Abstract
The signal recognition particle (SRP) is a ribonucleoprotein particle essential for the targeting of signal peptide-bearing proteins to the prokaryotic plasma membrane or the eukaryotic endoplasmic reticulum membrane for secretion or membrane insertion. SRP binds to the signal peptide emerging from the exit site of the ribosome and forms a ribosome nascent chain (RNC)–SRP complex. The RNC–SRP complex then docks in a GTP-dependent manner with a membrane-anchored SRP receptor and the protein is translocated across or integrated into the membrane through a channel called the translocon. Recently considerable progress has been made in understanding the architecture and function of SRP.
Keywords: membrane targeting/ribosome nascent chain/signal recognition particle/translocon
Introduction
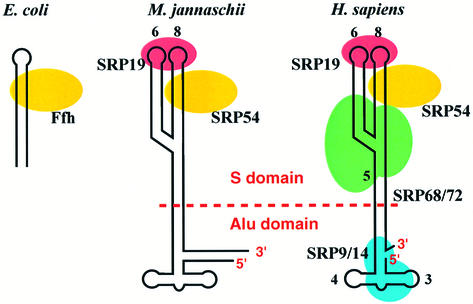
Signal recognition particle (SRP) is a ubiquitous ribonucleoprotein particle found in all three domains of life, but its complexity has increased considerably during evolution (Walter and Johnson, 1994; Rosenblad et al., 2003) (Figure 1). Mammalian SRP consists of 7SL RNA and six protein subunits: SRP54, SRP19, SRP68, SRP72, SRP14 and SRP9. 7SL RNA folds into a roughly Y-shaped double-stranded secondary structure (Walter and Blobel, 1983). Mammalian SRP can be divided into two functional domains by micrococcal nuclease: the Alu and S domains (Gundelfinger et al., 1983). The SRP14–SRP9 heterodimer binds to one end of 7SL RNA, forming the Alu domain (Walter and Johnson, 1994; Wild et al., 2002), whereas the forked region of 7SL RNA and the remaining four proteins form the S domain. The RNA component of archaebacterial SRP is similar in size and secondary structure to its mammalian counterpart, but homologues of only two mammalian SRP proteins, SRP54 and SRP19, have been identified so far in archaeal genomes (Bhuiyan et al., 2000; Zwieb and Eichler, 2002). Escherichia coli SRP is the simplest known SRP, consisting of 4.5S RNA and one protein, referred to as p48 or Ffh (fifty four homologue), a homologue of mammalian SRP54 (Poritz et al., 1990; Luirink et al., 1992; Keenan et al., 2001). SRP54/Ffh is the only protein component present in all SRPs, and hence plays an essential role in signal peptide and SRP receptor (SR) binding. SRP54 consists of an N-terminal four-helix bundle (N) domain, a GTPase (G) domain and a C-terminal α-helical methionine-rich (M) domain.
Fig. 1. Schematic representation of SRPs from three kingdoms of life: eubacteria (E.coli, left); archaebacteria (M.jannaschii; middle); and eukarya (Homo sapiens; right). Mammalian SRP can be separated by micrococcal nuclease into two functional domains: the S and Alu domains.
Function of SRP
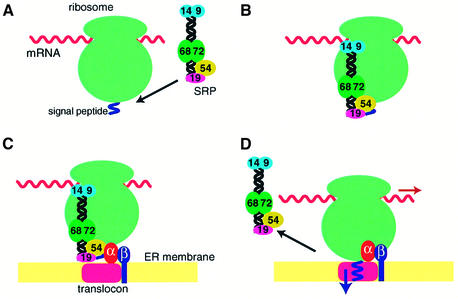
SRP binds through the M domain of SRP54 to the signal peptide of membrane or secretory proteins emerging from the ribosome and forms a ribosome nascent chain (RNC)–SRP complex (Zopf et al., 1990; Luirink et al., 1992; Lütcke et al., 1992) (Figure 2A and B). In eukaryotes, this causes a transient arrest of the polypeptide chain elongation (Walter et al., 1981) through an as yet unknown mechanism, but possibly through interaction of the Alu domain with the tRNA-binding A-site or the elongation factor-binding site at the interface between the small and large ribosomal subunits (Figure 2B). In the complex between canine SRP and rabbit reticulocyte or wheat germ ribosome, SRP54 can be chemically cross-linked to L23a and L35 proteins (homologues of yeast L25 and L35, respectively), located close to the exit site in the large ribosomal subunit (Pool et al., 2002). A photoactivatable cross-linking reagent, p-azidophenacyl bromide, attached to residues 17 and 25 within the N domain of E.coli Ffh can be cross-linked to L23 located at the exit site of the E.coli ribosome (Gu et al., 2003). The affinity of SRP54 for GTP is low but increases upon docking of SRP with the ribosome (Bacher et al., 1996). The RNC–SRP complex then associates with the SRP receptor (SR) anchored to the endoplasmic reticulum (ER) membrane (Rapiejko and Gilmore, 1997) (Figure 2C).
Fig. 2. Functional cycle of the mammalian SRP. (A) SRP binds through the M domain of SRP54 to the signal sequence of membrane and secretory proteins emerging from the exit site of the large ribosomal subunit. (B) The Alu domain promotes transient arrest of the polypeptide chain elongation through an as yet unknown mechanism. The affinity of SRP54 for GTP increases upon docking of SRP with the ribosome. (C) The RNC–SRP complex diffuses to the ER membrane and docks with the SR mainly through the interaction between SRP54 and SRα in the GTP-bound form. SRβ in the GTP-bound form interacts with the RNC complex and induces the transfer of the signal peptide to the translocon. (D) SRP54 and SRα mutually activate their GTPases, and SRP dissociates from the SR upon hydrolysis of GTP, allowing the elongation of the polypeptide to resume.
Mammalian SR is a heterodimer consisting of SRα and SRβ. SRα is homologous to the bacterial SRP receptor protein, FtsY, that consists of a highly charged N-terminal domain and N and G domains similar to those found in SRP54 (Freymann et al., 1997; Montoya et al., 1997). SRβ belongs to the Arf subfamily of GTPases and is anchored to the ER membrane through its N-terminal trans-membrane helix (Miller et al., 1995). The two SR subunits form a stable complex when GTP is bound to SRβ, whereas they may dissociate upon GTP hydrolysis (Legate et al., 2000; Schwartz and Blobel, 2003). The E.coli SRP and FtsY stably interact with each other when both Ffh and FtsY are in the GTP-bound form (Miller et al., 1994). This interaction induces reciprocal activation of the GTPases of Ffh and FtsY (Powers and Walter, 1995), and the SRP dissociates from FtsY upon GTP hydrolysis. As suggested by the high degree of sequence conservation of both SRP54 (Ffh) and SRα (FtsY) throughout evolution, the GTP-dependent interaction between SRP54 and SRα plays an important regulatory role in the SRP–SR interaction in eukaryotes. Both SRP54 and SRα have an intrinsically low affinity for GTP. The GTP affinity of SRP54 increases significantly when SRP binds to a signal sequence of a nascent peptide and forms a stable RNC–SRP complex (Bacher et al., 1996). When the RNC–SRP complex binds to SR, the GTP affinity of SRα also increases. SRP54 and SRα reciprocally stimulate their GTPases (Bacher et al., 1999), and GTP hydrolysis leads to a dissociation of SRP from its receptor (Rapiejko and Gilmore, 1997). The RNC complex binds to SRβ in the GTP-bound form even in the absence of SRα or SRP and stimulates its GTPase activity (Bacher et al., 1999). In the presence of GDP, the signal peptide can be cross-linked to SRP54, whereas in the presence of GMP-PNP, a significant proportion of the signal peptide is cross-linked to the translocon Sec61α (Fulga et al., 2001). When a mutant of SRβ, which preferentially binds xanthosine nucleotide, is used, the addition of XDP blocks the transfer of the signal peptide from SRP54 to Sec61α. Hence, the GTP binding to SRβ is essential for the release of the signal peptide from SRP and its transfer to the translocon (Fulga et al., 2001). Upon dissociation of SRP from SR, the elongation of the polypeptide chain resumes and the growing polypeptide chain is co-translationally translocated across or integrated into the ER membrane (Figure 2D). The cryo-electron microscopic structure of the translocon bound to translating ribosome visualized a membrane pore formed by the Sec61 complex (Beckmann et al., 2001).
Structure and assembly of the S domain
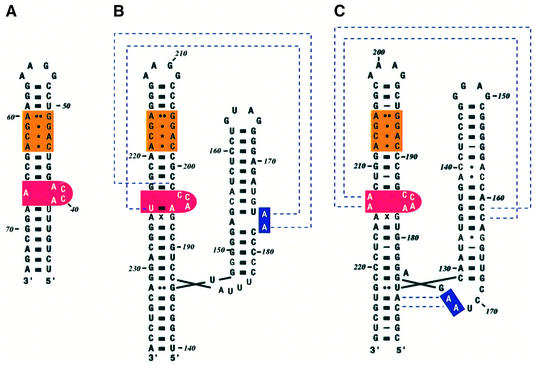
The secondary structure of domain IV of E.coli 4.5S RNA and the S domains of Methanococcus jannaschii and human 7SL RNA is shown in Figure 3. The assembly of mammalian SRP was studied in vitro using protein and RNA components isolated from canine pancreatic SRP (Walter and Blobel, 1983). SRP19 associates with 7SL RNA on its own, but canine or human SRP54 are unable to bind to 7SL RNA unless SRP19 is present. The binding site for SRP19 in 7SL RNA was mapped to the tips of helices 6 and 8 by α-sarcin cleavage (Siegel and Walter, 1988). The SRP68–SRP72 heterodimer binds to the region near the three-way junction of the S domain RNA (Siegel and Walter, 1988) through SRP68 (Lütcke et al., 1993). However, the mapping of the precise binding site for SRP68–SRP72 is hampered by difficulties in overproducing SRP68–SRP72. The symmetric and asymmetric loops in helix 8 (Figure 3B and C) show strong similarities to those in domain IV of E.coli 4.5S RNA, which is the binding site for Ffh (Figure 3A). The symmetric loop (orange in Figure 3) consists of four non-Watson–Crick base pairs (AG, GG, CA and AC), and these four base pairs are conserved in E.coli, M.jannaschii and human, as well as in most species sequenced thus far (Rosenblad et al., 2003). The ACC tri-nucleotide found in the long strand of the asymmetric loop (red in Figure 3) is another feature strongly conserved in the majority of SRP RNAs. The structure of the conserved domain IV of E.coli 4.5S RNA studied by NMR and crystallography shows that the asymmetric loop has considerable flexibility in free RNA (Schmitz et al., 1999; Jovine et al., 2000). The crystal structure of the M domain of E.coli Ffh in complex with 4.5S RNA shows that when the M domain binds to the minor groove of the symmetric loop through its helix–turn–helix motif, the structure of the symmetric loop remains unchanged whereas the asymmetric loop becomes well ordered (Batey et al., 2000). The bases of the ACC tri-nucleotide of the asymmetric loop become stacked continuously and form a platform, which interacts extensively with the M domain. The molecular recognition between Ffh and 4.5S RNA therefore consists of a rigid body docking of the symmetric loop and an induced fit of the asymmetric loop (Schmitz et al., 1999; Jovine et al., 2000; Batey et al., 2001).
Fig. 3. Secondary structure of SRP RNAs. (A) Escherichia coli, (B) M.jannaschii and (C) H.sapiens. The E.coli 4.5S RNA is 114 nucleotide long and forms an extended hairpin structure. The S domain of M.jannaschii and human 7SL RNA has two branches referred to as helices 6 and 8. Helix 8 bears similarities to domain IV of 4.5S RNA, the binding site for Ffh. The symmetric loop (orange) has the same four non-Watson–Crick base pairs, and the asymmetric loop (red) contains the ACC tri-nucleotide. The dotted lines for M.jannaschii RNA show the interaction between adenines in helix 6 with the backbone in helix 8 found in the SRP19–RNA complex (Hainzl et al., 2002). The dotted lines for human RNA show the A minor motif interactions found in the ternary complex (Kuglstatter et al., 2002).
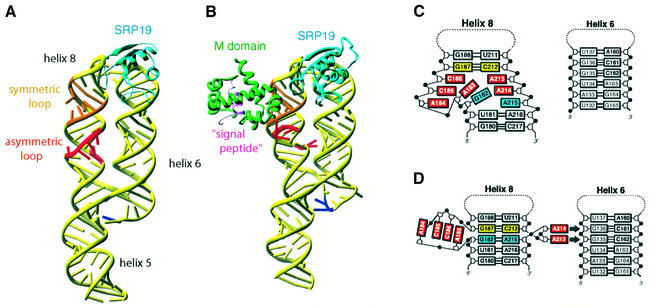
In contrast to E.coli Ffh, human SRP54 or its M domain are unable to bind to helix 8 of 7SL RNA in the absence of SRP19. How does SRP19 enable SRP54 to bind to 7SL RNA? The crystal structures of human 7SL RNA in complex with SRP19 alone and with SRP19 and the M domain of SRP54 together suggest a mechanism whereby SRP19 facilitates the binding of SRP54 (Kuglstatter et al., 2002; Oubridge et al., 2002). A large conformational change of the S domain RNA upon binding of SRP19 was revealed by the crystal structure of M.jannaschii SRP19 in complex with the S domain of either human or M.jannaschii 7SL RNA (Hainzl et al., 2002; Oubridge et al., 2002) (Figure 4A). SRP19 interacts with the major groove of the helix 6 tetra-loop, as first shown in the SRP19–helix 6 complex structure (Wild et al., 2001), and the minor groove of the helix 8 tetra-loop. These interactions induce extensive interactions between the tetra-loops of helices 6 and 8, in good agreement with the chemical probing experiment by Rose and Weeks (2001). For example, in human 7SL RNA, A149 of helix 6 and A201 of helix 8 form a symmetric A–A base pair between the N1 atom and the 6-exocyclic amino group (Oubridge et al., 2002). Equivalent interactions are also found in the M.jannaschii cognate complex (Hainzl et al., 2002). Mutation of A149 greatly weakens the binding of SRP19 to 7SL RNA, showing that the RNA–RNA interaction stabilizes the binding of SRP19 (Zwieb, 1992). SRP19 clamps the tetra-loops of helices 6 and 8 and allows these two helices to lie side by side; helix 8 stacks co-axially onto helix 5 (Figure 4A). The bases of the shorter strand in the asymmetric loop continuously stack, but the longer strand retains considerable flexibility, indicated by high temperature factors (Figure 4C). Helix 6 acts as a splint for helix 8 and reduces the flexibility of the short strand of the asymmetric loop and partially organizes the binding site for the M domain (Hainzl et al., 2002; Oubridge et al., 2002). The human ternary complex structure has revealed a striking conformational change of the asymmetric loop upon binding of the M domain (Kuglstatter et al., 2002) (Figure 4B). The asymmetric loop collapses and the G187–C212 base pair (yellow) becomes directly stacked onto the sheared G182–A215 base pair (blue) (Figure 4D). The AACC tetra-nucleotide of the longer strand of the asymmetric loop protrudes from helix 8 and forms a platform similar to that observed in the Ffh M domain–4.5S RNA complex (Batey et al., 2000). This structure is stabilized by an extensive network of interactions with the M domain. A213 and A214 flip out from helix 8 and form type II and type I A minor motif interactions (Doherty et al., 2001; Nissen et al., 2001) with the G135–C162 and G136–C161 base pairs in helix 6. The A minor motifs, found in large RNAs such as rRNA and group I intron, are proposed to be important tertiary interactions (Doherty et al., 2001; Nissen et al., 2001). Wild et al. (2002) postulated the formation of the A minor motif in the binary complex, but our RNA chemical probing experiment (C.Oubridge, C.Isel and K.Nagai, unpublished results) shows that it forms upon binding of the M domain, in perfect agreement with the crystal structure of the binary and ternary complexes (Kuglstatter et al., 2002; Oubridge et al., 2002). As the RNA platform cannot stably form in the absence of the M domain, the collapse of helix 8 and formation of the RNA platform and the A minor motifs must be highly cooperative. The formation of the A minor motifs is only possible when helix 6 is brought adjacent to helix 8 by the binding of SRP19 to the tetra-loops of helices 6 and 8. This mechanism explains how SRP19 enables SRP54 to bind to helix 8.
Fig. 4. Conformational changes of the S domain of human 7SL RNA during assembly. (A) The structure of the S domain of human 7SL RNA in complex with M.jannaschii SRP19 (PDB code:1L9A; Oubridge et al., 2002). SRP19 binds to the tetra-loops of helices 6 and 8, clamping them and allowing them to lie side by side. This interaction reduces the flexibility of the asymmetric loop in helix 8 and partially pre-organizes the binding site for SRP54. (B) The binding of the M domain of SRP54 induces a striking conformational change in the structure of the asymmetric loop (PDB code: 1MFQ; Kuglstatter et al., 2002). (C) In the binary complex, the bases of the shorter strand of the asymmetric loop continuously stack, but the longer strand still retains substantial flexibility. (D) In the ternary complex, the asymmetric loop collapses and the G187–C212 base pair (yellow) becomes directly stacked onto the G182–A215 base pair (blue) (Kuglstatter et al., 2002). The bases of A184, C185, C186 and A183 continuously stack, forming a structure reminiscent of the RNA platform seen in the E.coli Ffh–4.5S RNA complex (Batey et al., 2000). A213 and A214 flip out and form A minor motif interactions with the G135–C162 and G136–C161 base pairs of helix 6. This figure was generated using Ribbons (http://sgce.cbse.uab.edu/ribbons/). For an animation of the SRP S domain assembly, see http://www.mssm.edu/students/jovinl02/research/srp_assembly_movie.html
The same mechanism may not apply to archaebacterial SRP in which SRP54 is able to bind to 7SL RNA even in the absence of SRP19, although its binding is stabilized by SRP19 (Bhuiyan et al., 2000; Diener and Wilson, 2000). The asymmetric loop in helix 8 of M.jannaschii 7SL RNA contains the ACC tri-nucleotide found in E.coli 4.5S RNA and human 7SL RNA. The bases of these three nucleotides with high temperature factors are splayed out and interact with a symmetry-related molecule in the binary complex crystal (Hainzl et al., 2002). However, binding of the M domain of SRP54 will undoubtedly induce a significant structural change in the asymmetric loop and allow these three nucleotides to form an RNA platform, as observed in E.coli 4.5S RNA and human 7SL RNA (Batey et al., 2000; Kuglstatter et al., 2002). In the M.jannaschii binary complex (Hainzl et al., 2002), the bases of A176 and A177 from helix 6 interact with the ribose atoms of U224 and C198, respectively (Figure 3B), but the conformational change of the asymmetric loop accompanying the M domain binding may allow A176 and A177 to form A minor motifs possibly with G223–C198 and G222–C199, respectively. The asymmetric loop of Archaeoglobus fulgidus 7SL RNA is significantly different from the three mentioned above in that the longer strand does not contain the ACC tri-nucleotide. The DEPC (diethylpyrocarbonate) modification pattern of the asymmetric loop is similar between the binary complexes with SRP19 or SRP54 and the SRP54–SRP19 ternary complex (Diener and Wilson, 2000), suggesting that the asymmetric loop in helix 8 of A.fulgidus 7SL RNA might form an RNA platform-like structure upon binding of SRP19 alone. These results show that the important framework of the SRP54 (Ffh)–RNA interaction is largely conserved during evolution, but fine details of the assembly mechanism may be different from species to species.
The Alu domain
The Alu domain of mammalian SRP consists of an SRP9–SRP14 heterodimer bound to the 5′ and 3′ ends of 7SL RNA (Figure 1) with two branched helices 3 and 4. The crystal structure of the SRP9–SRP14 dimer has revealed a strong similarity between SRP14 and SRP9, both of which form a three-stranded antiparallel β-sheet flanked on one side by two α-helices (Birse et al., 1997). Within the heterodimer, SRP9 and SRP14 are related by a pseudo-dyad axis and form a continuous six-stranded β-sheet. The positively charged concave surface of the β-sheet forms the major binding site for the Alu domain RNA. The binding of SRP9–SRP14 to an Alu domain RNA fragment strengthens the pairing of the loops of helices 3 and 4, which results in the folding of the RNA into a highly compact structure (Figure 5A). The majority of the RNA–protein contacts are made through backbone atoms of the sugar–phosphate backbone between helices 3 and 4 that form a U-turn (Weichenrieder et al., 2000). The structure of SRP9–SRP14 was also solved at low resolution in complex with an 88 nucleotide RNA construct whose natural 5′ and 3′ ends are joined with a single U. In addition to the contact with the U-turn, the protein also makes contact with the internal loop region of helix 5 of a symmetry-related molecule. Based on this structure and chemical protection data, Weichenrieder et al. (2000) proposed that the gap naturally found between the 5′ and 3′ ends would allow helix 5 to fold back and wrap around the heterodimer. The structure of the Alu domain model bears no strong resemblance to tRNA or elongation factors and hence does not immediately suggest a molecular mechanism for elongation arrest (Wild et al., 2002). The Alu domain of archaebacterial SRP RNA has a similar secondary structure, but no homologues of SRP9 or SRP14 have been identified so far in the archaebacterial genome sequence. The secondary structure of the Alu domain of Bacillus subtilis SRP RNA shows similarity to those of M.jannaschii or mammalian SRP RNA (Walter and Johnson, 1994), and it has been shown that a histone-like protein, HBsu, interacts with this region of RNA (Nakamura et al., 1999). It is therefore possible that the Alu domain of archaebacterial SRP may contain proteins with no or very weak homology to SRP9–SRP14.
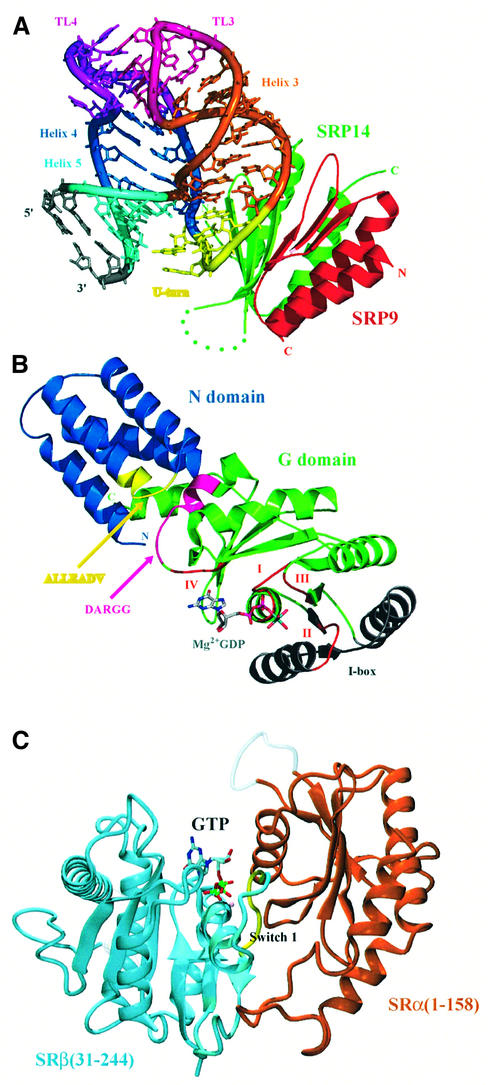
Fig. 5. The crystal structure of the mammalian Alu domain, the NG domain of T.aquaticus Ffh and part of the SR. (A) The crystal structure of SRP9–SRP14 in complex with a small fragment of the Alu domain RNA (PDB code: 1E8O; Weichenrieder et al., 2000). Green, SRP14; red, SRP9; yellow, U-turn; dark blue, helix 4 (H1.2); light blue, helix 5; orange, helix 3. (B) The crystal structure of the Ffh NG domain in the GDP-Mg2+-bound form (PDB code: 1NG1; Freymann et al., 1999). The conserved GTP-binding motifs (I, II, III and IV) are shown in red. The conserved ALLEADV (N domain) and DARGG (G domain) motifs and the I box proposed to be important in coupling the binding of signal peptide, GTP and SRs are highlighted in yellow, purple and grey (Newitt and Bernstein, 1997; Freymann et al., 1999). The actual sequence of the ALLEADV motif in T.aquaticus Ffh is ‘ALMDADV’. (C) The crystal structure of yeast SRβ (blue) in complex with the N-terminal SRβ-interacting (SRX) domain of SRα (brown) (PDB code: 1NRJ; Schwartz and Blobel, 2003). A tightly bound GTP is found at the interface between the two subunits, and the switch I of the GTPase domain of SRβ interacts extensively with SRX (courtesy of Tom Schwartz). (A) and (B) were generated using PyMOL (http://pymol.sourceforge.net/).
The role of SRP54 in signal peptide and receptor binding
The sequence alignment of SRP54 (Ffh) and SRα (FtsY) shows that these two proteins share a homologous region designated the N and G domains (Figure 5B). SRP54 has a C-terminal extension, the M domain, which binds RNA and the signal peptide (Zopf et al., 1990; Lütcke et al., 1992), whereas SRα (FtsY) has a highly charged N-terminal extension. SRα has an N-terminal domain, which interacts with SRβ (Young et al., 1995; Schwartz and Blobel, 2003). The crystal structures of the NG domain of Thermus aquaticus Ffh and E.coli FtsY have revealed that the N domain consists of a four-helix bundle that is associated with the GTPase (G) domain (Freymann et al., 1997; Montoya et al., 1997). The GTPase domains of SRP54 and FtsY are similar to other small GTPases except that they have an insertion of two β-strands and two α-helices including a so-called I box, which is implicated in the SRP–SR interaction (Freymann et al., 1997; Montoya et al., 1997; Leipe et al., 2002). The M domain of SRP54 can be cross-linked to the signal peptide and is considered to be the major site for the signal peptide binding (Zopf et al., 1990; Lütcke et al., 1992). The structure of the M domain from E.coli and T.aquaticus Ffh and human SRP54 (Keenan et al., 1998; Clemons et al., 1999; Batey et al., 2000; Kuglstatter et al., 2002) shows that the M domain forms a hydrophobic groove into which a peptide loop or an α-helix from a symmetry-related M domain is bound, possibly mimicking signal peptide binding (Figure 4B). These structures contain a poorly ordered region, which may become ordered upon binding of real signal peptides. The structure of the NG domain from T.aquaticus Ffh solved in the apo, GDP-bound and GDP-Mg2+-bound forms suggests that the N and G domains could move with respect to each other upon nucleotide binding and this movement may be important in coupling signal peptide binding and release, GTP binding and hydrolysis, and SR binding (Freymann et al., 1997, 1999; Ramirez et al., 2002) (Figure 5B). The N domain contains a highly conserved sequence motif, ALLEADV, including the C-terminal end of its second helix and most of the loop between its second and third α-helices. This motif is in close contact at the NG interface with the highly conserved DARGG motif in the G domain α4 helix and its preceding loop (Ramirez et al., 2002). Mutations in the ALLEADV motif alter the affinity for signal peptides as estimated by elongation arrest assay (Newitt and Bernstein, 1997). The structure of a complex between Ffh and FtsY will reveal how these proteins interact with each other in the GTP-bound form and how this will trigger reciprocal activation of Ffh and FtsY GTPases. The G domains of Ffh and FtsY are structurally related to dimeric ATP-utilizing proteins such as nitrogenase iron protein (NIP) and dithiobiotin synthetase (Montoya et al., 2000). Based on the structure of an NIP dimer, Montoya et al. (2000) proposed a model for the Ffh–FtsY complex in which the switch I and II regions and the I box are involved in the interaction. This may be consistent with GTP-dependent complex formation and reciprocal activation of Ffh and FtsY GTPases, but elucidation of the detailed mechanism requires the crystal structure of the complex.
Recently, Schwartz and Blobel (2003) reported the crystal structure of yeast SRβ bound to the N-terminal interaction domain of SRα (SRX), which is connected to the NG domain with a protease-sensitive region. SRβ contains the canonical features of the Ras superfamily of small GTPases and is structurally closest to the Arf subfamily (Figure 5C). Clear density for GTP is seen at the active site buried at the interface. The switch I region forms an intricate network of hydrogen bonds mainly involving main chain amide and carbonyl groups with SRX of SRα. The two subunits interact strongly when GTP is bound to SRβ, whereas they are likely to dissociate upon hydrolysis of GTP (Legate et al., 2000; Schwartz and Blobel, 2003). The protein targeting cycle is regulated by a concerted action of three GTPases, namely SRP54, SRα and SRβ. The formation of the SRP–SR complex leads to docking of the ribosome with the translocon, release of the signal peptide from SRP54 and its transfer to the Sec61 complex, hydrolysis of SRP54- and SRα-bound GTP that leads to dissociation of the SRP–SR complex, resumption of polypeptide chain elongation and hydrolysis of SRβ-bound GTP that leads to weakening or dissociation of the SRα–SRβ complex. The key question is in what order these events take place and what triggers each of these sequential steps.
Conclusions
In the last 5 years, considerable progress has been made in elucidating the structure of the components involved in SRP-mediated protein targeting. These structures have begun to provide important insights into the molecular mechanism of protein targeting.
Acknowledgments
Acknowledgements
This work was supported by the Medical Research Council, UK, and HFSP. A.K. was supported by Böhringer Ingelheim funds, E.M. by a Gates studentship, and L.J. by EMBO and HFSP long-term fellowships.
References
- Bacher G., Lütcke,H., Jungnickel,B., Rapoport,T.A. and Dobberstein,B. (1996) Regulation by the ribosome of the GTPase of the signal-recognition particle during protein targeting. Nature, 381, 248–251. [DOI] [PubMed] [Google Scholar]
- Bacher G., Pool,M. and Dobberstein,B. (1999) The ribosome regulates the GTPase of the β-subunit of the signal recognition particle receptor. J. Cell Biol., 146, 723–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey R.T., Rambo,R.P., Lucast,L., Rha,B. and Doudna,J.A. (2000) Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science, 287, 1232–1239. [DOI] [PubMed] [Google Scholar]
- Batey R.T., Sagar,M.B. and Doudna,J.A. (2001) Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle. J. Mol. Biol., 307, 229–246. [DOI] [PubMed] [Google Scholar]
- Beckmann R., Spahn,C.M.T., Eswar,N., Helmers,J., Penczek,P.A., Sali,A., Frank,J. and Blobel,G. (2001) Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell, 107, 361–372. [DOI] [PubMed] [Google Scholar]
- Bhuiyan S., Gowda,K., Hotokezaka,H. and Zwieb,C. (2000) Assembly of archaeal signal recognition particle from recombinant components. Nucleic Acids Res., 28, 1365–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birse D.E., Kapp,U., Strub,K., Cusack,S. and Åberg,A. (1997) The crystal structure of the signal recognition particle Alu RNA binding heterodimer SRP9/14. EMBO J., 16, 3757–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemons W.M., Gowda,K., Black,S.D., Zwieb,C. and Ramakrishnan,V. (1999) Crystal structure of the conserved subdomain of human protein SRP54M at 2.1 Å resolution: evidence for the mechanism of signal peptide binding. J. Mol. Biol., 292, 697–705. [DOI] [PubMed] [Google Scholar]
- Diener J.L. and Wilson,C. (2000) Role of SRP19 in assembly of the Archaeoglobus fulgidus signal recognition particle. Biochemistry, 39, 12862–12874. [DOI] [PubMed] [Google Scholar]
- Doherty E.A., Batey,R.T., Masquida,B. and Doudna,J.A. (2001) A universal mode of helix packing in RNA. Nat. Struct. Biol., 8, 339–343. [DOI] [PubMed] [Google Scholar]
- Freymann D.M., Keenan,R.J., Stroud,R.M. and Walter,P. (1997) Structure of the conserved GTPase domain of the signal recognition particle. Nature, 385, 361–364. [DOI] [PubMed] [Google Scholar]
- Freymann D.M., Keenan,R.J., Stroud,R.M. and Walter,P. (1999) Functional changes in the structure of the SRP GTPase on binding GDP and Mg2+ GDP. Nat. Struct. Biol., 6, 793–801. [DOI] [PubMed] [Google Scholar]
- Fulga T.A., Sinning,I., Dobberstein,B. and Pool,M.R. (2001) SRβ coordinates signal sequence release from SRP with ribosome binding to the translocon. EMBO J., 20, 2338–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S.Q., Peske,F., Wieden,H.J., Rodnina,M.V. and Wintermeyer,W. (2003) The signal recognition particle binds to protein L23 at the peptide exit of the Escherichia coli ribosome. RNA, 9, 566–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundelfinger E.D., Krause,E., Melli,M. and Dobberstein,B. (1983) The organization of the 7SL RNA in the signal recognition particle. Nucleic Acids Res., 11, 7363–7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainzl T., Huang,S.H. and Sauer-Eriksson,E. (2002) Structure of the SRP19–RNA complex and implications for signal recognition particle assembly. Nature, 417, 767–771. [DOI] [PubMed] [Google Scholar]
- Jovine L., Hainzl,T., Oubridge,C., Scott,W.G., Li,J., Sixma,T.K., Wonacott,A., Skarzynski,T. and Nagai,K. (2000) Crystal structure of the Ffh and EF-G binding sites in the conserved domain IV of Escherichia coli 4.5S RNA. Structure, 8, 527–540. [DOI] [PubMed] [Google Scholar]
- Keenan R.J., Freymann,D.M., Walter,P. and Stroud,R.M. (1998) Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell, 94, 181–191. [DOI] [PubMed] [Google Scholar]
- Keenan R.J., Freymann,D.M., Stroud,R.M. and Walter,P. (2001) The signal recognition particle. Annu. Rev. Biochem., 70, 755–775. [DOI] [PubMed] [Google Scholar]
- Kuglstatter A., Oubridge,C. and Nagai,K. (2002) Induced structural changes of 7SL RNA during the assembly of human signal recognition particle. Nat. Struct. Biol., 9, 740–744. [DOI] [PubMed] [Google Scholar]
- Legate K.R., Falcone,D. and Andrews,D.W. (2000) Nucleotide-dependent binding of the GTPase domain of the signal recognition particle receptor β-subunit to the α-subunit. J. Biol. Chem., 275, 27439–27446. [DOI] [PubMed] [Google Scholar]
- Leipe D.D., Wolf,Y.I., Koonin,E.V. and Aravind,L. (2002) Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol., 317, 41–72. [DOI] [PubMed] [Google Scholar]
- Luirink J., High,S., Wood,H., Giner,A., Tollervey,D. and Dobberstein,B. (1992) Signal-sequence recognition by an Escherichia coli ribonucleoprotein complex. Nature, 359, 741–743. [DOI] [PubMed] [Google Scholar]
- Lütcke H., High,S., Römisch,K., Ashford,A.J. and Dobberstein,B. (1992) The methionine-rich domain of the 54 kDa subunit of signal recognition particle is sufficient for the interaction with signal sequences. EMBO J., 11, 1543–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lütcke H., Prehn,S., Ashford,A.J., Remus,M., Frank,R. and Dobberstein,B. (1993) Assembly of the 68- and 72-kD proteins of signal recognition particle with 7S RNA. J. Cell Biol., 121, 977–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.D., Bernstein,H.D. and Walter,P. (1994) Interaction of E.coli Ffh/4.5S ribonucleoprotein and Fts Y mimics that of mammalian signal recognition particle and its receptor. Nature, 367, 657–659. [DOI] [PubMed] [Google Scholar]
- Miller J.D., Tajima,S., Lauffer,L. and Walter,P. (1995) The β subunit of the signal recognition particle receptor is a transmembrane GTPase that anchors the α subunit, a peripheral membrane GTPase, to the endoplasmic reticulum membrane. J. Cell Biol., 128, 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya G., Svensson,C., Luirink,J. and Sinning,I. (1997) Crystal structure of the NG domain from the signal-recognition particle receptor FtsY. Nature, 385, 365–369. [DOI] [PubMed] [Google Scholar]
- Montoya G., te Kaat,K., Moll,R., Schäfer,G. and Sinning,I. (2000) The crystal structure of the conserved GTPase of SRP54 from the archaeon Acidianus ambivalens and its comparison with related structures suggests a model for the SRP–SRP receptor complex. Structure, 8, 515–525. [DOI] [PubMed] [Google Scholar]
- Nakamura K., Hahagi,S., Yamazaki,T. and Yamane,K. (1999) Bacillus subtilis histone-like protein, HBsu, is an integral component of a SRP-like particle that can bind the Alu domain of small cytoplasmic RNA. J. Biol. Chem., 274, 13569–13576. [DOI] [PubMed] [Google Scholar]
- Newitt J.A. and Berstein,H.D. (1997) The N-domain of the signal recognition particle 54-kDa subunit promotes efficient signal sequence binding. Eur. J. Biochem., 245, 720–729. [DOI] [PubMed] [Google Scholar]
- Nissen P., Ippolito,J.A., Ban,N., Moore,P.B. and Steitz,T.A. (2001) RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc. Natl Acad. Sci. USA, 98, 4899–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oubridge C., Kuglstatter,A., Jovine,L. and Nagai,K. (2002) Crystal structure of SRP19 in complex with the S domain of SRP RNA and its implication for the assembly of the signal recognition particle. Mol. Cell, 9, 1251–1261. [DOI] [PubMed] [Google Scholar]
- Pool M.R., Stumm,J., Fulga,T.A., Sinning,I. and Dobberstein,B. (2002) Distinct modes of signal recognition particle interaction with the ribosome. Science, 297, 1345–1348. [DOI] [PubMed] [Google Scholar]
- Poritz M. A, Bernstein,H.D., Strub,K., Zopf,D., Wilhelm,H. and Walter,P. (1990) An E.coli ribonucleoprotein containing 4.5S RNA resembles mammalian signal recognition particle. Science, 250, 1111–1117. [DOI] [PubMed] [Google Scholar]
- Powers T. and Walter,P. (1995) Reciprocal stimulation of GTP hydrolysis by two directly interacting GTPases. Science, 269, 1422–1424. [DOI] [PubMed] [Google Scholar]
- Ramirez U.D., Minasov,G., Focia,P.J., Stroud,R.M., Walter,P., Kuhn,P. and Freymann,D.M. (2002) Structural basis for mobility in the 1.1 Å crystal structure of the NG domain of Thermus aquaticus Ffh. J. Mol. Biol., 320, 783–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapiejko R. and Gilmore,R. (1997) Empty site forms of the SRP54 and SRα GTPases mediate targeting of ribosome-nascent chain complexes to the endoplasmic reticulum. Cell, 89, 703–713. [DOI] [PubMed] [Google Scholar]
- Rose M.A. and Weeks,K.M. (2001) Visualizing induced fit in early assembly of the human signal recognition particle. Nat. Struct. Biol., 8, 515–520. [DOI] [PubMed] [Google Scholar]
- Rosenblad M.A., Gorodkin,J., Knudsen,B., Zwieb,C. and Samuelsson,T. (2003) SRPDB: Signal Recognition Particle Database. Nucleic Acids Res., 31, 363–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz U., Behrens,S., Freymann,D.M., Keenan,R.J., Lukavsky,P., Walter,P. and James,T.L. (1999) Structure of the phylogenetically most conserved domain of SRP RNA. RNA, 5, 1419–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz T. and Blobel,G. (2003) Structural basis for the function of the β subunit of the eukaryotic signal recognition particle receptor. Cell, 112, 793–803. [DOI] [PubMed] [Google Scholar]
- Siegel V. and Walter,P. (1988) Binding sites of the 19-kDa and 68/72-kDa signal recognition particle (SRP) proteins on SRP RNA as determined in protein–RNA footprinting. Proc. Natl Acad. Sci. USA, 85, 1801–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P. and Blobel,G. (1983) Disassembly and reconstitution of signal recognition particle. Cell, 34, 525–533. [DOI] [PubMed] [Google Scholar]
- Walter P. and Johnson,A.E. (1994) Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol., 10, 87–119. [DOI] [PubMed] [Google Scholar]
- Walter P., Ibrahimi,I. and Blobel,G. (1981) Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol., 91, 545–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichenrieder O., Wild,K., Strub,K. and Cusack,S. (2000) Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature, 408, 167–173. [DOI] [PubMed] [Google Scholar]
- Wild K., Sinning,I. and Cusack,S. (2001) Crystal structure of an early protein–RNA assembly complex of the signal recognition particle. Science, 294, 598–601. [DOI] [PubMed] [Google Scholar]
- Wild K., Weichenrieder,O., Strub,K., Sinning,I. and Cusack,S. (2002) Towards the structure of the mammalian signal recognition particle. Curr. Opin. Struct. Biol., 12, 72–81. [DOI] [PubMed] [Google Scholar]
- Young Y.C., Ursini,J., Legate,K.R., Miller,J.D., Walter,P. and Andrews,D.W. (1995) An amino-terminal domain containing hydrophobic and hydrophilic sequences binds the signal recognition particle receptor α subunit to the β subunit on the endoplasmic reticulum. J. Biol. Chem., 270, 15650–15657. [DOI] [PubMed] [Google Scholar]
- Zopf D., Bernstein,H.D., Johnson,A.E. and Walter,P. (1990) The methionine-rich domain of the 54 kd protein subunit of the signal recognition particle contains an RNA binding site and can be crosslinked to a signal sequence. EMBO J., 9, 4511–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C. (1992) Recognition of a tetranucleotide loop of signal recognition particle RNA by protein SRP19. J. Biol. Chem., 267, 15650–15656. [PubMed] [Google Scholar]
- Zwieb C. and Eichler,J. (2002) Getting on target: the archaeal signal recognition particle. Archaea, 1, 27–34. [DOI] [PMC free article] [PubMed] [Google Scholar]