Abstract
Background
Cell-specific expression of the gene that encodes brain-derived neurotrophic factor (BDNF) is required for the normal development of peripheral sensory neurons and efficient synaptic transmission in the mature central and peripheral nervous system. The control of BDNF gene expression involves multiple tissue and cell-specific promoters that are differentially regulated. The molecular mechanisms that are responsible for tissue and cell-specific expression of these promoters are still incompletely understood.
Results
The cloning and analysis of three additional zebrafish (Danio rerio) BDNF gene exons and two associated promoters, is reported. Among them are two exons that generate a novel tripartite mature transcript. The exons were located on the transcription unit, whose overall organization was determined by cloning, Southern blot hybridization and sequence analysis, and compared with the pufferfish (Fugu rubripes) and mammalian BDNF loci, revealing a conserved but more compact organization. Structural and functional analysis of the exons, their adjacent promoters and 5' flanks, showed that they are expressed cell-specifically. The promoter associated with the 5' exon of the tripartite transcript is GC-rich, TATA-less and the 5' flank adjacent to it contains multiple Sp1, Mef2, and AP1 elements. A fusion gene containing the promoter and 1.5 KB of 5' flank is directed exclusively to skeletal muscle of transiently transfected embryos. The second promoter, whose associated 5' exon contains a 25-nucleotide segment of identity with a mammalian BDNF gene exon, was transiently expressed in yolk of the early embryo. RT-PCR analysis of total RNA from whole juvenile fish and adult female skeletal muscle revealed tissue-specific expression of the 5' exons but the novel exon could not be detected even after two rounds of nested PCR.
Conclusion
The zebrafish BDNF gene is as complex as the mammalian gene yet much more compact. Its exons are expressed in an independently regulated and cell-specific fashion. An initial structural and functional analysis has shown that the regions controlling zebrafish BDNF gene expression have been cloned and identified. They can now be subjected to detailed molecular and genetic analyses to identify the cellular mechanisms by which the transcription factors that act on these regions control BDNF gene expression.
Background
The neurotrophins play crucial functions in the interaction of muscles with their innervating neurons during development and in the adult vertebrate organism. The neurotrophin family includes Nerve Growth Factor (NGF), Neurotrophin-3 (NT-3), Neurotrophin-4/5 (NT-4/5), Neurotrophin-6 (NT6) and Brain-derived Neurotrophic Factor (BDNF). Three neurotrophins, BDNF, NT-3, and NT-4/5 are known to contribute to the development, maintenance, and functioning of the neuromuscular system. Each of these three neurotrophins is expressed differentially in muscle during development, in the adult and in response to injury. NT-3 is the most abundantly expressed of the three neurotrophins during embryonic muscle development, followed by NT-4/5 and BDNF [1,2]. After birth, NT-3 and NT-4/5 are down-regulated in muscle whereas BDNF continues to be expressed at low levels [1]. BDNF mRNA is expressed in myocytes in the adult muscle [3]. Essentially nothing is known about BDNF gene expression in fish muscle. We detected BDNF mRNA in the muscle layer of the fin of the 4 days old zebrafish larvae using in situ hybridization [4], but the identity of the expressing cells has not yet been definitvely established.
The first demonstration that BDNF potentiates neuromuscular transmission was reported by Lohof AM, et al. [5]. BDNF transiently stabilizes silent NM synapses during the period of transition from polyneuronal to mononeuronal innervation of skeletal muscle fibers [6]. BDNF and CNTF cooperatively affect efficiency of embryonic xenopus neuromuscular synapses in culture [7,8]. BDNF stimulates transmitter release from neuromuscular synapses [9] and enhances the efficiency of transmission at the neuromuscular synapse [10]. BDNF, together with NT-3, can restore neuregulin levels and a normal distribution of AchR at the neuromuscular junction blocked with curare [11]. BDNF, in conjunction with fibronectin, increases spontaneous synaptic currents in the cultured embryonic xenopus neuromuscular junction [12]. BDNF down-regulates AchR clustering, most likely by binding to TrkB expressed in myocytes [13]. BDNF downregulates agrin levels in muscle but increases neuregulin in ventral horn spinal motor neurons at the time when they form neuromuscular synapses [14].
BDNF production is activity dependent [11,15] Muscle activity is required to maintain BDNF mRNA levels in rat soleus muscle [16,17]. Vigorous exercise increases BDNF mRNA levels above baseline [17]. Neuromuscular blockade in culture also decreases BDNF mRNA levels. In response to nerve injury BDNF mRNA is slowly but substantially increased in Schwann cells and muscle fibers [18]. BDNF expression is also increased in cardiac myocytes following ischemic injury [19].
The molecular mechanisms that induce BDNF gene expression in muscle during development, maintain basal expression in the adult muscle and dynamically regulate BDNF mRNA in muscle in response to neuromuscular activity and injury are not known. To understand the contribution of BDNF gene transcription to this regulation we have begun to examine the structure, organization, and expression of the zebrafish BDNF gene. The mammalian BDNF gene is known to have at least four promoters whose cell-specific and developmental activity is differentially regulated [20-26]. The transcripts derived from these promoters have a common organization. They all consist of two exons: one of the 5' non-coding exons followed by the coding exon, which is common to all transcripts.
We previously cloned a zebrafish BDNF transcript that appeared to contain two exons and thus suggested that the zebrafish and mammalian BDNF genes have similar organizations [4]. We recently cloned the zebrafish BDNF gene and identified the previously cloned 5' exon in the genomic clone [27]. The exon was named exon 1c. Sequence analysis confirmed that the position of the intron that separates exon 1c from the common coding exon is identical in fish and man. In addition, the size of the intron is similar in fish and mouse. Exon 1c contains a 39 bp sequence that is 93% identical to a segment in exon III of the rat BDNF gene. Transient expression in developing zebrafish embryos of the promoter upstream from exon 1c using an EGFP-F reporter revealed expression in a number of cell types, including skeletal muscle, epithelial cells, notochord, and vascular cells.
I here report the cloning, structural analysis, and transcriptional activity in transiently transfected developing zebrafish embryos of three additional 5' exons and their associated promoters. Two of the 5' exons were found in the same transcript. This transcript thus contains three exons, a novel structure of a BDNF transcript not previously reported. These two exons could not be found by homology searches in the mammalian genome but were detected in the pufferfish genome. The promoter upstream from the 5' exon of the tripartite transcript was linked to the EGFP-F reporter. The fusion gene was exclusively expressed in terminally differentiated myocytes of developing zebrafish embryos. The 5' flank immediately upstream from the promoter is GC-rich and contains multiple Sp1-like sequences as well as several potential Mef-2 binding sites. These transcription factors are known to be important for the expression of genes in myocytes. Although the endogenous exon and promoter were expressed in embryonic muscle the endogenous exon could not be detected in juvenile fish or adult female muscle by RT-PCR. Possible explanations for this discrepancy are discussed.
Results
The zebrafish BDNF gene has at least four 5' non-coding exons
The organization of all neurotrophin genes so far examined is similar [28,29]. It was therefore expected that the zebrafish BDNF gene would have multiple 5' exons. To identify additional 5' exons, 5' RACE was employed. To maximize the chances of finding all existing exons total RNA extracted from 10 days old larvae was used as the template. The relative abundance of BDNF transcripts in total RNA dramatically increases in the first 7 days of development and plateaus by 10 days pf [4].
Initially, 48 of the 5' RACE clones were screened for inserts by restriction enzyme digest (figure 1). To identify 5' RACE clones that contained novel 5' non-coding exons the nucleotide sequences of representatives of groups of clones with similar sizes were determined. The sequences were compared with the sequences of subclones of the genomic BDNF clone c241 which we had previously identified and partially sequenced [27]. Five of the ten sequenced 5' RACE clones contained the already known and previously cloned 5' exon [4]. This exon was named exon 1c because it contains a 39 nucleotide segment in which 37 nucleotides are identical with a segment in rat exon III, suggesting the two exons are orthologous (see table 1 and figure 2a) [25].
Figure 1.
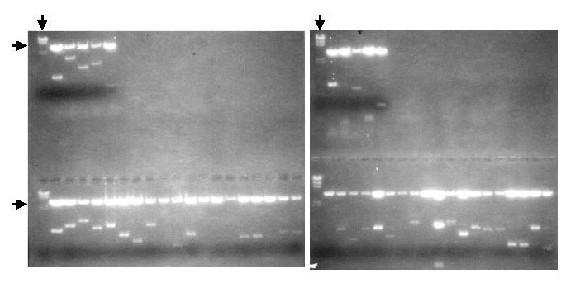
Agarose Gel Electrophoresis of Restriction Enzyme Digests of 5' RACE Clones. DNA was extracted from 48 clones obtained from 5' RACE of total RNA from 10 days old larvae. The clones were digested with EcoRI and SpeI and the inserts separated from the cloning vector by gel electrophoresis through two-tier 1.2 % agarose gels containing ethidium bromide. The DNA was visualized under UV light. The horizontal arrows mark the cloning vector. The vertical arrows point to HindIII digests of phage lambda.
Figure 2.
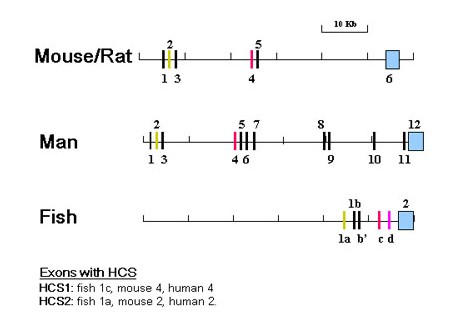
Organization of BDNF Genes. The boxes represent exons and the lines introns, 5' flanks and 3' flank. Exons containing highly conserved sequences (HCS) have the same colors. The positions of the mouse BDNF gene exons were taken from sequence posted in the NCBI databank http://www.ncbi.nlm.nih.gov/entrez/, accession AY057907. The rat exons were published by Timmusk et al. [25] and Bishop et al. [26]. The sequences posted in the NCBI databank (Accession numbers S76799, S76760, S76759, S76758, S76757, X67107, X67106, X67108) were compared with the mouse and found to be >98% identical. Their positions in the rat gene were assumed to be identical to those of the mouse exons. The positions of the human BDNF gene exons were taken from the sequence posted in the NCBI databank, accession number AF411339. The pufferfish BDNF gene sequence was taken from the Ensembl Genome Data Resource of the Wellcome Trust Sanger Centre http://www.sanger.ac.uk/), scaffold_551 and scaffold_1408. Exons 1b, b', 1c, and 2 of the zebrafish BDNF gene were mapped on the sequence of clone CH211-251J8, posted in the NCBI databank, accession number AL935207. Zebrafish and pufferfish gene exon sequences were >75% identical including presumptive exon 1d.
Two clones contained 248 bp inserts. The first 53 nucleotides were found close to the 5' end of a 9 kb genomic HindIII fragment of c241 that contains exon 1c close to its centre [27]. The next 195 nucleotides were found 905 nucleotides downstream, suggesting that they represented an internal exon. Classical splice acceptor and donor di-nucleotides surround this 195 bp exon (see table 2 – additional file 2), consistent with an internal position in a BDNF transcript. The shorter more 5' exon was named exon 1b, and the more 3' exon exon 1b' (see figure 2).
All known BDNF gene 5' exons are directly spliced to the common coding exon, generating mature messages that contain two exons, a single 5' non-coding and the common coding exon. Finding an internal exon was thus unexpected and surprising.
A single clone contained a 441 bp insert. The sequence of the insert was not found in the available c241 subclone sequences. To determine if it might be a BDNF 5' exon it was compared with all known sequences using BLAST. A segment of 29 nucleotides that was identical with a segment in rat exon II was found, suggesting the insert was a 5' BDNF gene exon. This segment was named highly conserved sequence 2, or HCS2 (see table 1 – additional file 1). The exon was named exon 1a.
Exon 1a was located outside the sequenced regions of the genomic clone c241 [27]. To identify its location in c241, a HindIII restriction digest of c241 was subjected to southern blot hybridization analysis using a probe specific for exon 1a. A 5.7 KB HindIII fragment specifically hybridized to the probe. This fragment was subcloned into the HindIII site of the EGFP vector by plus/minus green cloning. A 2 KB region containing exon 1a was sequenced. Comparison of the genomic and 5' RACE sequences indicated that the novel 5' portion of the insert was present as a continuous sequence in the genomic clone. Moreover, a classical splice donor site followed the 5' end (table 2 – see additional file 2). The insert thus represented a 5' non-coding exon of the zebrafish BDNF gene.
These cloning and sequencing data indicate that at least three distinct transcripts are derived from the zebrafish BDNF gene (figure 3).
Figure 3.
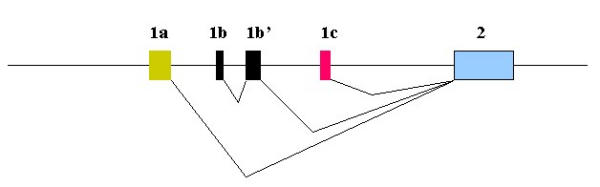
Splicing of Zebrafish BDNF Exons into Mature Transcripts. The boxes are exons and the horizontal line represents 5' flanks, introns, and 3' flank. Angled lines to indicate splicing into mature transcripts connect the exons. The depicted combinations were identified by 5' RACE and cDNA cloning. Additional combinations may exist.
A single 5' RACE clone did not exhibit any homology to the mammalian or zebrafish BDNF genes. A BLAST search identified the insert as the 5' end of β-actin mRNA.
Organization of the Zebrafish and mammalian BDNF Genes are similar
The locations of the exons identified by 5' RACE in the BDNF transcription unit were mapped in three ways. First their nucleotide sequences were compared with that of clone CH211-251J8, which carries a 149 KB zebrafish genomic fragment (NCBI databank accession # AL935207). Exon 1b was found at nucleotides 1050–1101, exon 1b' at nucleotides 2006–2200, and exon 1c at nucleotides 6100–6397 (figure 2). Exon 1a was not found.
Second, the nucleotide sequences were compared to known BDNF exons by a BLAST search. A short segment of identity was found in exon 1a with exon 2 of the mouse, human, and rat genes, suggesting it is orthologous to these exons, and thus should lie upstream from exon 1b (see table 1 – see additional file 1). These analyses placed exon 1a upstream from exons 1b, b' and c and upstream from the 5' end of clone CH211-251J8.
Third, to confirm this upstream location, and to estimate the distance between exon 1a and 1b, long-range PCR was done with a sense primer (ex1aP3) in exon 1a and an anti-sense primer in exon 1b (sc1P18) using the genomic clone c241 as a template. A PCR product of 3.2 KB was obtained (see figure 4). No PCR product was obtained when sense and antisense primers were switched. The PCR product would be expected to be 2.4 if the exon 1a and 1b containing HindIII fragments were contiguous. The larger size suggested that a 0.8 KB HindIII fragment was present between the two. To test this possibility, the PCR product was digested with HindIII. Three fragments were generated, indicating the presence of an interposed fragment. Their sizes were consistent with the largest and smallest being derived from the exon 1b and 1a containing HindIII fragments, respectively, and the middle one being an interposed fragment. A HindIII subclone of c241 was found with an insert identical in size with the interposed fragment (figure 2). The subclone was named Dex1aDS. The insert of Dex1aDS was sequenced and its orientation determined by PCR using either primer ex1aP3 or sc1P18 paired with a primer (Dex1aDSP1) located within Dex1aDS.
Figure 4.
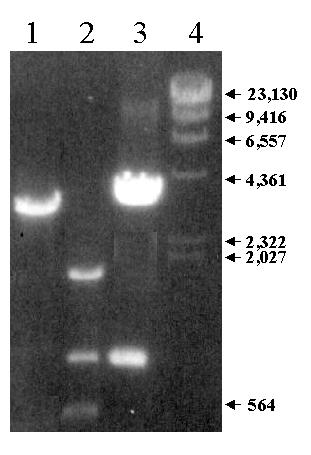
Identification of Interposed HindIII Fragment by Long Range PCR and Restriction Enzyme Digestion of HindIII Subclone of Genomic Clone c241. PCR was carried out using the zebrafish BDNF genomic clone c241 as a template. The primers were ex1aP3 and sc1P18. An aliquot of the PCR products (lane 1) was digested with HindIII (lane 2). A HindIII subclone of genomic clone c241 was also digested with HindIII. The PCR and digestion products were separated by electrophoresis on a 1.2% agarose gel containing ethidium bromide. Lane 1: PCR product. Lane 2: HindIII digest of PCR product. Lane 3: HindIII digest of subclone. Lane 4: HindIII digest of phage lambda. Sizes of fragments are shown in the right hand margin.
The sequencing and PCR data and the nucleotide sequence of clone CH211-251J8 were used to construct a HindIII map of the zebrafish BDNF gene (see figure 5).
Figure 5.
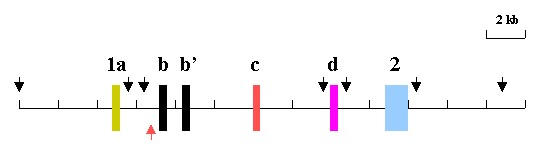
HindIII Map of Zebrafish BDNF Gene. The map is based on the nucleotide sequence of clone CH211-251J8 (NCBI databank accession number AL935207) as well as restriction digest, southern blot hybridization, PCR and sequence analyses of c241. See results and methods for details. The boxes are exons and the line 5' flanks, introns, and 3' flank. The black vertical arrows mark HindIII sites. The red arrow marks the EcoRI site at the 5' end of clone CH211-251J8.
Comparison with pufferfish gene suggests existence of an additional exon
The pufferfish BDNF gene is located in the scaffold_551.1 contig (Ensembl). Fugu rubripes and Danio rerio both belong to the infraclass teleostei. The two BDNF genes can therefore be expected to be well conserved. The nucleotide sequences of the two genes were searched for identities using Bestfit and Gap from the GCG sequence analysis package. These comparisons revealed three regions in the pufferfish gene whose nucleotide sequences were >78% identical with exons 1a, b, and c of the zebrafish gene. The relative positions of these exons to each other and the coding exons were identical in both genes. The distances between these exons and their distances to the coding exon were also very similar. An additional region of >78% identity was found between exon 1c and the coding exon at nucleotides 9592–9876 of the zebrafish sequence (nucleotide numbers of clone CH211-251J8). This region could represent exon 1d and hence might correspond to rat and mouse exon 5. Since this exon has not yet been cloned by 5' RACE its inclusion in figure 3 is tentative.
Collectively, the 5' RACE, mapping, PCR and sequence comparison data suggest that the zebrafish BDNF gene has at least 4 and possibly 5 non-coding exons if exon 1 d is included, and that at least one transcript has 3 exons (see figure 3).
Exons 1a and 1c contain highly conserved sequences
Nucleotide sequence analysis of the 5' RACE clones so far identified three additional 5' exons, namely exons 1a, 1 b and 1b'. A striking structural feature of exon 1a is a 25 nucleotide highly conserved segment (HCS2) that is identical to a segment of rat exon II (see table 1 [additional file 1] and figure 2). The function of HCS2 is unknown. This segment is also conserved in the pufferfish genome (scaffold_551; scaffold_1408). We previously reported the presence of a 39 bp segment within exon 1c in which 37 bps are identical in man and fish (see table 1 – additional file 1). The significance of this highly conserved segment (HCS1) is unknown.
In contrast to exons 1a and 1c, exons 1b and 1b' exhibit only random similarity with the known mammalian exons. A region of high similarity was found in the pufferfish gene in a location expected for exon 1b. Exon 1b' differs from any mammalian counterparts in that it is an internal non-coding exon. So far internal non-coding exons have only been described in other neurotrophin genes, at first in the mouse NGF gene [30,31] and subsequently in the NT-3 gene [32] and more recently in the NT-4/5 gene [33].
The 5' flank of exon 1b is GC-rich and contains multiple Sp1 and Mef2 binding site consensus sequences
The flanking regions of exons 1a and 1b were analyzed for potential transcription factor binding sites using the Transfac database and search engine [34]. The flank of exon 1b is GC-rich and contains multiple potential Sp1 binding sites close to the promoter. Two potential Mef2 sites were also found (see table 3 – additional file 3). The nucleotide sequence is strikingly similar to that of the 5' flank of the FGFR1 gene. The FGFR1 gene is expressed in myocytes in their terminal differentiation phase.
The 5' flank of exon 1a, in contrast, has a more classical TA-rich promoter region, and is devoid of Sp1 and Mef2 sites (see table 4 – additional file 4).
Fusion genes containing exon 1b/b' are specifically expressed in skeletal muscle
To confirm that the cloned promoters were functionally competent and to begin to identify regulatory regions within the promoters the cloned promoters were linked to the EGFP-F reporter (see figure 6). The fusion genes were injected into embryos and expression examined by fluorescent microscopy in live embryos.
Figure 6.
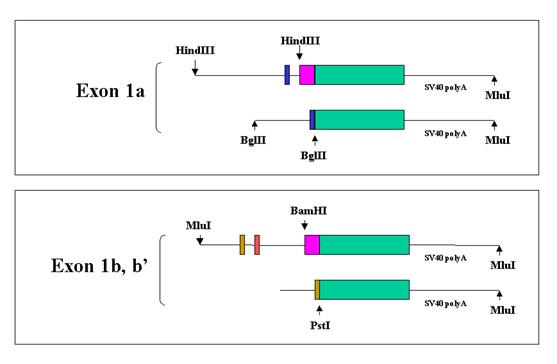
Expression Constructs. The constructs were assembled from cloned or amplified DNA. All junctions were sequenced to confirm correct assembly. The green box represents the reporter EGFP-F. The maroon box represents a 300 bp fragment of the zebrafish BDNF gene that begins 280 bps upstream from exon 2 and ends at nucleotide 21 of the coding exon and thus contains the splice acceptor site. Note that the5' exons are directly joined to reporter in the shorter constructs. All constructs share the 3' UT and polyadenylation signal of the IE of SV40.
Promoter 1b was linked to the EGFP-F reporter using a HindIII/BamHI fragment that contained 5' flank, exons 1b and b', 3 KB of intron 1b', and the coding exon splice acceptor site preceded by 250 nucleotides of intron 1c. This construct was named AJ. For expression studies, the vector sequences and all SV40 sequences except the SV40 3' UT and polyA addition site were removed by MluI digestion followed by purification on an agarose gel.
In two series of injections 350 embryos were injected with AJ. The 2-day survival rate was 60%. 56 of the surviving embryos expressed the reporter 2 days after fertilization. Over 90% of the expressing cells were skeletal muscle cells. Expression was mostly seen in trunk muscles, rarely close to the tail, and never in the head (figure 4). In a few embryos AJ was also expressed in notochord and vascular cells. In a rare embryo expression was seen in neurons and blood cells.
To further narrow down the sequences that are responsible for expression in myocytes exon 1b was directly linked to the reporter using a PstI site 10 nucleotides downstream from the transcription start site (see clone CH211-251J8). In this construct, which was named AJ/Pst, intron 1b, exon 1b' and most of intron 1c, totalling 3.6 KB, are eliminated. In a series of 250 injections with a survival rate of 55% and an expression rate of 30% AJ/Pst was also specifically expressed in myocytes (see figure 7).
Figure 7.
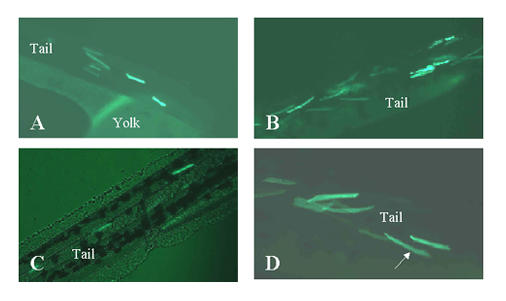
Expression of promoter 1b fusion construct in myocytes. The exon 1b/1b' constructs shown in the lower panel of figure 6 were injected into zebrafish embryos at the 1–4 blastomere stages. Live embryos and larvae were observed at various developmental stages under a fluorescent microscope to detect expression of the EGFP-F reporter. Panel A: 2 days old embryo. Panels B–D: 3 days old larvae. The arrow in panel D points to a labeled myocyte.
Fusion genes containing exon 1a are transiently expressed in yolk
Promoter 1a was linked to the EGFP-F reporter in a similar fashion (figure 6). The 5' flank extended to -3.5 KB. However, only 179 bps of intron 1a were used. Thus the total composite intron size was 325 bps. This construct, which was named DOA5 was injected into sets of 150 and 250 embryos. The survival rate was 45% and 55%. 40% of the combined survivors expressed the reporter. All had an unusual pattern of expression. There was intense labelling of round structures of variable size and labelling intensity by 10 h pf (see figure 8). These structures did not conform to any cellular or subcellular shapes. They were close to the surface of the embryos. Because of limited spatial resolution it was not clear whether they were in the yolk or in cells on the surface of the embryo. These structures tended to be present in limited sections of the embryo but generally did not follow a specific pattern. However, in a few embryos there were more ordered patterns. In one embryo there was a line of labelled structures coinciding with the embryonic midline. In the same embryo there was an anterior semilunar patch arranged symmetrically about the midline. By 20 h pf most of the labelled structures had moved into the embryo in various and seemingly random locations. By 2 days pf only a few remained, and by 3 days pf no labelled structures or cells were seen in 99% of the embryos. A very rare embryo had expression in the notochord or transient expression in several myocytes. The same expression was found with a construct in which the exon was directly linked to the reporter at a BglII site. This construct has no intron.
Figure 8.
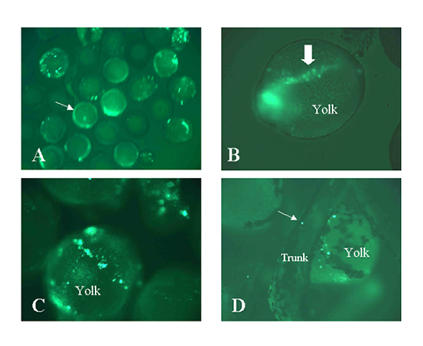
Expression of promoter 1a fusion construct: labelling of yolk platelets. The exon 1a constructs shown in the upper panel of figure 6 were injected into zebrafish embryos at the 1–4 blastomere stages. Live embryos were observed at various stages of development under a fluorescent microscope to detect the EGFP-F reporter. Panel A: low magnification view of multiple embryos 10 hrs after fertilization (1–5 somite stages). The speckles are yolk platelets labeled by EGFP-F. They are slightly out of focus due to bulk motion of the water in which the embryos are suspended. The arrow points to a developing embryo on top of the yolk. The embryo exhibits background fluorescence. Panel B: A 10 hrs old embryo in which some of the yolk platelets line up with the developing trunk and head. The arrow points to the trunk. The head, to the left, is out of focus. Panel C: 10 hrs old embryo. The line is not coincident with the body axis in this embryo. Panel C: 2 days old embryo. Some platelets remain in the yolk. The arrow points to a platelet in the trunk.
Expression of endogenous exons is tissue-specific
The fusion genes containing exon 1b and its 5' flank were expressed only in skeletal muscle. To determine whether the endogenous exon were also expressed cell-specifically, total RNA was extracted from the skeletal muscle of an adult female and analyzed for exon 1a using RT-PCR. To increase specificity and sensitivity, two rounds of PCR were done with nested primers. Diluting the cDNA template 100-fold prior to amplification with β-actin primers checked the quality of the cDNA. β-actin mRNA was readily amplified in the first round of PCR indicating the cDNA was of good quality. Surprisingly, exon 1b could not be detected even after two rounds of PCR (see figure 12). The primers were competent as they yielded product when the 5'RACE clone was used as a template.
Exon 1b could be spliced directly to exon 2. The resulting bipartite transcript would yield a PCR product of 93 bps. No PCR products in that size-range were seen. A set of nested antisense primers that is 300 bps further downstream also gave negative results. In contrast to exon 1b, exon 1c was detected in muscle RNA after the second round of PCR. Total RNA from whole juvenile fish was also analyzed, as it should contain all exons that are expressed at this stage of development. Both exon 1a and 1c were found. The amplifications with exon 1b primers yielded minor amounts of DNA that did not co-migrate with the controls. It is possible that they represent transcripts in which exon 1b is spliced to exons other than 1b'.
Discussion
The data presented in this report show that the overall organization of the BDNF gene is similar in fish and man. However, fish and human genes differ significantly in the size of the transcription unit, the number of exons, and the splicing patterns of the exons. In contrast, the zebrafish and pufferfish BDNF genes are highly similar in all regions that could be compared. The pufferfish gene is flanked on the 5' side by a kinesin gene that lies 30 kb upstream from the BDNF coding exon and on the 3' side by a Lin7 gene, less than 5 kb downstream from the coding exon. The fish transcription unit thus appears to span between 15 and 30 KB. In contrast, the mouse and human genes are between 60 and 80 KB long, at least twice and possibly four times as large. The difference in size is due to much larger introns in the mammalian genes.
A second difference between fish and mammalian genes is the generation of a tripartite transcript from the fish gene. Such tripartite transcripts have not yet been described in mouse, rat, or human. A more thorough examination of all human transcripts may eventually uncover tripartite messages in the human.
A third difference between fish and mammalian genes is the much greater diversity of exons in the human gene, even when compared with the mouse. However, neither in the zebrafish nor the mouse has an exhaustive search for all possible exons been conducted yet. We have recently found additional exons in a second round of 5'RACE. However, they have not yet been sequenced or mapped on the transcription unit.
The 5' exons of the rat and mouse genes and their associated promoters are expressed in a tissue and cell-specific manner [25]. The data presented here as well as data presented in a previous report [35] indicate that the zebrafish BDNF gene promoters are also expressed in a tissue-specific manner. When linked to the EGFP-F reporter exon 1b was primarily expressed in skeletal muscle cells. Visible expression did not start until the myocytes exhibited striations. However, there may a significant time lag between onset of transcription of the transfected DNA, EGFP-F aggregation and insertion into the membrane. Both are required for efficient fluorescence. Thus, the precise onset of transcription of the transgene is uncertain. It is also possible that the EGFP-F reporter is quite stable and persists beyond a period of transient transcription.
The exclusive expression of the promoter 1b construct in skeletal muscle differs markedly from the previously reported expression of a promoter 1c construct [27]. The previously tested reporter 1c construct was identical to the promoter 1b construct injected here except for the promoter sequences. It was expressed in a number of cell types including notochord, epithelial and endothelial cells, and skeletal muscle.
The 5' flank of exon 1b was searched with TransFac for potential transcription factor binding sites [34]. Potential binding sites for three transcription factors that have been shown to play key roles in muscle gene expression were found, Sp-1, Mef2 and SRF [36]. Four potential Sp1 and two Mef2 bindings sites were found within 300 bps of the transcription start. A potential SRF binding site is also close to the transcription start site. Farther upstream two AP-1 sites were found. These Sp-1 and Mef2 sites may play a role in muscle expression of the BDNF gene. The proximal 5' flank of the FGFR1 gene has a similar clustering of SP-1, Mef2, and SRF sites [36]. The FGFR1 gene is expressed in terminally differentiated muscle, although at a lower level than in proliferating myoblasts [36]. The expression depends on Sp-1 sites in the proximal promoter and the binding of Sp-1 and Sp-3 to these sites. The absence of Sp-1 and Sp-3 from fully differentiated myocytes may explain the down-regulation in terminally differentiated muscle.
Unexpectedly, RT-PCR did not detect exon 1b in total RNA extracted from skeletal muscle of an adult female (see figure 9). It is possible that exon 1b is expressed in skeletal myoblasts only transiently during the period of innervation by primary and secondary motor neurons. We did not assay RNA from these early stages of development. It is also possible that exon 1b is expressed primarily in proliferating myoblasts. The expression in skeletal myocytes observed in the reported experiments could represent persistence of the EGFP-F reporter protein past the time of downregulation of transcription in myocytes. Or our construct could lack a suppressor that turns the endogenous promoter off in terminally differentiated and innervated myocytes. It is also possible that our construct is expressed ectopically. However, the similarity of its 5' flank with that of the FGFR1 gene is striking. A more detailed analysis of the time course of expression of the endogenous gene and cellular localization of BDNF transcripts in muscle is needed to address these possibilities.
Figure 9.
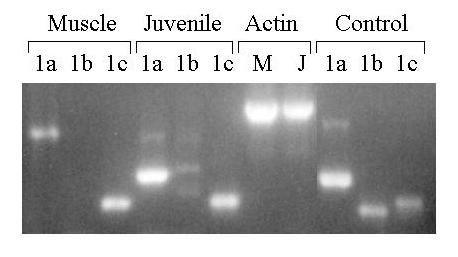
Expression of BDNF 5' exons in juvenile zebrafish and skeletal muscle of adult female zebrafish. Total RNA was extracted from two whole juvenile zebrafish and skeletal muscle of an adult female zebrafish. The RNA was analyzed by two rounds of nested RT-PCR for the presence of exons 1a, 1b/b' and 1c. Both outside and inside primers spanned at least one intron. An aliquot of each cDNA preparation was diluted 1:100 and amplified with beta-actin primers to assess the quality of the cDNA preparations. The beta-actin primers span at least one intron. The 5' RACE clones were used as control templates and size markers. Amplified DNA was separated according to size by electrophoresis on a 1.8% agarose gel containing ethidium and visualized under UV light.
In contrast to the situation with exon 1b, exon 1c was detected by RT-PCR in adult female muscle RNA (see figure 9). The RT-PCR data do not identify the cells that express exon 1c. Muscle contains blood vessels and mixed nerves. Endothelial cells are known to express BDNF [37], and so do Schwann cells [38]. These cells may contribute to the exon 1c transcripts. Is exon 1c expressed in myocytes? The RT-PCR data strongly suggest so. In addition, a construct that is driven by the exon 1c promoter is expressed in various types of muscle cells [27]. However, it is also expressed in a number of non-muscle cells. Thus, its expression is much broader than the exclusive muscle expression of the exon 1b construct. When the exon 1c construct was extended in the 5' direction to include exon 1b and its flank, expression was still broad (data not shown, manuscript in preparation). The Sp1 and Mef2 elements thus are not capable of suppressing expression in other cell types.
It should be noted that both the exon 1b and exon 1c expression constructs contained the SV40 IEG 3'UT and polyadenylation signals downstream from the EGFP-F reporter rather than BDNF sequences (see figure 6). It is becoming clear that their presence affects expression (manuscript in preparation). However, their effect may be quantitative rather than qualitative. In any event, exon 1c is the only exon that was detected in adult female muscle by RT-PCR. Exon 1b appears to play a more limited role during development, and possibly in muscle injury and regeneration.
The exon 1a construct was not expressed in any identifiable embryonic or larval cells at the 1-somite stage yet there was intense labelling of yolk platelets. The platelets are close to the surface. They are congregated preferentially near the midline. It is not clear whether the platelets become labelled in the yolk or within cells. Their location close to the surface suggests they become labelled in the yolk syncitial layer (YSL). The YSL begins to form at the 1k-cell stage, or 3 h after fertilization, when transcription of zygotic genes begins. By the dome stage the YSL has completely covered the yolk cell and effectively separates the embryo from the yolk cell. The peripheral blastomeres of the YSL appear to maintain cytoplasmic bridges with the yolk cell. These peripheral blastomeres may continue to take up platelets. They must also transcribe the exon 1a construct. This possibility is consistent with our previous in situ hybridization analyses, which revealed BDNF expression in the peripheral YSL [35]. Some of the labeled platelets end up in cells of the embryo and persist up to 2 days of development. These platelets must represent the earliest onset of transcription of the injected construct, possibly at the onset of zygotic gene transcription.
It is not clear why this particular construct consistently and strongly labels yolk platelets. We routinely get expression of other constructs in the yolk of a small number of embryos, presumably when there is unintended injection or leakage into the yolk. However, in all these embryos there is diffuse labeling of the membrane of the yolk cell rather than labeling of platelets. Since they all encode the same reporter and share the same 3' UT, the difference must originate from the non-coding 5' regions of the construct, either the 5' flank or 5' UT.
Conclusions
The zebrafish BDNF gene is much more compact than the mammalian gene and rivals in compactness that of the vertebrate with the smallest known genome, the puffer fish. The more compact size makes the zebrafish more amenable to comprehensive molecular genetic analysis than the mammalian gene. The zebrafish has multiple differentially regulated promoters and 5' exons like its mammalian counterpart. Two of the four presently known 5' exons contain conserved segments that are identical in fish and man. The promoter containing a segment identical to rat exon II is transiently expressed in the yolk syncitial layer. The remaining two exons are novel and generate a tripartite transcript. The novel promoter is specifically expressed in myocytes of transfected embryos but the endogenous exon is not expressed in adult female muscle. The cloned BDNF gene and knowledge of its organization are invaluable tools for a future genetic analysis of the regulation of BDNF gene expression.
Methods
5' RACE
Amplified cDNA was generated using the Invitrogen GeneRacer kit according to the manufacturer's directions http://www.invitrogen.com. The template for cDNA preparation was total RNA extracted from 10 days old larvae using the guanidine thiocyanate method. cDNA was generated using random hexamer oligonucleotides. The cDNA was amplified with the GeneRacer primer and a BDNF primer, P1, complementary to 18 nucleotides near the 5' end of the coding exon. Half-nested PCR was then performed using the internal 5' GeneRacer primer and P1.
The amplified cDNA was cloned using the Invitrogen TOPO Cloning kit. DNA was extracted from individual clones. The inserts were released by EcoRI and SpeI digestion and sized using agarose gel electrophoresis. Inserts between 0.200 and 1.5 KB were seen in 48 digests. The inserts of 12 clones were sequenced.
Sequencing of Genomic DNA and Identification of Exons
The previously identified genomic BDNF clone [27] was digested with HindIII and HindIII fragments subcloned into the HindIII site of pEGFP. BDNF exon-containing subclones were identified by dot-blot hybridization using digoxigenin-labelled probes prepared from 5' RACE clones by PCR. The exon-containing regions of hybridizing subclones were sequenced and the genomic sequences compared with the cDNA sequences to identify transcription start sites and exon/intron junctions.
RT-PCR
Total RNA was extracted from two juvenile zebrafish and skeletal muscle of a single adult female using the method of Chomczynski [39]. The integrity of the RNA was checked by agarose gel electrophoresis. Approximately 1 μg of total RNA was reverse transcribed using random hexamers for priming of murine Maloney virus reverse transcriptase. Aliquots of the samples were amplified with nested BDNF exon-specific primers in two rounds of amplification. β-actin primers were used as internal quality controls. The amplified DNA was subjected to agarose gel electrophoresis.
Oligonucleotide Primer Sequences
P1: ATAGTAACGAACAGGATG
Dex1aDS1: TAGCTGTCATTCTCGTCA
Sc1P18: TTCCCAGAGGAACTCAGC
ex1aP3: GCACTTTGGACAGAGGCA
Exon 1a nested
Outside pair: GCACTTTGGACAGAGGCA/ATAGTAACGAACAGGATG
Inside pair: GGAGTCGTTGAACGCTGC/GTCTACCGGTCACTCTTCTAACCTGTTG
Exon 1b nested
Outside pair: ACATGGACTGAGGAGTAG/ATAGTAACGAACAGGATG
Inside pair: TCAATAACAAATCTGAGC/GTCTACCGGTCACTCTTCTAACCTGTTG
Exon 1c nested
Outside pair: CTCAATGCGCACTAC/ATAGTAACGAACAGGATG
Inside pair: GCTCAGTCATGGGAGTCC/GTCTACCGGTCACTCTTCTAACCTGTTG
β-actin: CAACGGCTCCGGCATGTG/TGCCAGGGTACATGGTGG
Construction of Fusion Genes
Starting materials were the previously cloned genomic subclones and a plasmid carrying EGFP-F (pEGFP-F from Clontech, Palo Alto, CA). EGFP-F is a derivative of GFP that exhibits enhanced fluorescence and possesses a farnesylation signal at the COOH-terminal. The farnesylation signal is derived from the src protein. Once farnesylated EGFP-F becomes anchored in the cell membrane. Standard methods of restriction enzyme digestion and ligation of selected fragments were applied to generate the various expression constructs. Junctions of heterologous fragments were sequenced to confirm correct construction of fusion genes.
Preparation of DNA And Microinjection of Embryos
Plasmids were digested with restriction enzymes to remove SV40 enhancers and other vector sequences. The desired restriction fragments were purified by agarose gel electrophoresis. DNA was dissolved in 100 mM KCl at a concentration of 10–50 μg/ml. Phenol red was added to visualize injected DNA solution. DNA was injected into a blastomere or into the cytoplasmic stream below the blastomere(s) at the 1–8 cell stages. Embryos were enzymatically dechorionated with Pronase and extensively washed in embryo medium prior to injection Embryo medium = 13 mM NaCl; 4.2 mM NaHCO3; 0.54 mM KCl; 0.025 mM Na2HPO4; 0.044 mM KH2PO4; 1.3 mM CaCl2; 1 mM MgSO4). Embryos were injected and maintained for 14 hours post fertilization on a bed of 0.7% agarose in embryo medium. Subsequently they were placed into deionized water in clear plastic dishes for observation with a Zeiss AxioVert 200 M fluorescent microscope or further growth. Pictures were taken with a digital camera and acquired with the AxioVision software package, version 3.0.6. The The images were processed with PowerPoint and Adobe PhotoDeluxe Business Edition.
Supplementary Material
Table 1 – Highly Conserved Sequences in non-coding regions. The highly conserved sequences (HCS) in the non-coding regions of the fish, rat, and human BDNF genes were aligned. The stars indicate nucleotides that are identical in all three species. Dashes denote gaps that were introduced to maximize alignment.
Table 2 – Exon/Intron junctions. Exon sequences are shown in blue capital letters and introns in lower case black letters. Splice acceptors are maroon and splice donors red.
Table 3 – Nucleotide Sequence and Potential Transcription Factor Binding Sites of the 5' Flank of Exon 1b. The nucleotide sequence of exon 1b was established by 5' RACE. The nucleotide sequence of the 5' flank was obtained from a HindIII subclone of c241. The nucleotide sequence was searched for potential transcription factor binding sites using Transfac [34]. The potential binding sites for Sp1 (maroon), Mef2 (aqua), AP-1 (yellow), and SRF (green) are shown. The 5' end of exon 1b is shown in uppercase and boldface letters. Key restriction enzyme sites are shown in red. The EcoRI site is at the 5' end of clone CH211-251J8.
Table 4 – Nucleotide Sequence and Potential Transcription Factor Binding Sites of the 5' Flank of Exon 1a. The nucleotide sequence of exon 1a was established by 5' RACE. The nucleotide sequence of the 5' flank was obtained from a HindIII subclone of c241. The nucleotide sequence was searched for potential transcription factor binding sites using Transfac [34]. The display was formatted by Transfac and contains active links. The 5' end of exon 1a is shown in boldface letters.
Acknowledgments
Acknowledgements
This work was supported by a Merit Review Grant from the Veterans Administration.
References
- Griesbeck O, Parsadanian AS, Sendtner M, Thoenen H. Expression of neurotrophins in skeletal muscle: quantitative comparison and significance for motoneuron survival and maintenance of function. J Neurosci Res. 1995;42:21–33. doi: 10.1002/jnr.490420104. [DOI] [PubMed] [Google Scholar]
- Ip FC, Cheung J, Ip NY. The expression profiles of neurotrophins and their receptors in rat and chicken tissues during development. Neurosci Lett. 2001;301:107–110. doi: 10.1016/S0304-3940(01)01603-2. [DOI] [PubMed] [Google Scholar]
- Liem RS, Brouwer N, Copray JC. Ultrastructural localisation of intramuscular expression of BDNF mRNA by silver-gold intensified non-radioactive in situ hybridisation. Histochem Cell Biol. 2001;116:545–551. doi: 10.1007/s00418-001-0349-z. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Heinrich G. Brain-derived neurotrophic factor gene expression in the developing zebrafish. Int J Dev Neurosci. 1997;15:983–997. doi: 10.1016/S0736-5748(97)00017-8. [DOI] [PubMed] [Google Scholar]
- Lohof AM, Ip NY, Poo MM. Potentiation of developing neuromuscular synapses by the neurotrophins NT-3 and BDNF. Nature. 1993;363:350–353. doi: 10.1038/363350a0. [DOI] [PubMed] [Google Scholar]
- Kwon YW, Gurney ME. Brain-derived neurotrophic factor transiently stabilizes silent synapses on developing neuromuscular junctions. J Neurobiol. 1996;29:503–516. doi: 10.1002/(SICI)1097-4695(199604)29:4<503::AID-NEU7>3.3.CO;2-#. [DOI] [PubMed] [Google Scholar]
- Stoop R, Poo MM. Synaptic modulation by neurotrophic factors: differential and synergistic effects of brain-derived neurotrophic factor and ciliary neurotrophic factor. J Neurosci. 1996;16:3256–3264. doi: 10.1523/JNEUROSCI.16-10-03256.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoop R, Poo MM. Synaptic modulation by neurotrophic factors. Prog Brain Res. 1996;109:359–364. doi: 10.1016/s0079-6123(08)62118-4. [DOI] [PubMed] [Google Scholar]
- Zhang X, Poo MM. Localized synaptic potentiation by BDNF requires local protein synthesis in the developing axon. Neuron. 2002;36:675–688. doi: 10.1016/s0896-6273(02)01023-1. [DOI] [PubMed] [Google Scholar]
- Boulanger L, Poo MM. Presynaptic depolarization facilitates neurotrophin-induced synaptic potentiation. Nat Neurosci. 1999;2:346–351. doi: 10.1038/7258. [DOI] [PubMed] [Google Scholar]
- Loeb JA, Hmadcha A, Fischbach GD, Land SJ, Zakarian VL. Neuregulin expression at neuromuscular synapses is modulated by synaptic activity and neurotrophic factors. J Neurosci. 2002;22:2206–2214. doi: 10.1523/JNEUROSCI.22-06-02206.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu WM, Liou HH, Wang CL. Collaboration of fibronectin matrix and neurotrophin in regulating spontaneous transmitter release at developing neuromuscular synapses in Xenopus cell cultures. Neurosci Lett. 2001;300:115–119. doi: 10.1016/S0304-3940(01)01567-1. [DOI] [PubMed] [Google Scholar]
- Wells DG, McKechnie BA, Kelkar S, Fallon JR. Neurotrophins regulate agrin-induced postsynaptic differentiation. Proc Natl Acad Sci U S A. 1999;96:1112–1117. doi: 10.1073/pnas.96.3.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb JA, Fischbach GD. Neurotrophic factors increase neuregulin expression in embryonic ventral spinal cord neurons. J Neurosci. 1997;17:1416–1424. doi: 10.1523/JNEUROSCI.17-04-01416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou JC, Yang RS, Fu WM. Regulation of quantal secretion by neurotrophic factors at developing motoneurons in Xenopus cell cultures. J Physiol. 1997;503 ( Pt 1):129–139. doi: 10.1111/j.1469-7793.1997.129bi.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Pinilla F, Ying Z, Opazo P, Roy RR, Edgerton VR. Differential regulation by exercise of BDNF and NT-3 in rat spinal cord and skeletal muscle. Eur J Neurosci. 2001;13:1078–1084. doi: 10.1046/j.0953-816x.2001.01484.x. [DOI] [PubMed] [Google Scholar]
- Gomez-Pinilla F, Ying Z, Roy RR, Molteni R, Edgerton VR. Voluntary exercise induces a BDNF-mediated mechanism that promotes neuroplasticity. J Neurophysiol. 2002;88:2187–2195. doi: 10.1152/jn.00152.2002. [DOI] [PubMed] [Google Scholar]
- Zhang JY, Luo XG, Xian CJ, Liu ZH, Zhou XF. Endogenous BDNF is required for myelination and regeneration of injured sciatic nerve in rodents. Eur J Neurosci. 2000;12:4171–4180. doi: 10.1046/j.1460-9568.2000.01312.x. [DOI] [PubMed] [Google Scholar]
- Hiltunen JO, Laurikainen A, Vakeva A, Meri S, Saarma M. Nerve growth factor and brain-derived neurotrophic factor mRNAs are regulated in distinct cell populations of rat heart after ischaemia and reperfusion. J Pathol. 2001;194:247–253. doi: 10.1002/path.878. [DOI] [PubMed] [Google Scholar]
- Hayes VY, Towner MD, Isackson PJ. Organization, sequence and functional analysis of a mouse BDNF promoter. Brain Res Mol Brain Res. 1997;45:189–198. doi: 10.1016/S0169-328X(96)00254-9. [DOI] [PubMed] [Google Scholar]
- Nakayama M, Gahara Y, Kitamura T, Ohara O. Distinctive four promoters collectively direct expression of brain-derived neurotrophic factor gene. Brain Res Mol Brain Res. 1994;21:206–218. doi: 10.1016/0169-328X(94)90251-8. [DOI] [PubMed] [Google Scholar]
- Timmusk T, Lendahl U, Funakoshi H, Arenas E, Persson H, Metsis M. Identification of brain-derived neurotrophic factor promoter regions mediating tissue-specific, axotomy-, and neuronal activity-induced expression in transgenic mice. J Cell Biol. 1995;128:185–199. doi: 10.1083/jcb.128.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi A, Nakaoka R, Amano K, Yukimine M, Andoh T, Kuraishi Y, Tsuda M. Differential activation of brain-derived neurotrophic factor gene promoters I and III by Ca2+ signals evoked via L-type voltage-dependent and N-methyl-D-aspartate receptor Ca2+ channels. J Biol Chem. 2000;275:17269–17275. doi: 10.1074/jbc.M909538199. [DOI] [PubMed] [Google Scholar]
- Marmigere F, Rage F, Tapia-Arancibia L, Arancibia S. Expression of mRNAs encoding BDNF and its receptor in adult rat hypothalamus. Neuroreport. 1998;9:1159–1163. doi: 10.1097/00001756-199804200-00037. [DOI] [PubMed] [Google Scholar]
- Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, Persson H. Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron. 1993;10:475–489. doi: 10.1016/0896-6273(93)90335-o. [DOI] [PubMed] [Google Scholar]
- Bishop JF, Mueller GP, Mouradian MM. Alternate 5' exons in the rat brain-derived neurotrophic factor gene: differential patterns of expression across brain regions. Brain Res Mol Brain Res. 1994;26:225–232. doi: 10.1016/0169-328X(94)90094-9. [DOI] [PubMed] [Google Scholar]
- Huynh G, Heinrich G. Brain-derived neurotrophic factor gene organization and transcription in the zebrafish embryo. Int J Dev Neurosci. 2001;19:663–673. doi: 10.1016/S0736-5748(01)00046-6. [DOI] [PubMed] [Google Scholar]
- Gotz R, Raulf F, Schartl M. Brain-derived neurotrophic factor is more highly conserved in structure and function than nerve growth factor during vertebrate evolution. J Neurochem. 1992;59:432–442. doi: 10.1111/j.1471-4159.1992.tb09389.x. [DOI] [PubMed] [Google Scholar]
- Ebendal T. Function and evolution in the NGF family and its receptors. J Neurosci Res. 1992;32:461–470. doi: 10.1002/jnr.490320402. [DOI] [PubMed] [Google Scholar]
- Selby MJ, Edwards R, Sharp F, Rutter WJ. Mouse nerve growth factor gene: structure and expression. Mol Cell Biol. 1987;7:3057–3064. doi: 10.1128/mcb.7.9.3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RH, Selby MJ, Rutter WJ. Differential RNA splicing predicts two distinct nerve growth factor precursors. Nature. 1986;319:784–787. doi: 10.1038/319784a0. [DOI] [PubMed] [Google Scholar]
- Leingartner A, Lindholm D. Two promoters direct transcription of the mouse NT-3 gene. Eur J Neurosci. 1994;6:1149–1159. doi: 10.1111/j.1460-9568.1994.tb00613.x. [DOI] [PubMed] [Google Scholar]
- Salin T, Timmusk T, Lendahl U, Metsis M. Structural and functional characterization of the rat neurotrophin-4 gene. Mol Cell Neurosci. 1997;9:264–275. doi: 10.1006/mcne.1997.0625. [DOI] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, Kloos DU, Land S, Lewicki-Potapov B, Michael H, Munch R, Reuter I, Rotert S, Saxel H, Scheer M, Thiele S, Wingender E. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum T, Huynh G, Heinrich G. Brain-derived neurotrophic factor and TrkB tyrosine kinase receptor gene expression in zebrafish embryo and larva. Int J Dev Neurosci. 2001;19:569–587. doi: 10.1016/S0736-5748(01)00041-7. [DOI] [PubMed] [Google Scholar]
- Parakati R, DiMario JX. Sp1- and Sp3-mediated transcriptional regulation of the fibroblast growth factor receptor 1 gene in chicken skeletal muscle cells. J Biol Chem. 2002;277:9278–9285. doi: 10.1074/jbc.M108411200. [DOI] [PubMed] [Google Scholar]
- Donovan MJ, Lin MI, Wiegn P, Ringstedt T, Kraemer R, Hahn R, Wang S, Ibanez CF, Rafii S, Hempstead BL. Brain derived neurotrophic factor is an endothelial cell survival factor required for intramyocardial vessel stabilization. Development. 2000;127:4531–4540. doi: 10.1242/dev.127.21.4531. [DOI] [PubMed] [Google Scholar]
- Meyer M, Matsuoka I, Wetmore C, Olson L, Thoenen H. Enhanced synthesis of brain-derived neurotrophic factor in the lesioned peripheral nerve: different mechanisms are responsible for the regulation of BDNF and NGF mRNA. J Cell Biol. 1992;119:45–54. doi: 10.1083/jcb.119.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table 1 – Highly Conserved Sequences in non-coding regions. The highly conserved sequences (HCS) in the non-coding regions of the fish, rat, and human BDNF genes were aligned. The stars indicate nucleotides that are identical in all three species. Dashes denote gaps that were introduced to maximize alignment.
Table 2 – Exon/Intron junctions. Exon sequences are shown in blue capital letters and introns in lower case black letters. Splice acceptors are maroon and splice donors red.
Table 3 – Nucleotide Sequence and Potential Transcription Factor Binding Sites of the 5' Flank of Exon 1b. The nucleotide sequence of exon 1b was established by 5' RACE. The nucleotide sequence of the 5' flank was obtained from a HindIII subclone of c241. The nucleotide sequence was searched for potential transcription factor binding sites using Transfac [34]. The potential binding sites for Sp1 (maroon), Mef2 (aqua), AP-1 (yellow), and SRF (green) are shown. The 5' end of exon 1b is shown in uppercase and boldface letters. Key restriction enzyme sites are shown in red. The EcoRI site is at the 5' end of clone CH211-251J8.
Table 4 – Nucleotide Sequence and Potential Transcription Factor Binding Sites of the 5' Flank of Exon 1a. The nucleotide sequence of exon 1a was established by 5' RACE. The nucleotide sequence of the 5' flank was obtained from a HindIII subclone of c241. The nucleotide sequence was searched for potential transcription factor binding sites using Transfac [34]. The display was formatted by Transfac and contains active links. The 5' end of exon 1a is shown in boldface letters.


