Abstract
The p53 tumor suppressor protein participates in multiple cellular processes including cell cycle checkpoints and programmed cell death. In cell lines, loss of p53 function is associated with increased genetic instability including aneuploidy, gene amplification, and point mutation. Although similar genetic instability often accompanies the progression of malignancy in tumors, its role in tumor initiation in normal cells is not clear. To study whether or not loss of p53 leads to genetic instability in normal cells in vivo, we have examined mechanisms of loss of heterozygosity (LOH) at the Aprt (adenine phosphoribsyltransferase) and flanking loci in normal fibroblasts and T lymphocytes of p53-deficient mice. Somatic cell variants that arose in vivo as a consequence of genetic or epigenetic alterations abolishing Aprt function were selected and expanded in vitro by virtue of their resistance to 2,6-diaminopurine (DAP). We observed that p53 null mice produced about three times as many DAP-resistant fibroblast colonies than wild-type mice, but the frequency of DAP-resistant T lymphocyte colonies was not significantly changed. Mitotic recombination, but not point mutation, partly accounted for the increase in the frequency of DAP-resistant fibroblasts. Most significantly, chromosome loss/duplication and interstitial deletion, which were extremely rare events in the wild-type mice, represented a significant proportion of LOH events in both fibroblasts and T lymphocytes of p53 null mice. Also, increased interstitial deletion was observed in fibroblasts of p53 heterozygous mice. These data suggest that increased genetic variation, including chromosome instability, starts at the initiation stage of tumorigenesis when functional p53 is absent or reduced.
The tumor suppressor gene TP53 plays a key role in the regulation of cell cycle progression and apoptosis (1, 2). Its action also has been implicated in nucleotide excision repair (3), base excision repair (4), and recombinational repair (5, 6). These cellular functions of p53 can presumably prevent the accumulation of cells harboring deleterious genomic alterations. Consistent with this proposition, cells in culture that lack p53 exhibit increased genomic instability, evidenced by increased aneuploidy (7–9), gene amplification (10, 11), point mutation (12), and homologous recombination (13). An elevated frequency of loss of heterozygosity (LOH) at the TK locus was also observed in a human lymphoblastoid cell line with mutant TP53 (14, 15).
Mutation or loss of p53 appears to be a late event in colorectal cancer (16), but an early event in skin carcinogenesis (17). The manner by which loss of p53 function can exert a deleterious effect on the cell can range from induction of abnormal cellular proliferation, failure of apoptosis, accumulation of genetic alterations, or enhanced angiogenesis at late stages of malignant progression (18).
One approach to understanding the role of p53 mutations in the initiation of carcinogenesis is the evaluation of genetic instability in pathologically normal tissues of p53−/− mice. By using the lacI transgene as a reporter, two groups have characterized point mutations in lacI in multiple tissues of p53−/− mice. No change in either the frequency or the spectrum of point mutations was detected (19, 20), raising questions about the proposed role of p53 in the maintenance of genomic integrity. However, the lacI reporter strategy used in these studies has the following shortcomings. First, the lacI transgene may behave differently than endogenous genes (21). Second, rodent cells are less efficient in global nucleotide excision repair than human cells (22); thus lacI may not be a sensitive reporter of the changes, if any, in p53−/− mice. Third, it cannot detect genetic alterations at the chromosomal level, such as chromosome loss/duplication, mitotic recombination, and large deletions. This latter shortcoming is underscored by the observation that pro-B cells and thymocytes from p53−/− scid mice displayed a significant degree of aneuploidy during the premalignant phase (23), indicating that certain types of genetic instability may start to accumulate at an early stage of carcinogenesis in the absence of p53.
We have used the Aprt (adenine phosphoribosyltransferase) gene as a reporter of in vivo LOH in human T cells (24) and mouse fibroblasts (25). In both cases, we were able to distinguish between multiple pathways of LOH and to demonstrate that mitotic recombination accounted for 70–80% of the Aprt LOH events analyzed. Chromosome loss/duplication and deletion were extremely rare.
To study whether or not the loss of p53 may affect genomic stability at both gene and chromosomal levels in normal tissues, 129 × C3H Aprt+/− mice were crossed into a p53-deficient background, and the frequency and mechanistic spectrum of the 2,6-diaminopurine (DAP)-resistant variants of skin fibroblasts and T lymphocytes were characterized. We observed an increased level of chromosomal abnormalities, including chromosome loss/duplication and interstitial deletion, in both cell types. Because fibroblasts and T-cells are known to exhibit an elevated predisposition to sarcomas and lymphomas, respectively, in p53-deficient mice (26, 27), increased chromosome instability likely plays a role in the initiation of carcinogenesis in these cells.
Materials and Methods
Mice.
p53+/− mice in either 129/Sv and N5C3H/HeOuJ background were purchased from The Jackson Laboratory (27). p53+/− mice of 129/Sv strain were crossed to 129/Sv Aprt+/− to generate p53+/−Aprt+/− double heterozygotes. The N5C3H/HeOuJ p53+/− mice were backcrossed to C3H/HeJ for three generations. The N8C3H/HeJ p53+/− mice were crossed to 129/Sv p53+/− Aprt+/− to generate (129 × C3H)F1 hybrids that are Aprt+/− and p53+/+, p53+/−, or p53−/−. Some p53+/+ and p53+/− mice were also obtained from the crosses between 129/Sv p53+/− Aprt+/−, and C3H/HeJ mice.
Cell Culture.
Single skin fibroblasts were prepared from each ear as described (25). They were seeded at the density of 8 × 105 per 100-mm dish in selection medium (DAP, 50 μg/ml). At the same time, 1 × 104 cells were plated in drug-free medium with feeder cells to estimate colony-forming efficiency (CFE). The feeder cells were primary skin cells irradiated with 10,000 rads of gamma rays. The selection medium was changed every 4 days. The dishes for estimation of CFE, which were incubated without change of medium, were scored at day 12. At days 13–15, DAP-resistant colonies were counted and isolated from dishes containing selection medium.
The preparation and culture of spleen T lymphocytes were as reported (28). To recover DAP-resistant variants, primed lymphocytes were seeded at 2 × 104 per well in 96-well plates containing growth medium and DAP (50 μg/ml). Four cells were seeded per well in the presence of feeder cells (2 × 104 irradiated lymphocytes) to estimate CFE. Positive wells were scored at day 10.
Molecular and Cytogenetic Studies.
Characterization of the DAPr variants is described elsewhere (25). Allele-specific PCR determined whether or not the DAP-resistant phenotype was caused by physical loss of Aprt+. Clones with physical loss of Aprt+ were designated as class I and were further characterized for LOH of flanking simple sequence repeat (SSR) markers. Some class I clones were further characterized by fluorescence in situ hybridization (FISH) with whole chromosome 8 painting probes and an Aprt gene probe (pMAprt11). Clones without physical loss of Aprt+ were designated as class II and were assayed for APRT activity, and their Aprt genomic DNA was sequenced, as described previously (25).
Results
Comparison of CFE.
Both skin fibroblasts and T lymphocytes from p53−/− mice have a higher CFE than their p53+/+ counterparts. Whereas an average CFE of 1.6 ± 0.1% was observed for p53+/+ skin fibroblasts, it was elevated 1.6-fold, to 2.6 ± 0.2%, in p53 null mice (P < 0.001). To rule out the possibility that the increased CFE was caused by the re-seeding of migrating cells from early colonies, CFE was also estimated with 96-well flat-bottom plates. With 20 skin fibroblasts and 1 × 104 feeder cells per well, the p53-deficient cells still exhibited an increased CFE, 1.5-fold greater than that of the p53+/+ cells (data not shown). The CFE of fibroblasts from p53+/− mice, 1.8 ± 0.1%, was not significantly different from that of p53+/+ mice. T cells of p53−/− mice were observed to have 3-fold higher CFE than their p53+/+ counterparts, 4.3 ± 0.4% and 15.1 ± 1.3%, respectively (P < 0.001). The increased CFE we observed for skin fibroblasts and T cells was consistent with the enhanced in vitro proliferation reported for fibroblasts and epithelial cells of p53−/− mice (29).
Frequency of DAP-Resistant Variants in p53−/− Mice.
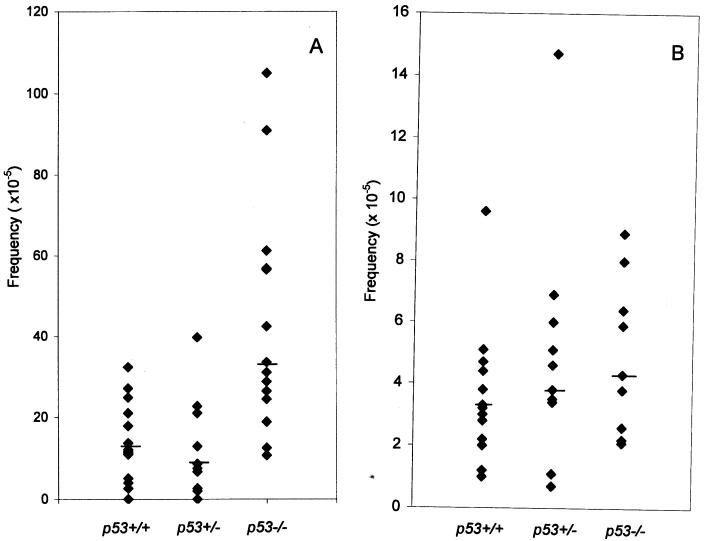
After adjustment for CFE, the frequency of DAPr fibroblast variants was estimated for each ear (Fig. 1A). Each group of mice showed a wide range in the frequency of DAPr cells in individual ears, but the median frequency was about 3-fold higher in p53−/− than in p53+/+ mice, 33.5 × 10−5 and 11.4 × 10−5, respectively (P < 0.002, Wilcoxon rank sum test). The frequency of DAPr fibroblast variants from p53+/− mice was not significantly different from that of p53+/+ mice (P = 0.56). Experiments using 96-well plates showed similar results (data not shown).
Figure 1.
Frequency of DAPr variants in p53+/+, p53+/−, and p53−/− mice; with fibroblasts (A), each point represents one ear; with T cells (B), each point represents one spleen. The bars indicate the median values.
The frequency of DAPr T cells also varied among individual p53+/+ mice, ranging from less than 1 × 10−5 to 73 × 10−5 in a group of 17 mice. Although the median frequency of 4 × 10−5 in p53−/− mice was slightly higher than that of the 3 × 10−5 median frequency observed in p53+/+ mice, the difference was not significant (P = 0.64, Fig. 1B).
Mitotic Recombination as the Primary Cause of LOH in p53+/+ and p53−/− Mice.
As reported earlier, DAPr variants can be classified into class I or class II by simultaneous amplification of the targeted (Aprtneo) and nontargeted (Aprt+) alleles (25). Clones exhibiting physical loss of Aprt+ (no amplification of Aprt+) were designated as class I, and those retaining Aprt+ as class II. Class I clones were further genotyped with SSR markers flanking Aprt to determine their putative mechanistic origin, e.g., mitotic recombination, chromosome loss or interstitial deletion. As shown in Tables 1 and 2, about 80% of the DAPr fibroblast variants isolated from p53+/+ mice were caused by mitotic recombination. Point mutation and epigenetic inactivation accounted for the remaining 20%. Chromosome loss/duplication and interstitial deletion were not observed in 88 clones.
Table 1.
Classification of DAP-resistant variants by allele-specific PCR
| Fibroblasts
|
T cells
|
|||||
|---|---|---|---|---|---|---|
| p53+/+ (n = 35) | p53+/− (n = 18) | p53−/− (n = 19) | p53+/+ (n = 19) | p53+/− (n = 9) | p53−/− (n = 11) | |
| Class I | 96 | 50 | 112 | 73 | 70 | 79 |
| Class II | 22 | 19 | 74 | 23 | 8 | 14 |
| Total no. clones | 118 | 69 | 186 | 96 | 78 | 93 |
| Fraction (%) of class I clones | 81 | 72 | 60 | 76 | 90 | 85 |
n, Number of mice.
Table 2.
Classification of class I clones by SSR genotyping
| Fibroblasts
|
T cells
|
|||||
|---|---|---|---|---|---|---|
| p53+/+ (n = 35) | p53+/− (n = 18) | p53−/− (n = 19) | p53+/+ (n = 19) | p53+/− (n = 8) | p53−/− (n = 11) | |
| A. Mitotic recombination | 88 | 42 | 93 | 57 | 65 | 63 |
| B. Chromosome loss with or without reduplication | 0 | 0 | 7 | 0 | 0 | 2 |
| C. Deletion/gene conversion | 0 | 5 | 6 | 1 | 1 | 11 |
| Total no. clones | 88 | 47 | 106 | 58 | 66 | 76 |
| Fraction (%) of B and C | 0 | 11 | 12 | 2 | 2 | 17 |
n, Number of mice.
The DAPr T cell variants recovered from p53+/+ mice display a similar mutational spectrum as the DAPr fibroblasts (Tables 1 and 2). A majority (76%) of the DAPr T cell variants were of the class I type, and an overwhelming majority of the class I clones (98%) exhibited LOH at the most distal, but not the most centromeric, locus (Table 2), suggesting mitotic recombination for their origin. Putative interstitial deletion/gene conversion (retention of heterozygosity at both D8Mit155 and D8Mit56) was detected in only one T cell clone. The mutational spectrum of the DAPr T cells we observed is consistent to that reported for 8-azaadenine-resistant T cells in Aprt+/− mice (30).
To determine whether or not deficiency of p53 may modulate the pathways of LOH, we characterized DAPr fibroblasts and T cells recovered from p53−/− mice. For fibroblasts, the majority of clones were of class I (60%). However, the fraction of class II variants was significantly increased (P < 0.005) in comparison to p53+/+ mice. In contrast, for T cells, the fractions of class I and class II clones were similar to those in p53+/+ mice (Table 1).
Genotyping with SSR markers showed that most class I variants from p53−/− mice exhibited LOH at all loci distal to Aprt, but not at loci near the centromere (Table 2), indicative of mitotic recombination. To confirm mitotic recombination as a mechanism of LOH, representative clones were analyzed by FISH with whole chromosome 8 painting probes and an Aprt gene probe, pMAprt11. All metaphases examined exhibited two normal copies of chromosome 8 and heteromorphic centromeres characteristic of each parental strain (25). FISH analysis with pMAprt11 DNA showed Aprt sequences on both copies of chromosome 8. Taking into consideration the increased overall frequency of the DAPr fibroblasts in p53−/− mice, the frequency of the fibroblast variants derived from mitotic recombination was calculated to be about 2-fold higher in p53−/− mice (2 × 10−4 vs. 1 × 10−4).
We reported previously that there were more than the expected number of mitotic crossovers in the interval between 59 and 67 centimorgans (cM) in 129 × C3H hybrids of p53+/+ mice (25). To test whether this is also true for the p53−/− mice, 61 class I clones isolated from p53−/− mice were characterized for mitotic crossover breakpoints. Although meiotic map predicts that seven clones would have a crossover in the 59- to 67-cM interval, 13 clones with a crossover in this interval were observed (P < 0.02). This observation indicates that the distribution of mitotic crossover breakpoints was similar in p53+/+ and p53−/− mice.
Taken together, mitotic recombination still predominates as the pathway of LOH in both fibroblasts and T cells of p53−/− mice. Thus, lack of p53 has minimal effect, if any, on mitotic recombination in T cells, although it may cause a modest increase in fibroblasts.
Prevalence of Somatic Variants with Chromosomal Abnormalities in p53−/− Mice.
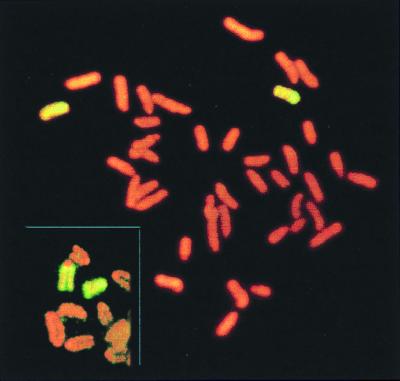
Molecular characterization of class I DAPr clones showed that two categories of clones were over-represented in p53−/− mice as compared with p53+/+ controls (Table 2). First, seven fibroblast clones and two T cell clones exhibited LOH at all loci tested, including D8Mit155, which is 1 cM from the centromere. This particular pattern of LOH could have arisen either by chromosome loss/duplication or by a mitotic crossover between the centromere and D8Mit155. Whole chromosome 8 painting of one such clone, 758R2A, showed two apparently normal copies of chromosome 8 in diploid metaphase spreads. However, the characteristic heteromorphic centromeric marker of the C3H strain was absent from each chromosome 8 (Fig. 2), indicating that C3H/HeJ chromosome 8, which contributes the Aprt+ allele, was lost and the 129/Sv parental chromosome 8 was duplicated, presumably by nondisjunction. Unless p53 deficiency enhances crossover between the centromere and D8Mit155, it is most likely that all these clones were derived from chromosome loss/duplication.
Figure 2.
Cytogenetic evidence that LOH was caused by chromosome loss followed by duplication in a class I clone. Genotyping showed loss of all C3H alleles of chromosome 8. Whole chromosome painting revealed two copies of chromosome 8 that are characteristic of the strain 129. Typical chromosome 8 of strains 129 and C3H are shown in inset, with strain C3H exhibiting a large centromeric spot.
Second, 6 fibroblast and 11 T cell class I clones remained heterozygous at the most centromeric locus, D8Mit155, and at the most telomeric locus, D8Mit56, but exhibited LOH at a variable number of loci flanking Aprt (Table 3). These DAPr variants were presumably caused by interstitial deletion or gene conversion. Clones with LOH extending to at least one flanking SSR marker were probably caused by interstitial deletion. Thus, the majority of class I clones in both p53+/+ and p53−/− mice are caused by mitotic recombination, whereas chromosome loss/duplication and interstitial deletion, which are rare in p53+/+ mice, account for more than 10% of the class I clones in p53−/− mice.
Table 3.
Intervals of interstitial LOH in representative clones
| No. of clones
|
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
p53−/−
|
p53+/−
|
||||||||||||||
| cM* | D8Mit | Fibroblasts
|
T cells
|
Fibroblasts
|
T cells
|
||||||||||
| 1 | 1 | 1 | 3 | 1 | 1 | 3 | 2 | 4 | 1 | 2 | 1 | 1 | 1 | ||
| 1 | 155 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● |
| 59 | 321 | ○ | ● | ● | ● | ● | ○ | ● | ND | ● | ● | ● | ○ | ○ | ● |
| 67 | Aprt | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ | ○ |
| 67 | 14 | ○ | ○ | ○ | ● | ○ | ● | ○ | ● | ● | ○ | ● | ● | ○ | ND |
| 72 | 280 | ● | ND | ● | ● | ○ | ● | ● | ● | ● | ● | ● | ● | ● | ND |
| 73 | 56 | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● | ● |
●, Retention of heterozygosity; ○, LOH; ND, not determined.
Distance from centromere.
Characterization of Class II Variants.
The CFE corrected frequency of DAPr T cells is not significantly higher in p53−/− mice than in p53+/+ mice. However, the frequency of DAPr fibroblasts, especially class II variants, was much higher in p53−/− mice. To determine whether the increase was because of increased Aprt point mutation, all five exons, introns 1, 3, and 4, and part of the promoter region of Aprt of 16 clones were sequenced. Only one clone revealed a point mutation, suggesting that most of the class II variants arose by other mechanisms.
We next determined the resistance of the class II fibroblast variants to 2-fluoroadenine (FA), an adenine analog that is more toxic than DAP to cells with residual APRT activity. Although all of the class I variants can grow in FA (2 μg/ml), only 1 of 18 class II variants survived the FA, suggesting that most of the class II variants were not completely APRT-deficient.
Representative fibroblast clones of class II were assayed for their residual APRT activity. Clones recovered from p53−/− mice were observed to retain higher residual APRT activity than those from p53+/+ mice. Whereas only 4 of 18 clones from p53+/+ mice have more than 40% of the APRT activity of the parental Aprt+/− cells, all 9 clones from p53−/− mice exhibited as much APRT activity (P < 0.01), indicating that p53−/− cells with APRT activity are more resistant to the toxicity of DAP.
Characterization of DAP-Resistant Variants from p53 Heterozygous Mice.
Mice of p53+/− genotype did not exhibit an increased frequency of DAPr cells compared with p53+/+ mice. Sixty-nine DAPr fibroblast variants isolated from p53+/− mice were characterized by allele-specific PCR. Fifty (72%) were designated as class I, and 19 (28%) as class II. Interestingly, 5 of the 47 class I clones examined exhibited interstitial LOH. These five clones were independently derived and recovered from three different mice (Tables 2 and 3). Because none of the 88 class I fibroblast clones recovered from p53+/+ mice exhibited interstitial LOH, the p53+/− mice may differ from p53+/+ mice in the generation or surveillance of this category of variants. The fact that fibroblast clones with interstitial deletion in p53−/− mice are not more common than in p53+/− mice suggests that complete deficiency of p53 protein is not a prerequisite for the generation of interstitial deletion. Consistent with that notion is that none of the five clones exhibited LOH at the p53 locus (data not shown). Probably a mere reduction of p53 dosage is enough for the generation of fibroblast variants with interstitial deletion. This notion is consistent with the finding that tumors from p53 heterozygous mice frequently retain a functional, wild-type p53 allele (31). Also, the transcriptional activity of p53 is less efficient in p53 heterozygous mice in response to radiation (32). In contrast to the overrepresentation of variants with interstitial deletion in fibroblasts of p53+/− mice, the spectrum of mutational pathways of the DAPr T cells recovered from p53+/− mice appeared to be similar to that in p53+/+ mice (Table 2).
Discussion
In Vivo Chromosome Instability and p53 Deficiency.
We compared the spontaneous frequency of DAPr fibroblasts and T cell variants in Aprt+/− mice with different p53 genotypes and characterized the spectrum of mutational pathways of these DAPr variants. The DAPr variants from p53-deficient mice exhibited more complex and diverse mutational pathways compared with those from p53+/+ mice. First, the frequency of fibroblast variants, but not T cell variants, was increased in p53−/− mice. The increase was, in part, a consequence of an enhanced level of mitotic recombination, but not of point mutation. Second, in both fibroblasts and T cells, chromosome loss/duplication and interstitial deletion/gene conversion emerged as prominent mechanisms of LOH in p53−/− mice. Third, interstitial deletion appears to be more common in fibroblast cells of p53+/− mice compared with wild-type mice. We conclude that chromosomal abnormalities, which are extremely rare in p53+/+ mice, account for a significant fraction of LOH in p53−/− and p53+/− mice.
Increased interstitial deletion in fibroblasts of p53+/− mice supports the notion of haploinsufficiency of p53 in heterozygotes (31, 32). It appears that, at least in fibroblasts, a mere reduction of p53 dosage is enough for the generation and propagation of somatic variants with chromosomal deletion. Thus, two functional p53 alleles may be required for the proper maintenance of chromosomal integrity in some tissues.
The increase of chromosomal abnormalities was not reflected in the overall frequency of DAPr variants due to the fluctuation in the frequency of variants derived from mitotic recombination. Mitotic recombination still predominated in p53−/− mice, accounting for more than half of the total variants, thus making a moderate increase (10–20%) in frequency indistinguishable in a small sample.
Regulation of Cell Cycle Checkpoint, Apoptosis, and DNA Repair by p53.
Characterization of the mutational pathways exhibited by DAPr variants in p53−/− mice serves to test hypotheses about possible roles for p53 in vivo. First, the increase of chromosomal abnormalities in p53−/− mice supports the notion of p53 playing a key role in regulation of cell cycle checkpoints and/or apoptosis. For example, failure of a spindle checkpoint would result in increased aneuploidy. Failure of apoptosis might result in an accumulation of interstitial deletions or other chromosomal abnormalities. It is possible that p53+/+ cells with two double strand breaks (DSBs) in one chromosome most likely undergo apoptosis. But when apoptosis is compromised by the absence or reduction of p53, they might be more likely to survive and allow joining of fragment ends. Misjoining may produce deletions or other chromosomal abnormalities.
It has been reported that p53 may suppress homologous recombination via the inhibition of RecA/Rad51 (5). Increased mitotic recombination observed in fibroblasts in p53−/− mice is consistent with that notion, so that repair of DSBs is shifted, to some extent, from end joining to homologous recombination repair in fibroblasts in the absence of p53. But it is also possible that cells without p53 may be more likely to survive DSBs because of the failure of apoptosis, thus allowing more recombinants to form. The dramatic increase of interstitial deletion in both fibroblasts and T cells of p53−/− mice suggest that direct end-joining still plays an important role in the repair of DSBs in the absence of p53.
Although the frequency of class II variants was elevated about 5-fold, from 2.4 × 10−5 in p53+/+ mice to 1.3 × 10−4 in p53−/− mice, most were not caused by point mutation. The lack of increased point mutation is consistent with the studies using lacI transgene as a reporter (19, 20). If lacI reflects only the efficiency of global nucleotide excision repair, Aprt as a reporter should be capable of detecting changes in transcription-coupled repair as well. Our results suggest that the effect of p53 on transcription-coupled repair of spontaneous DNA damage is minimal, if any, in the mouse, and is in agreement with the report that p53 mutant human fibroblasts exhibit normal transcription-coupled repair (33).
Cytotoxicity of DAP and p53.
The relative scarcity of variants with demonstrable Aprt point mutations and the retention of high levels of residual APRT activity in class II variants suggest that mechanisms other than Aprt point mutation may be responsible for the increased frequency of class II variants of p53−/− mice. The apparent increased frequency of class II variants in p53−/− mice may have been due to differential recovery in vitro. p53−/− cells with residual APRT activity appeared to have higher tolerance to the toxic affect of DAP, thus allowing more DAPr colonies. DAP is known to exert its toxic effects by blocking RNA synthesis, and it can also lead to abnormal nucleotide pools (34). The latter are known to be inducers of p53 (35, 36). Thus, the cytotoxic affect of DAP may be mediated by p53 to some extent, so that some p53−/− cells with residual APRT can divide to form colonies in the presence of DAP.
Chromosome Instability and Tumorigenesis.
The increased chromosome instability in T lymphocytes and fibroblasts of p53−/− mice may be pertinent to the high incidence of lymphomas, including T cell lymphoma and sarcomas in these mice. It is likely that increased chromosome instability is one of the major contributing factors to the increased predisposition to malignancy. Interestingly, chromosome instability seems to correlate to the observed tumor spectrum in p53+/− mice, as it is known that sarcomas replace lymphomas as the predominant tumor type in p53+/− mice (27). Correspondingly, we observed an increased interstitial deletion in fibroblasts, but not in T cells, of p53+/− mice. Consistent with this correlation is the report that p53−/− mice did not display an increased mutation frequency at the Dlb-1 locus within the cells of intestinal epithelium (37), whereas p53−/− mice are known not to be particularly predisposed to intestinal carcinoma (38).
In summary, we observed an increased level of chromosome abnormalities in normal tissues of p53 null and heterozygous mice. It should be noted that some types of chromosomal abnormalities like gene amplification and translocation (unless it disrupts Aprt) would not be detected by our assay. Assuming more cells with DSBs survived and the DSBs were not properly repaired in the absence or reduction of p53, an increase in the prevalence of translocation should also be expected. Chromosomal translocations are known to be a major cause of activation of protooncogenes. A population of cells with diverse chromosomal abnormalities should be favorably disposed to the initiation and progression of tumorigenesis.
Acknowledgments
This work was supported by National Institutes of Health Grants DK38185 and ES05652.
Abbreviations
- LOH
loss of heterozygosity
- Aprt
adenine phosphoribosyltransferase
- DAP
2,6-diaminopurine
- p53
p53 tumor suppressor gene
- CFE
colony-forming efficiency
- SSR
simple sequence repeat
- FISH
fluorescence in situ hybridization
- cM
centimorgan
- DSB
double strand break
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Levine A J. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal M L, Taylor W R, Chernov M V, Chernova O B, Stark G R. J Biol Chem. 1998;273:1–4. doi: 10.1074/jbc.273.1.1. [DOI] [PubMed] [Google Scholar]
- 3.Wang X W, Yeh H, Schaeffer L, Roy R, Moncollin V, Egly J M, Wang Z, Friedberg E C, Evans M K, Taffe B G, et al. Nat Genet. 1995;10:188–195. doi: 10.1038/ng0695-188. [DOI] [PubMed] [Google Scholar]
- 4.Offer H, Wolkowicz R, Matas D, Blumenstein S, Livneh Z, Rotter V. FEBS Lett. 1999;450:197–204. doi: 10.1016/s0014-5793(99)00505-0. [DOI] [PubMed] [Google Scholar]
- 5.Sturzbecher H W, Donselmann B, Henning W, Knippschild U, Buchhop S. EMBO J. 1996;15:1992–2002. [PMC free article] [PubMed] [Google Scholar]
- 6.Willers H, McCarthy E E, Wu B, Wunsch H, Tang W, Taghian D G, Powell S N. Oncogene. 2000;19:632–639. doi: 10.1038/sj.onc.1203142. [DOI] [PubMed] [Google Scholar]
- 7.Bischoff F Z, Yim S O, Pathak A, Grant G, Siciliano M J, Giovanella B C, Strong L C, Tainsky M A. Cancer Res. 1990;50:7979–7984. [PubMed] [Google Scholar]
- 8.Bouffler S D, Kemp C J, Balmain A, Cox R. Cancer Res. 1995;55:3883–3889. [PubMed] [Google Scholar]
- 9.Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude G F. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- 10.Yin Y, Tainsky M A, Bischoff F Z, Strong L C, Wahl G M. Cell. 1992;70:937–948. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 11.Livingstone L R, White A, Sprouse J, Livanos E, Jacks T, Tlsty T D. Cell. 1992;70:923–933. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 12.Havre P A, Yuan J, Hedrick L, Cho K R, Glazer P M. Cancer Res. 1995;55:4420–4424. [PubMed] [Google Scholar]
- 13.Mekeel K L, Tang W, Kachnic L A, Luo C M, DeFrank J S, Powell S N. Oncogene. 1997;14:1847–1857. doi: 10.1038/sj.onc.1201143. [DOI] [PubMed] [Google Scholar]
- 14.Honma M, Hayashi M, Sofuni T. Mutat Res. 1997;374:89–98. doi: 10.1016/s0027-5107(96)00223-0. [DOI] [PubMed] [Google Scholar]
- 15.Xia F, Wang X, Wang Y H, Tsang N M, Yandel D W, Kelsey K T, Liber H L. Cancer Res. 1995;55:12–15. [PubMed] [Google Scholar]
- 16.Fazeli A, Steen R G, Dickinson S L, Bautista D, Dietrich W F, Bronson R T, Bresalier R S, Lander E S, Costa J, Weinberg R A. Proc Natl Acad Sci USA. 1997;94:10199–10204. doi: 10.1073/pnas.94.19.10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziegler A, Jonason A S, Leffell D J, Simon J A, Sharma H W, Kimmerlman J, Remington L, Jacks T, Brash D E. Nature (London) 1994;372:773–776. doi: 10.1038/372773a0. [DOI] [PubMed] [Google Scholar]
- 18.Dameron K M, Volpert O V, Tainsky M A, Bouck N. Science. 1994;265:1582–1584. doi: 10.1126/science.7521539. [DOI] [PubMed] [Google Scholar]
- 19.Sands A T, Suraokar M B, Sanchez A, Marth J E, Donehower L A, Bradley A. Proc Natl Acad Sci USA. 1995;92:8517–8521. doi: 10.1073/pnas.92.18.8517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nishino H, Knoll A, Buettner V L, Frisk C S, Maruta Y, Havvik J, Sommer S S. Oncogene. 1995;11:263–270. [PubMed] [Google Scholar]
- 21.Mirsalis J C, Monforte J A, Wineger R A. Annu Rev Pharmacol Toxicol. 1995;35:145–164. doi: 10.1146/annurev.pa.35.040195.001045. [DOI] [PubMed] [Google Scholar]
- 22.Vijg J, Mullaart E, van der Schans G P, Lohman P H, Knook D L. Mutat Res. 1984;132:129–138. doi: 10.1016/0167-8817(84)90007-5. [DOI] [PubMed] [Google Scholar]
- 23.Guidos C J, Williams C J, Grandal I, Knowles G, Huang M T F, Danska J S. Gene Dev. 1996;10:2038–2054. doi: 10.1101/gad.10.16.2038. [DOI] [PubMed] [Google Scholar]
- 24.Gupta P K, Sahota A, Bye S, Boyadjiev S, Shao C, O'Neill P, Albertini R J, Stambrook P J, Tischfield J A. Cancer Res. 1997;57:1188–1193. [PubMed] [Google Scholar]
- 25.Shao C, Deng L, Henegariu O, Liang L, Raikwar N, Sahota A, Stambrook P J, Tischfield J A. Proc Natl Acad Sci USA. 1999;96:9230–9235. doi: 10.1073/pnas.96.16.9230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donehower L A, Harvey M, Slagle B L, McArthur M J, Montgomery C A, Butel J S, Bradley A. Nature (London) 1992;356:215–221. doi: 10.1038/356215a0. [DOI] [PubMed] [Google Scholar]
- 27.Jacks T, Remington L, Williams B O, Schmitt E M, Halachmi S, Bronson R T, Weinberg R A. Curr Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- 28.Meng Q, Skopek T R, Walker D M, Hurley-Leslie S, Chen T, Zimmer E M, Walker V E. Environ Mol Mutagen. 1998;32:236–243. [PubMed] [Google Scholar]
- 29.Tsukada T, Tomooka Y, Takai S, Ueda Y, Nishikawa S, Yagi T, Tokunaga T, Takeda N, Suda Y, Abe S, et al. Oncogene. 1993;8:3313–3322. [PubMed] [Google Scholar]
- 30.Van Sloun P P H, Wijnhoven S W P, Kool H J M, Slater R, Weeda G, van Zeeland A A, Lohman P H M, Vrieling H. Nucleic Acids Res. 1998;26:4888–4894. doi: 10.1093/nar/26.21.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venkatachalam S, Shi Y P, Jones S N, Vogel H, Bradley A, Pinkel D, Donehower L A. EMBO J. 1998;17:4657–4667. doi: 10.1093/emboj/17.16.4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gottlieb E, Haffner R, King A, Asher G, Gruss P, Lonai P, Oren M. EMBO J. 1997;16:1381–1390. doi: 10.1093/emboj/16.6.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ford J M, Hanawalt P C. Proc Natl Acad Sci USA. 1995;92:8876–8880. doi: 10.1073/pnas.92.19.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weckbecker G, Cory J G. Adv Enzyme Regul. 1989;28:125–144. doi: 10.1016/0065-2571(89)90068-x. [DOI] [PubMed] [Google Scholar]
- 35.Chernova O B, Chernov M V, Agarwal M L, Taylor W R, Stark G R. Trends Biochem Sci. 1995;20:431–434. doi: 10.1016/s0968-0004(00)89094-5. [DOI] [PubMed] [Google Scholar]
- 36.Linke S P, Clarkin K C, Di Leonardo A, Tsou A, Wahl G M. Genes Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- 37.Clarke A, Howard L A, Harrison D J, Winton D J. Oncogene. 1997;14:2015–2018. doi: 10.1038/sj.onc.1201040. [DOI] [PubMed] [Google Scholar]
- 38.Clarke A, Cummings M C, Harrison D J. Oncogene. 1995;11:1913–1920. [PubMed] [Google Scholar]