Abstract
The ERGOTM (http://ergo.integratedgenomics.com/ERGO/) genome analysis and discovery suite is an integration of biological data from genomics, biochemistry, high-throughput expression profiling, genetics and peer-reviewed journals to achieve a comprehensive analysis of genes and genomes. Far beyond any conventional systems that facilitate functional assignments, ERGO combines pattern-based analysis with comparative genomics by visualizing genes within the context of regulation, expression profiling, phylogenetic clusters, fusion events, networked cellular pathways and chromosomal neighborhoods of other functionally related genes. The result of this multifaceted approach is to provide an extensively curated database of the largest available integration of genomes, with a vast collection of reconstructed cellular pathways spanning all domains of life. Although access to ERGO is provided only under subscription, it is already widely used by the academic community. The current version of the system integrates 500 genomes from all domains of life in various levels of completion, 403 of which are available for subscription.
INTRODUCTION
During the last few years, the genomes of nearly 100 organisms have been completely sequenced while several hundred other genome projects are currently at various level of completion according to the GOLD database (1). It has become evident that the single most important tool for interpreting newly sequenced genomes is the effective integration and analysis of existing genomic sequence data on a comparative level. The success of the comparative analysis is directly dependent on the efficiency of integration, which in turn will be determined by the diversity of the organisms, the quality of their annotations and the level of detail in cellular reconstructions.
The ERGOTM bioinformatics suite has been designed at Integrated Genomics Inc. (IG) in order to accommodate such data integration, to provide the tools necessary to support the comparative analysis of genomes and the generation of sophisticated metabolic and cellular reconstructions (Appendix, Fig. A1). Emerging from PUMA and WIT, which were previously developed at Argonne National Laboratories (2,3), ERGOTM is a third generation bioinformatics suite offered exclusively from IG at: http://ergo.integratedgenomics.com/ERGO/.
The ERGO system represents the development of a genome analysis strategy into a multi-dimensional environment, which supports both automatic and manual genome-wide curation. Rather than just repackaging known information, ERGO integrates genomic information with biochemical data, literature and high-throughput analysis into a comprehensive user-friendly network of metabolic and non-metabolic pathways. In contrast to conventional systems, the ERGO user can take into account sequence similarity, protein and gene context clustering, occurrence profiles, regulatory and expression data, as well as functional hierarchies in order to achieve a set of the best possible functional predictions. In fact, using the ERGO system, a major part of the metabolism of an organism can be reconstructed entirely in silico (4). The cyclical nature of the integration of these information types continually elevates our knowledge and understanding of the complex dynamics residing in living organisms.
ERGO: A VIEW TO A GENOME
General description
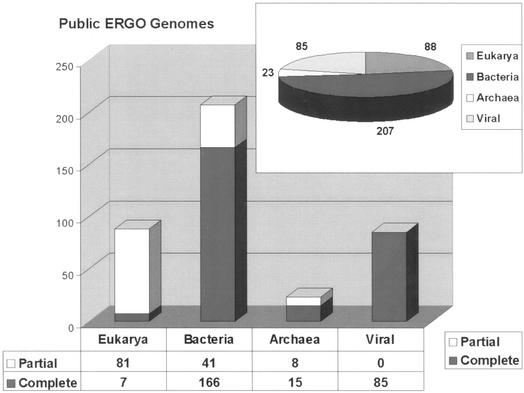
The current version of ERGO contains over 500 genomes at various stages of sequencing completion, 403 of which are publicly available for access through subscription. Both of those lists are growing on a bi-weekly basis (For a detailed list of the available genomes on ERGO, send your request to contact@integratedgenomics.com). From the list of the 403 genomes, 207 are bacterial (166 of which are complete), 23 archaeal (15 of which are complete), 88 eukaryal (7 of which are complete) and 85 are viral (all of which are complete) (Fig. 1). It should be mentioned here, that completely sequenced doesn't necessary imply one single contig, but rather according to our definition, a level of completion where more than 95% of the genes are identified (5).
Figure 1.
The number of complete and gapped genomes, available through the public ERGOTM bioinformatics suite.
These genomes consist of more than 735 636 Open Reading Frames (ORFs), of which ∼64% have a functional description. This level of function prediction rises close to 70% for the smaller set of published complete bacterial genomes, revealing that the function of approximately only 30% of genes remains relatively unknown for those well studied organisms.
The ERGO system integrates many different types of data, which are summarized in Table 1. These include mostly genomic and pathway related data. Currently under development is the integration of regulatory, essentiality and expression data (in fact these data are already available on the non-public version of the system). The genomic data include genome contigs, locations of ORFs and their translations, locations of RNAs, locations of insertion elements, functional assignments (along with their history records) and a number of proprietary gene clustering tools. The primary tools involve clustering of the ORFs according to sequences similarity (i.e. orthologs, paralogs and protein clusters) or gene context (i.e. chromosomal and fusion clusters). The ortholog clusters are essentially bi-directional best hits across different genomes, while paralog clusters are homologs within the same genome. Currently, for the set of 403 genomes there are over 26 000 ortholog clusters connecting more than 35% of the ORFs in ERGO and over 60,000 paralog families clustering more than 36% of the ORFs. Protein family clustering represents a new clustering technology being developed at IG. It is based on the highly manually curated ORF database of ERGO and is an attempt to produce protein families where all ORFs share strong sequence homology and have the same predicted function. More than 60% of the ORFs in ERGO are currently connected to these sets of clusters. The principal of chromosomal and fusion clustering and their importance in function prediction has been previously reported (6,7).
Table 1. Summary of data types in ERGO.
| Genomic data |
| DNA sequence data into contigs (from over 400 genomes) |
| ORFs and their Location (graphical visualization of ORFs on a contig) |
| Translation of ORFs |
| Pre-computed sequence similarities for each ORF (against the entire database) |
| Functional assignments of proteins (with their history records) |
| RNA assignments |
| Identification and localization of insertion elements (ISs) |
| Ortholog clusters |
| Paralog clusters |
| Protein family clusters |
| Chromosomal clusters |
| Fusion clusters |
| Pathway data |
| Chemical structures |
| Enzyme records |
| Metabolic pathways |
| Non-metabolic pathways |
| Cellular overviews (networks of metabolic and non-metabolic pathways) |
| Functional hierarchies (functional roles organized into gene ontologies) |
| Regulatory data |
| Essentiality data |
| Expression data |
GENOME ANALYSIS WITH ERGO (FUNCTION PREDICTION AND METABOLIC RECONSTRUCTION)
Loading a new genome into ERGO
In order to incorporate a genome into ERGO, all the potential ORFs must first be identified. This is accomplished with a set of IG-proprietary software tools. Sequence similarities are then calculated for all the newly predicted ORFs against the entire non-redundant set of ORFs in ERGOTM using the FASTA algorithm. The DNA sequence, the predicted ORFs, their coordinates and their calculated similarities are then loaded into the ERGO system in preparation for the analysis.
Annotations
In general, up to three levels of detail can be applied for functional annotation of the ORFs in the ERGO system: two before the completion of the organism's metabolic reconstruction and one after (see below). The first round of annotations is fully automated and is performed with a variety of IG-proprietary algorithms in order to predict the function of as many genes as possible. This round of annotations is largely based on the existence of ortholog and protein family clusters.
The second round involves a detailed manual expert analysis. This includes a manual inspection of the automatically assigned functions, as well as an exhaustive manual study of every single gene, by employing the combined use of both proprietary and publicly available tools. Since functional annotations have been traditionally based on similarity to genes of known function, ERGO provides online access to sequence similarity tools such as BLASTP or PSI-BLAST searches that are submitted to the NCBI server (8). In addition to these, queries can be submitted to more sensitive sequence similarity search tools such as the motif/pattern databases Pfam (9), PROSITE (10), ProDom (11), or COGs (12) (see ERGO ORF page). Furthermore, along with the fast growing numbers of sequenced genomes, additional methods that rely on gene context rather than on sequence similarity have also been developed (6,7). A number of IG-proprietary tools that explore the predictive power of chromosomal clustering and fusion events are also employed to assign a putative function, even in the absence of adequate sequence similarity.
Overall, the combined use of these tools, along with detailed manual curation supported by the ERGO system, results in a significant increase in the function prediction coverage (on average at the level of 10–20% for every genome project), as compared to most of the publicly available annotations. We have recently demonstrated the predictive power of this combinatorial approach, by using the genome of Thermotoga maritima as a showcase (13).
One of ERGOs most significant features is its comparative annotations environment that provides quality checks for both the automatic annotations and manual analysis. To this end, a user may request to compare all different annotations available for the genes of a particular genome. These annotations come either from other users of the ERGO system or from external databases (whose annotations have been already integrated into ERGO). By default all the function predictions from SWISS-PROT and TrEMBL (14), or PIR (15) are included for all genomes, as well as those based on Pfam and COGs (see Fig. A2 and A3 from the Appendix).
ERGO ORF page
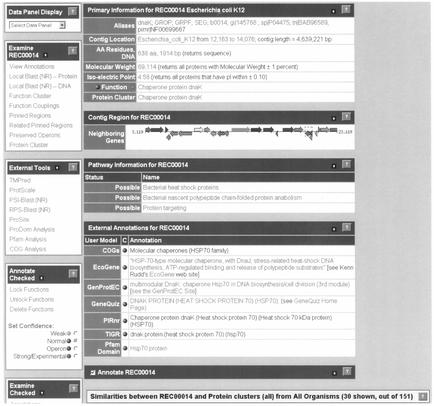
Most of the above queries are possible through the ERGO ORF curation page, the first part of which is displayed on Figure 2. Great effort has been expended to render this page essentially a workbench of curation and analysis of a single gene or its protein family, thus minimizing the need for performing time-consuming external database queries. As an example, the ORF page of the Escherichia coli dnaK gene is presented on Figure 3. Starting from the top, the user can see the ID of the ORF and the name of the organism. Below this, the Primary Information of this particular ORF is presented in a table. This includes a number of general features such as: (a) Aliases of this gene including different gene names or links to other databases such as SWISS-PROT and TrEMBL, or PIR, that have information about this gene; (b) Contig Location for this gene, which provides links to either a graphical viewer of this contig (or chromosomal region if it is a long contig) (see Appendix, Fig. A4), or to a page which presents information about the reported contig location in tabular form; (c) AA Residues, DNA provide links to pages that have the deduced amino acid or nucleotide sequence of this particular ORF; (d) predicted Molecular Weight or Iso-electric Point based on the EMBOSS Software Suite (16) and (e) predicted Function of this gene and predicted function of the Protein cluster to which it is a member. Below the Primary information table, there is a graphical Contig Region display for this ORF. This display provides the user with information regarding the genes in a 20 kb neighborhood around the query ORF, which is always displayed in the middle of the contig and colored in red. Below the Contig Region display the Pathway Information table provides a hyperlinked list of potential cellular pathways in which this function may play a role. Annotations derived from other users working on the system or other databases with publicly available information about the query gene are provided in the External Annotations table. The ERGO permits a user to interact with the system and introduce information related to the function of the gene. After opening the Annotation box, a user can add manually a new function or a comment for this ORF. Finally, the pre-computed similarities of this ORF against the rest of the database are presented further down this page (Fig. A5 from the Supplementary Material).
Figure 2.
ORF page in ERGO.
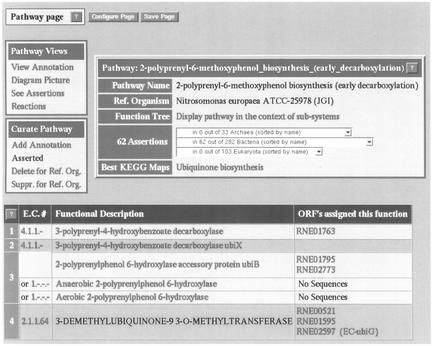
Figure 3.
Pathway page in ERGO.
Additional tools to analyze the query ORF are presented in a series of menus on the left of the page. The top menu, provides a list of links to IG-proprietary tools that are available for the analysis of the ORF. The following tools may be available: (a) View Annotations provides information related to the annotation history and previous comments made for this ORF; (b) Local Blast (NR) provides results in a Blast query of the current ORF's protein sequences against the ERGO non-redundant database; (c) Functional Cluster searches for other proteins that cluster with the query protein on the basis of functional annotation; (d) Functional Couplings identifies other functions that are ‘coupled’ to the function of the current protein; (e) Paralog Cluster presents the paralog cluster for the current protein; (f) Pinned Regions examine the chromosomal regions in other organisms that have the same structural and functional properties as the query protein (Fig. A6 from the Supplementary Material); (g) Related Pinned Regions explores the pinned regions of proteins orthologous to the query sequence; (h) Possible Fusions identifies the possible fusion events that may have occurred to form this protein; (i) Preserved Operons explores potential functional ‘operons’ that this protein may participate in as compared to other organisms and (j) Protein Cluster examines the protein cluster that this protein is a member.
External Tools menu provides the hyperlinks for direct submission of the query sequence to the set of public tools mentioned above. Below the external tools, there is a secondary ORF curation menu that provides quick access for processing the annotations by either locking them (in case multiple users are using the same username), or assigning different confidence levels for them (visualized as different background colors on the annotations).
Finally, the last menu on the page works with the list of pre-computed similarities (shown on Fig. A5 in the Supplementary Material) to allow analysis and comparison of groups of proteins. This menu includes tools that perform multiple alignments (protein or DNA) using CLUSTALW (17), or domain analysis using ProDom (11) in addition to the tools that display protein and DNA sequences in FastA formats.
ERGO pathway collection and their assertions
As soon as the genes are assigned with functions, they are automatically connected to their corresponding cellular pathways. The level of detail and coverage at this step is directly related to the number of pathways present in the ERGO system. Currently, the IG-pathways database (IG-Pathdb) contains over 5000 cellular pathways (the majority of which are metabolic). The metabolic pathway collection originates from the EMP database (18), with significant further corrections and development at IG Inc. Each metabolic pathway entry stores information about metabolites, reactions and corresponding enzymatic functions. The non-metabolic pathways, unlike the metabolic ones, represent either lists of functionally related genes (i.e. genes of the large ribosomal subunit, or genes of the type IV protein secretion) or general lists of process related functions (i.e. general transcription activators or Phage proteins).
Similar to the annotation process, there are at least two rounds of pathway assertions. During the first round, only the pathways that have all their steps (functions) connected to at least one gene will be assigned. Each function can be a part (step) of several different or alternative pathways. At the second round, an expert user can manually perform a ‘reality check’ to the set of asserted pathways (particularly, to the alternative ones), or assert additional ones, according to the literature data concerning the organism's ‘life style’, as well as its biochemistry and genetics. Once all possible pathways are asserted for this particular organism, then all possible connections are made across the asserted pathways. This leads to the design of complicated pathway networks, which is the ‘in silico’ functional reconstruction of the organism (4) (Supplementary Material, Fig. A1).
It then becomes possible to ask which functions should be expected to be present in this organism and yet have escaped identification (see below in the pathway page). This brings us to the third and final step of annotations, which entails a directed and reverse (as compared to the first two rounds) approach. Along this highly laborious step, the query is the function predicted to be present, and the target is the gene expected to be found, as opposed to the first two rounds where the query was the gene that had been predicted to exist and the target was the function that remained unidentified.
Thus, ERGO provides an ideal framework not only to identify and connect all possible functions to genes, but also to predict which functions should also be present and further facilitate the discovery of their corresponding genes.
ERGO pathways page
The ERGO pathway page presents information about a single pathway. As an example the path-way ‘2-polyprenyl-6-methoxyphenol_biosynthesis_(early_decarboxylation)’ in Nitrosomonas (upper part of ubiquinone biosynthesis in Bacteria) is presented on Figure 3. In the center of the page, the user can see the Pathway Name, the name of the reference (or current working) organism, a link to the functional hierarchy of this organism that places this pathway in the context of its functional reconstruction. On a more global comparative level, ERGO also displays the total number and names of all other organisms for which this pathway has been asserted. To simplify the comparisons, the organisms are classified into the three domains of life.
On the left of this table, two smaller menu tables provide access to Pathway Views and its Curation. The first menu provides hyperlinked access to the Annotation history of the pathway and its Diagram Picture (if it is a metabolic pathway) (Supplementary Material, Fig. A7). The See Assertions hyperlink submits a query to identify the genes for all of the steps of this pathway in other organisms. The second menu allows user specific Curation of the pathways, such as assertion or deletion.
Finally, below these menus, there is the pathway table. From here, the steps (functional roles) of the pathway with descriptions of all functions (through the EC# hyperlinks) can be accessed. In the right column of this table, the ORFs with the corresponding functions are linked. If an ORF encoding a particular functional role has not been identified in this organism but genes performing this role have been identified in other organisms, no ORF is displayed for that step. If the pathway has been asserted for this particular organism then this implies that the gene for this particular step should have been there. Therefore, the genes for those ‘empty’ steps in the asserted pathways are identified as locally missing genes (i.e. step 2, Fig. 3). Sometimes there is biochemical evidence for the existence of a particular enzymatic activity but no genes in any organism have been identified that perform this role. For those functional roles, ‘No Sequences’ is displayed in the pathway table (part of step 3, Fig. 3).
Up to a few years ago, bioinformatics tools could extend only to the limits of sequence similarity, and therefore were only as good as the pre-existing knowledge of gene functions based on traditional biochemical verification. In other words, bioinformatics tools could not predict the genes for new functions, if a gene encoding the function had not previously been cloned from any organism. During the last few years, new technology has been developed (mostly based on gene-context) that allows novel function predictions. One of those methods is based on the observation by Jacob and Monod (1961) that genes encoding consecutive biochemical reactions tend to localize close on the genome in some bacterial genomes, forming operons. Therefore, based on the tendency of functionally related genes to cluster along the chromosome it is now possible to expand our ability to predict functions beyond the realm of mere sequence similarity and to systematically identify missing components of known biochemistry.
Since approximately only a third of the genes of an average bacterial genome are functionally clustered, a large number of genomes are needed for the method to work. To demonstrate this, all prokaryotic organisms in ERGO were split into three different groups: (a) organisms that have their genome completely sequenced and published in the public domain (73 genomes); (b) organisms that have more than 90% of their genome sequenced (141 genomes); (c) and finally the complete set of ERGO prokaryotic organisms (230 genomes) (Fig. 4). Assuming that we identify the maximum amount (i.e. 100%) of functional connections (chromosomal cluster units) with the largest number of organisms, we can identify only 81% of those with the set of the above 90% sequenced organisms and a mere 14% when the analysis is restricted to the completely sequenced organisms. Thus, integrating a large collection of prokaryotic genomes with cellular pathways and chromosomal clustering provides a powerful tool for predicting functions for ‘missing’ genes as well as genes with weak homology (19,20).
Figure 4.
Comparison of the numbers of identified functional connections across three sets of organisms in ERGO: those that are completely sequenced and their genome in published (blue), those that are over 95% completion (red), and finally the complete set of prokaryotic organisms. Genomes stand for number of organisms, ORFs (K) displays the total number of ORFs in each group (in thousands), and PCBBs (Pair of Coupled Bi-directional Best Hits) describe the unit of the chromosomal cluster.
One can suggest a functional role for an unknown ORF by cross-referencing the chain of biochemical reactions with an ORF cluster in any genome. With 70% of biochemical functions shared between the kingdoms (IG, in press), a number of eukaryotic missing genes can also be predicted using bacterial and archaeal orthologs.
Metabolic reconstruction
Metabolic reconstruction (MR) refers to the deduction of the core functionality of a whole organism from sequence data. This technology permits the blending of sequence data with factual biochemical knowledge and the strain's physiology into a balanced model of cellular functionality. MR yields integrated information about the metabolism of a sequenced organism including probable biochemical and physiological characteristics. As a result, this information can be used to improve annotation of ORFs, to predict new functions (both for individual ORFs and for entire pathways), or even for genetic engineering purposes.
For a practitioner, metabolic reconstruction is an extremely powerful research tool, as it provides an amount of information comparable to the results of decades of wet-lab experimentation. A reconstruction model is particularly invaluable for production strains, oftentimes poorly studied and lacking proper literature coverage. Since each reaction from IG database can be represented in the form of a stoichiometric matrix, the MR model serves as a basis for static stoichiometric modeling and in silico simulation (21). Such simulations are an important component of rational strain development as they help in solving practical problems such as flux distribution, energy balancing, optimization of growth or nutrients utilization, etc.
A web-based metabolic overview of an organism is designed within the ERGO system, stored as a set of diagrams (Supplementary Material, Fig. A8). From the graphical overview, a queryable functional hierarchy is then automatically deduced (Supplementary Material, Fig. A9). This is essentially an ERGO-Ontology that provides a controlled vocabulary for annotations and better understanding of gene function (22). Currently, four general reconstructions exist within ERGO: a bacterial, a fungal, a plant and a human general overview. These general functional hierarchies provide a template for high-throughput comparative analysis of genomes.
Further development of metabolic reconstruction will be directed towards minimization of human efforts for curation of metabolic overviews, which can be achieved by means of stoichiometric analysis of metabolic networks.
COMPARATIVE GENOMICS WITH ERGO
Two key technologies have been developed within the ERGO bioinformatics suite, to facilitate the comparative analysis across whole genomes: WorkBenchTM and GenomeWalkTM.
The first one, WorkBenchTM is essentially a robust protein-clustering algorithm. Its goal is to provide fast and efficient identification of the shared and unique clusters of genes between different genomes. The second, GenomeWalkTM provides a graphical whole genome comparison environment that facilitates the identification of unique chromosomal regions between phylogenetically related genomes.
The importance of both tools has been demonstrated in the comparative analysis of different Xylella (23,24) and Fusobacteria (25) strains.
ERGO SYSTEM INFORMATION
ERGO is a web-based analysis package, and as such, the user capacity is limited only by the data processing power, memory and bandwidth available to the server, allowing many users to access and analyze data simultaneously. Standard CGI technology is used to access, retrieve and edit data. Database services are provided by a PostgreSQL database backend. Currently, almost 40 gigabytes of genomic data and more than 400 organism genomes are available for browsing. Access to the ERGO suite of tools is made available on a subscription basis or stand-alone servers can be purchased. Subscription information can be found at http://ergo.integratedgenomics.com/ERGO.
FUTURE PLANS OF ERGO
Two important trends are driving the development of ERGO. First, there is an exponential growth in the availability of new genomes, particularly eukaryotic genomes. One of our goals is to further develop ERGO into a key system for the characterization of the eukaryotic gene pool. We anticipate that ERGO will contain over a thousand genomes within the next three years.
The second driving force is the growing availability of expression profiles (from microarray data and proteomics). These data are supplemented with ‘conditional essentiality’ data, protein–protein interaction data and data from gene regulation. The large volumes publicly available for microarray and proteomic data will become a major source of clues for the clarification of gene function.
The development of a comprehensive genome analysis suite requires growth of the ERGO databases and discovery environment to incorporate new forms of genomic and proteomic data. It is our goal to develop ERGO into a central repository of key biological data leading to the elucidation of function for more genes and ultimately for the better understanding of the underlying cellular complexity.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank our colleagues Allen Bartman, Axel Bernal, Matt Daugherty, Josh England, Uy Ear, Galina Grechkina, Lynn Jablonski, Pete Jablonski, Jean-Louis Lassez, Tamara Los, Athanasios Lykidis, Gary Reznik, Eugene Selkov, Shiliang Wang and Lihua Zhu, as well as the Sequencing, Assembly, Microarray and Applications groups of Integrated Genomics for their contribution to the ERGO development.
REFERENCES
- 1.Bernal A., Ear,U. and Kyrpides,N. (2001) Genomes OnLine Database (GOLD): a monitor of genomes projects world-wide. Nucleic Acids Res., 29, 126–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Overbeek R., Larsen,N., Smith,W., Maltsev,N. and Selkov,E. (1997) Representation of function: the next step. Gene, 191, GC1–GC9. [DOI] [PubMed] [Google Scholar]
- 3.Overbeek R., Larsen,N., Pusch,G.D., D'Souza,M., Selkov,E.,Jr, Kyrpides,N., Fonstein,M., Maltsev,N. and Selkov,E. (2000) WIT—integrated system for high-throughput genome sequence analysis and metabolic reconstruction. Nucleic Acids Res., 28, 123–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Selkov E., Overbeek,R., Kogan,Y., Chu,L., Vonstein,V., Holmes,D., Silver,S., Haselkorn,R. and Fonstein,M. (2000) Functional analysis of gapped microbial genomes: amino acid metabolism of Thiobacillus ferrooxidans. Proc. Natl Acad. Sci. USA, 97, 3509–3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Overbeek R. (2000) Genomics: what is realistically achievable. Genome Biol., 1, Comment 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Overbeek R., Fonstein,M., D'Souza,M., Pusch,G.D. and Maltsev,N. (1999) The use of gene clusters to infer functional coupling. Proc. Natl Acad. Sci. USA, 96, 2896–2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enright A., Iliopoulos,I., Kyrpides,N. and Ouzounis,C. (1999) Protein interaction maps for complete genomes based on gene fusion events. Nature, 402, 86–90. [DOI] [PubMed] [Google Scholar]
- 8.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bateman A., Birney,E., Cerruti,L., Durbin,R., Etwiller,L., Eddy,S.R., Griffiths-Jones,S., Howe,K.L., Marshall,M. and Sonnhammer,E.L. (2002) The Pfam protein families database. Nucleic Acids Res., 30, 276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falquet L., Pagni,M., Bucher,P., Hulo,N., Sigrist,C.J., Hofmann,K. and Bairoch,A. (2002) The PROSITE database, its status in 2002. Nucleic Acids Res., 30, 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corpet F., Servant,F., Gouzy,J. and Kahn,D. (2002) ProDom and ProDom-CG: tools for protein domain analysis and whole genome comparisons. Nucleic Acids Res., 28, 267–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tatusov R.L., Natale,D.A., Garkavtsev,I.V., Tatusova,T.A., Shankavaram,U.T., Rao,B.S., Kiryutin,B., Galperin,M.Y., Fedorova,N.D. and Koonin,E.V. (2001) The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res., 29, 22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kyrpides N., Ouzounis,C.A., Iliopoulos,I., Vonstein,V. and Overbeek,R. (2000) Analysis of the Thermotoga maritima genome combining a variety of sequence similarity and genome context tools. Nucleic Acids Res., 28, 4573–4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bairoch A. and Apweiler,R. (2000) The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res., 28, 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu C.H., Huang,H., Arminski,L., Castro-Alvear,J., Chen,Y., Hu,Z.Z., Ledley,R.S., Lewis,K.C., Mewes,H.W., Orcutt,B.C. et al. (2002) The Protein Information Resource: an integrated public resource of functional annotation of proteins. Nucleic Acids Res., 30, 35–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rice P., Longden,I. and Bleasby,A. (2000) EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet., 16, 276–267. [DOI] [PubMed] [Google Scholar]
- 17.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Selkov E., Basmanova,S., Gaasterland,T., Goryanin,I., Gretchkin,Y., Maltsev,N., Nenashev,V., Overbeek,R., Panyushkina,E., Pronevitch,L. et al. (1996) The metabolic pathway collection from EMP: the enzymes and metabolic pathways database. Nucleic Acids Res., 24, 26–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Covert M.W., Schilling,C.H., Famili,I., Edwards,J.S., Goryanin,I.I., Selkov,E. and Palsson,B.O. (2001) Metabolic modeling of microbial strains in silico. Trends Biochem. Sci., 26, 179–186. [DOI] [PubMed] [Google Scholar]
- 20.Stevens R., Goble,C.A. and Bechhofer,S. (2000) Ontology-based knowledge representation for bioinformatics. Brief. Bioinform., 1, 398–414. [DOI] [PubMed] [Google Scholar]
- 21.Bhattacharyya A., Stilwagen,S., Ivanova,N., D'Souza,M., Bernal,A., Lykidis,A., Kapatral,V., Anderson,I., Larsen,N., Los,T. et al. (2002) Whole-genome comparative analysis of three phytopathogenic Xylella fastidiosa strains. Proc. Natl Acad. Sci. USA, 99, 12403–12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhattacharyya A., Stilwagen,S., Reznick,G., Feil,H., Feil,W., Lykidis,A., Anderson,I., Bernal,A., D'Souza,M., Ivanova,N. et al. (2002) Comparative functional reconstruction of three Xylella fastidiosa genomes. Genome Res., in press. [Google Scholar]
- 23.Kapatral V., Ivanova,N., Anderson,I., Mikhailova,N., Reznik,G., Gardner,W., Bhattacharayya,A., E., Larsen,N., D'Souza,M., Walunas,T. et al. (2002) Genome analysis of F. nucleatum spp vincentii and comparative analysis with F. nucleatum spp. Genome Res. [DOI] [PMC free article] [PubMed]
- 24.Daugherty M., Vonstein,V., Overbeek,R. and Osterman,A. (2001) Archaeal shikimate kinase, a new member of the GHMP-kinase family. J. Bacteriol., 183, 292–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Daugherty M., Polanuyer,B., Farrell,M., Scholle,M., Lykidis,A., de Crecy-Lagard,V. and Osterman,A. (2002) Complete reconstitution of the human coenzyme. A biosynthetic pathway via comparative genomics. J. Biol. Chem., 277, 21431–21439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.