Abstract
The University of California Santa Cruz (UCSC) Genome Browser Database is an up to date source for genome sequence data integrated with a large collection of related annotations. The database is optimized to support fast interactive performance with the web-based UCSC Genome Browser, a tool built on top of the database for rapid visualization and querying of the data at many levels. The annotations for a given genome are displayed in the browser as a series of tracks aligned with the genomic sequence. Sequence data and annotations may also be viewed in a text-based tabular format or downloaded as tab-delimited flat files. The Genome Browser Database, browsing tools and downloadable data files can all be found on the UCSC Genome Bioinformatics website (http://genome.ucsc.edu), which also contains links to documentation and related technical information.
INTRODUCTION
As the amount of genomic sequence in the public databases grows, annotation has become a critical element in data analysis and interpretation. The UCSC Genome Brower Database (http://genome.ucsc.edu) provides access to current and archived genome assemblies of selected organisms. One of the distinctive features of the database is the large collection of annotation data that accompanies each assembly. The annotations include mRNA and expressed sequence tag (EST) alignments, gene predictions, cross-species homologies, high-level maps, single nucleotide polymorphisms (SNPs) and many other types of data.
The vast size of vertebrate genome data sets presents challenges in efficient data storage and retrieval. In addition, the burgeoning number of versions of a particular genome demands a process that can rapidly integrate new data and annotations into the database while implementing creative solutions for maintaining and enhancing views of the data. Through software algorithmic refinements and optimizations to both the database and hardware, the UCSC Genome Browser viewer maintains the same interactive response time on the large Homo sapiens and Mus musculus genomes that its predecessor had on the much smaller Caenorhabditis elegans genome.
The Genome Browser Database—originally designed to support the Human Genome Project (1)—is readily extensible to accommodate the increasing number of genomic sequences in the public domain. Early assemblies of the human genome in the database were produced at UCSC from public data. Since the December 2001 freeze, the assemblies have been obtained directly from the National Center for Biotechnology Information (NCBI). A mouse genome based on data from the Mouse Sequencing Consortium was added early in 2002, with assemblies provided by the Mouse Genome Sequencing Consortium. The Genome Browser Database will support several additional genomes by 2003.
DATABASE ORGANIZATION
Sequence and annotation data for each genome assembly are stored in a MySQL relational database, which is quite efficient at retrieving data from indexed files. The database is loaded in large batches and is used primarily as a read-only database. To improve performance, each of the Genome Browser web servers has a copy of the database on its local disk.
The Genome Browser Database contains both positional tables with data based on genomic start–stop coordinates and non-positional tables with data independent of position. The coordinates in positional tables are defined using half-open zero-based ranges, i.e. the first 100 bases of a chromosome are represented as 0,100, while the next 100 bases are represented as 100,200 and so on. Half-open coordinates allow the length of a feature to be obtained by simply subtracting the start from the end and tend to minimize +/−1 errors during software development.
With such a large data set, care must be taken in the database organization to ensure good interactive performance in the Genome Browser, which creates each line of its graphical annotation image by querying the corresponding table in the database. The database is optimized to support the browser's range-based queries, with additional optimizations to accommodate the varying sizes of the positional tables in the database.
For smaller tables, it is often sufficient to create a few critical indices and do some initial presorting of the data before loading the database. For example, in the cpgIsland table, indices are created on chrom,chromStart and chrom,chromEnd. Presorting the data by chrom,chromStart significantly improves performance, particularly in tables where the indices are small enough to fit into RAM, thus reducing the number of disk seeks needed to load the data from the table into the Genome Browser.
Larger tables, such as the EST alignment table, require more complex schemes to keep interactive performance at an acceptable level. This is accomplished by first dividing up the data by chromosome into separate tables (e.g. chrN_est), which eliminates the need to index the chromosome field and increases the likelihood that the indices will fit into RAM. To further improve performance on large chromosome tables, a binning scheme suggested by Lincoln Stein and Richard Durbin has been implemented (2).
In addition to these tables, the database includes several non-positional tables that contain auxiliary data related to the mRNA sequences, e.g. DNA sequence, organism, or author. This information supplements the details pages for individual features in the graphical browser, but is not used in the display.
The document http://genome.ucsc.edu/goldenPath/gbdDescriptions.html contains a detailed description of the tables in the database.
To facilitate the programmatic interaction between the database and the graphical browser, UCSC uses the autoSql program written by Jim Kent (3). This program takes a data definition as input and creates a C structure, a SQL create statement and various C functions for data loading and memory management.
ANNOTATIONS
The Genome Browser Database provides dozens of aligned annotations for each genome assembly. Approximately half of the annotations are computed at UCSC and the remainder are contributed by external research scientists.
UCSC generates several annotations based on mRNA alignments. The mRNA and EST sequences are extracted from GenBank, and are aligned against the genome using the BLAST-like Alignment Tool (BLAT), a fast sequence alignment tool developed by Jim Kent (4). The data is filtered based on percentage identity and near best in genome to select only those alignments that best match the sequence. The spliced EST annotation is computed from the filtered data by analyzing the EST alignments for evidence of splicing.
The database contains a large collection of gene prediction annotations. The RefSeq Genes annotation is computed at UCSC from RefSeq mRNAs that have been aligned against the genome using BLAT and then filtered. The protein-coding portion of the mRNA is mapped to the genome and blocks separated by gaps of 5 or fewer bases are merged into exons. Gene prediction annotations contributed by external sources include Ensembl (5), Fgenesh++ (6), Genie (7), Acembly (contributed by Danielle and Jean Thierry-Mieg and Vahan Simonyan), and Genscan (8).
Several cross-species homology annotations are computed by UCSC and its collaborators, including BLAT and BLASTZ alignments of human and mouse genomes against the target genome, mRNA and EST alignments from other species, and synteny annotations. In cross-species annotations, RepeatMasker (9) and Tandem Repeats Finder (10) are first applied to the target genome to mask repetitive elements before the alignments are generated.
The database includes several high-level map annotations. Some examples of these include the Chromosome Bands annotation that approximates the locations of Giemsa-stained chromosomes bands at an 800-band resolution, the Sequence-Tagged Site (STS) Markers annotation showing the positions of several markers—many of which were used in constructing genome-wide genetic and physical maps—and the Fluorescent In Situ Hybridization (FISH) Clones annotation showing the location of FISH-mapped BAC clones from the BAC Resource consortium (11) on the genome.
Custom annotations are an important research feature supported by UCSC. Space constraints and confidentiality restrictions exclude many interesting annotations from the public version of the Genome Browser Database. The Genome Browser's custom annotation track feature allows individuals to upload formatted personal data for temporary display on the machine where the data resides. The custom annotation track is viewable only by users on that machine or via a custom URL that the users provide to other collaborators, thus making it an ideal mechanism for displaying and sharing private data. The document http://genome.ucsc.edu/customTrack.html contains complete information for constructing and uploading a custom annotation track.
VIEWING AND DOWNLOADING DATA
The UCSC Genome Bioinformatics website provides both graphical and text-based views of the sequence data and annotations.
The Genome Browser—accessed via the Genome Browser link on the home page—is a rapid interactive interface to the data at varying levels of detail. In response to a user's query, the Genome Browser displays a set of annotations tracks corresponding to the assembly and range dictated by the query and configured based on user preferences. Alternatively, the BLAT search tool can be used to quickly search for homologous regions to a DNA or protein sequence, which can then be displayed in the browser. The user can navigate through the assembly and zoom the image in or out using navigation buttons. A set of track controls allows the user to display each annotation in single line (dense) or expanded (full) mode or hide the track completely. Some tracks have filters to fine-tune the data displayed. Individual entries within a track have associated details pages that provide information about the annotation, related links to outside sites and database, and in some cases links to the genomic, mRNA and protein sequence.
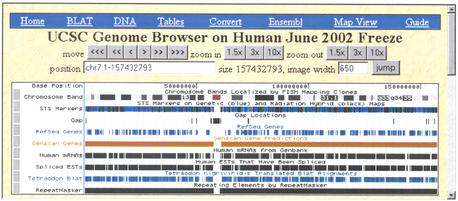
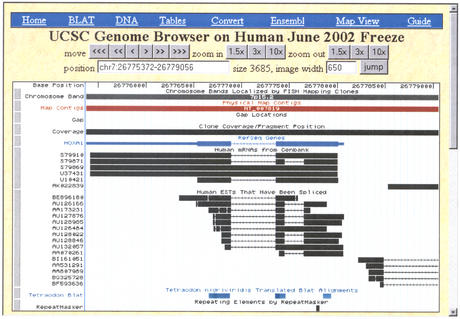
At the chromosome level (Fig. 1), the Genome Browser display provides an overview of the coverage and completeness for the region. At a zoomed in level (Fig. 2), the display can focus on a specific area of research interest, for example the examination of alternative splicing patterns of a gene. Most annotation tracks are displayed in a horizontal linear fashion, with the exception of a few ‘wiggle’ tracks that plot the feature's scores on a vertical axis.
Figure 1.
Screenshot of the Genome Browser annotation tracks display showing a view of an entire chromosome, human chr7 from the June 28, 2002 assembly. This level of detail provides an overview of the coverage and completeness for the region. The red section in the Chromosome Band track indicates the centromere. The shading in the Coverage track shows finished areas and coverage depth in draft areas. The Gap track indicates gaps in the assembly. The RefSeq Genes track shows gene density.
Figure 2.
A zoomed-in view of the same chromosome shown in Figure 1. The finer level of detail pinpoints the location of individual features in the annotation tracks. In the mRNA and RefSeq Genes gene prediction tracks, aligning regions (usually coding exons) are depicted by blocks connected by thin horizontal lines representing gaps (usually spliced-out introns). Thinner blocks on the leading and trailing ends of the aligning regions depict the 5′ and 3′ untranslated regions (UTRs). Arrowheads on the connecting lines show the direction of transcription.
The Genome Browser menu bar provides access to the BLAT search tool, the DNA sequence underlying the features in the annotation tracks graphic, a coordinate conversion tool for locating the position of a feature in a different release of a genome, and user documentation. The menu bar also contains direct links to the complementary annotation in the Ensembl Genome Browser and NCBI's Entrez MapView.
The Table Browser—accessed through the Tables link on the UCSC Genome Bioinformatics home page—furnishes an alternative text-based view of the data. This tool provides access to both positional and non-positional tables, and offers an enhanced level of query support that includes free-form SQL queries and restrictions based on field values. Output can be filtered to restrict which fields and lines are returned, and may be organized into one of several formats, including a simple tab-delimited file that can be loaded into a spreadsheet or database and then processed to produce the data format suitable for a custom annotation track.
The Downloads link on the UCSC Genome Bioinformatics home page offers a convenient interface for downloading current and archived sequence and annotation data. Data can also be downloaded via ftp at ftp://genome.cse.ucsc.edu/goldenPath/. Zipped data for each genome assembly is organized into three folders. BigZips contains the sequence data, which is packaged by chromosome and by contig. Files ending in suffix .fa contain the sequence in Fasta format for the contig layout. Files with suffix .agp contain an index that shows how the corresponding .fa file is built, with each line representing an actual sequence record or a gap. The Chromosomes folder contains the assembled sequence in Fasta format divided up by chromosome. The Database folder contains the genome annotation database tables in tab-delimited format.
A large portion of the Genome Browser Database is accessible using the Distributed Annotation Service (DAS) protocol (12). The UCSC DAS server is located at http://genome.ucsc.edu/cgi-bin/das. To accommodate the large size of some of the annotation tables, it is best to enable compression on DAS clients when accessing the UCSC DAS server.
In addition to the browsing tools, the UCSC Genome Bioinformatics home page provides links to the BLAT alignment tool, user and technical documentation, the Genome Browser mirror sites and archives, and acknowledgements to the many people who have contributed to the Genome Browser Database.
FUTURE DIRECTIONS
In the coming years, UCSC plans to add several new genomes to the Genome Browser Database, in addition to updating currently supported genomes as new assemblies become available. Enhanced searching capabilities will improve the query interface to the database. We will continue to incorporate annotations from the research community into our public Genome Browser, as well as support private access to custom annotation tracks created by users.
CONTACTING US
The mailing list genome@cse.ucsc.edu provides a forum for announcements of new releases and features, questions and discussion about the Genome Browser Database. Users may subscribe to this list at http://www.cse.ucsc.edu/mailman/listinfo/genome. To report problems accessing the website, servers, or mirror sites, or for correspondence inappropriate for the public forum, send email to genome-www@cse.ucsc.edu.
Acknowledgments
ACKNOWLEDGEMENTS
The UCSC Genome Browser Database project is funded by grants from the National Human Genome Research Institute (NHGRI) and the Howard Hughes Medical Institute (HHMI). We would also like to thank the many collaborators who have contributed annotation data to our project, as well as our users for their feedback and support.
REFERENCES
- 1.International Human Genome Sequencing Consortium (2001) Initial sequencing and analysis of the human genome. Nature, 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 2.Kent W.J., Sugnet,C.W., Furey,T.S., Roskin,K.M., Pringle,T.H., Zahler,A.M. and Haussler,D. (2002) The Human Genome Browser at UCSC. Genome Res., 12, 996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kent W.J. and Brumbaugh,H. (2002) autoSql and autoXml: Code Generators from the Genome Project. Linux J., 99, 68–77. [Google Scholar]
- 4.Kent W.J. (2002) BLAT—the BLAST-like alignment tool. Genome Res., 12, 656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hubbard T., Barker,D., Birney,E., Cameron,G., Chen,Y., Clark,L., Cox,T., Cuff,J., Curwen,V., Down,T., Durbin,R., Eyras,E., Gilbert,J., Hammond,M., Huminiecki,L., Kasprzyk,A., Lehvaslaiho,H., Lijnzaad,P., Melsopp,C., Mongin,E., Pettett,R., Pocock,M., Potter,S., Rust,A., Schmidt,E., Searle,S., Slater,G., Smith,J., Spooner,W., Stabenau,A., Stalker,J., Stupka,E., Ureta-Vidal,A., Vastrik,I. and Clamp,M. (2002) The Ensembl genome database project. Nucleic Acids Res., 30, 38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salamov A.A. and Solovyev,V.V. (2000) Ab initio gene finding in Drosophila genomic DNA. Genome Res., 10, 516–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kulp D., Haussler,D., Reese,M.G. and Eeckman,F.H. (1996) A generalized hidden Markov model for the recognition of human genes in DNA. Ismb, 4, 134–142. [PubMed] [Google Scholar]
- 8.Burge C. and Karlin,S. (1997) Prediction of complete gene structures in human genomic DNA. J. Mol. Biol., 268, 78–94. [DOI] [PubMed] [Google Scholar]
- 9.Smit A.F. (1999) Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev., 9, 657–663. [DOI] [PubMed] [Google Scholar]
- 10.Benson G. (1999) Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res., 27, 12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheung V.G., Nowak,N., Jang,W., Kirsch,I.R., Zhao,S., Chen,X.N., Furey,T.S., Kim,U.J., Kuo,W.L. and Livier,M. (2001) Integration of cytogenetic landmarks into the draft sequence of the human genome. Nature, 409, 953–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dowell R.D., Jokerst,R.M., Day,A., Eddy,S.R. and Stein,L. (2001) The Distributed Annotation System. BMC Bioinformatics, 2, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]