and enzymes. More recent work has emphasized plant and parasite paralogous families and proteins characteristic of prokaryotic transmissable genetic elements such as CRISPR (8), temperate phage and conjugative transposons.
Validation of TIGRFAMs-based annotation has been attempted in two ways. First, HMM search results versus complete genomes are stored in relational database tables and subjected to quality control queries. The same region of the same protein should not belong to two different equivalog families. Equivalogs should appear once per genome for most genomes, excepting known cases of multiple isozymes. Second, TIGRFAMs models are tested by use. Models have been used for some time to make preliminary protein name assignments during microbial annotation at the Institute for Genomic Research (TIGR). Subsequent manual review of these annotations has provided steady feedback that has led to the improvement of many models.
We try to maintain existing models as we develop new ones. We would like to invite input from users of the database, including both suggestions to improve existing models and contributions of curated alignments. Contact information is available through the web page at http://www.tigr.org/TIGRFAMs/.
Acknowledgments
ACKNOWLEDGEMENT
TIGRFAMs is supported by Department of Energy grant number DEFC0201ER63203.
REFERENCES
- 1.Haft D.H., Loftus,B.J., Richardson,D.L., Yang,F., Eisen,J.A., Paulsen,I.T. and White,O. (2001) TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res., 29, 41–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fitch W.M. (1970) Distinguishing homologous from analogous proteins. Syst. Zool., 19, 99–113. [PubMed] [Google Scholar]
- 3.Nelson K.E., Clayton,R.A., Gill,S.R., Gwinn,M.L., Dodson,R.J., Haft,D.H., Hickey,E.K., Peterson,J.D., Nelson,W.C., Ketchum,K.A. et al. (1999) Evidence for lateral gene transfer between archaea and bacteria from genome sequence of Thermotoga maritima. Nature, 399, 323–329. [DOI] [PubMed] [Google Scholar]
- 4.Hayashi T., Makino,K., Ohnishi,M., Kurokawa,K., Ishii,K., Yokoyama,K., Han,C.G., Ohtsubo,E., Nakayama,K., Murata,T., Tanaka,M., Tobe,T., Iida,T., Takami,H., Honda,T., Sasakawa,C., Ogasawara,N., Yasunaga,T., Kuhara,S., Shiba,T., Hattori,M. and Shinagawa,H. (2001) Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res., 28, 11–22. [DOI] [PubMed] [Google Scholar]
- 5.Ashburner M., Ball,C.A., Blake,J.A., Botstein,D., Butler,H., Cherry,J.M., Davis,A.P., Dolinski,K., Dwight,S.S. and Eppig,J.T. (2000) Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nature Genet., 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eddy S.R. (1998) Profile hidden Markov models. Bioinformatics, 14, 755–763. [DOI] [PubMed] [Google Scholar]
- 7.Bateman A. and Haft,D.H. (2002) HMM-based databases in InterPro. Brief. Bioinform., 3, 236–245. [DOI] [PubMed] [Google Scholar]
- 8.Jansen R., Embden,J.D., Gaastra,W. and Schouls,L.M. (2002) Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol., 43, 1565–1575. [DOI] [PubMed] [Google Scholar]