Abstract
Research on human inherited diseases provides a powerful tool to identify an intrinsically important subset of genes vital to healthy functioning of the organism. Progressive myoclonus epilepsies (PMEs) are a group of rare inherited disorders characterized by the association of epilepsy, myoclonus and progressive neurological deterioration. Significant progress has been made in elucidating the molecular background of PMEs. Here, progress towards understanding the molecular pathogenesis of PMEs is reviewed using the most common single cause of PME, Unverricht–Lundborg disease, as an example. Mutations in the gene encoding cystatin B (CSTB), a cysteine protease inhibitor, are responsible for the primary defect in Unverricht–Lundborg disease. CSTB-deficient mice, produced by targeted disruption of the mouse Cstb gene, display a phenotype similar to the human disease, with progressive ataxia and myoclonic seizures. The mice show neuronal atrophy, apoptosis and gliosis as well as increased expression of apoptosis and glial activation genes. Although significant advances towards understanding the molecular basis of Unverricht–Lundborg disease have been achieved, the physiological function of CSTB and the molecular pathogenesis of the disease remain unknown.
Keywords: apoptosis/cystatin B/EPM1/neurodegeneration/progressive myoclonus epilepsy
Introduction
Progressive myoclonus epilepsies (PMEs) are a hetero geneous group of inherited disorders defined by the association of myoclonus, epilepsy and progressive neurological deterioration (Berkovic et al., 1986; Marseille Consensus Group, 1990). Despite a common name, PMEs differ in clinical features, aetiology and pathogenesis. Five disease entities or disease groups, Unverricht–Lundborg disease, Lafora’s disease, neuronal ceroid lipofuscinoses (NCLs), mitochondrial disorders and sialidoses, account for the majority of PME cases in the world. In addition, a number of quite rare disorders can cause the PME phenotype.
Recent advances in molecular genetics have significantly increased the understanding of the basic mechanisms involved in the PMEs. Positional cloning has been used successfully to identify genes underlying the major forms of PME (Table I). In addition, several genes for the more rare forms have been identified, e.g. the DRPLA gene underlying dentatorubral–pallidoluysian atrophy found predominantly in Japan (Koide et al., 1994; Nagafuchi et al., 1994). The research now aims at understanding the function of the proteins encoded by the PME genes as well as revealing the underlying disturbed metabolic pathways. In this review, progress towards these goals is described using Unverricht–Lundborg disease, the most common single cause of PME, as an example.
Table I. The major forms of PME and their underlying genes.
| Disorder | Locus/chromosome | Gene product | References |
|---|---|---|---|
| Unverricht–Lundborg disease | EPM1/21q22.3 | Cystatin B (CSTB) | Pennacchio et al. (1996) |
| Cysteine protease inhibitor | |||
| Lafora’s disease | EPM2A/6q24 | Laforin; dual-specificity phosphatase | Minassian et al. (1998); Serratosa et al. (1999); Ganesh et al. (2000) |
| NCL | |||
| Infantile | CLN1/1p32 | Palmitoyl-protein thioesterase 1 (PPT1) | Vesa et al. (1995) |
| Late infantile | CLN2/11p15 | Tripeptidyl peptidase 1 (TPP1) | Sleat et al. (1997); Rawlings and Barrett (1999); Vines and Warburton (1999) |
| Finnish variant late infantile | CLN5/13q22 | Novel membrane protein (CLN5) | Savukoski et al. (1998) |
| Variant late infantile | CLN6/15q21–23 | Novel membrane protein (CLN6) | Gao et al. (2002); Wheeler et al. (2002) |
| Juvenile | CLN3/16p12 | Novel membrane protein (CLN3) | International Batten Disease Consortium (1995) |
| Northern epilepsy | CLN8/8p23 | Novel membrane protein (CLN8) | Ranta et al. (1999) |
| MERRF | MTTK/mtDNA | tRNALys | Shoffner et al. (1990); Yoneda et al. (1990) |
| Sialidosis | NEU1/6p21 | Neuraminidase 1 (NEU1) | Bonten et al. (1996); Pshezhetsky et al. (1997) |
Unverricht–Lundborg disease (EPM1; OMIM254800) is an autosomal recessive inherited disorder characterized by onset at the age of 6–15 years, severe incapacitating stimulus-sensitive progressive myoclonus, tonic–clonic epileptic seizures and characteristic abnormalities in the electroencephalogram (EEG) (Koskiniemi et al., 1974a,b; Norio and Koskiniemi, 1979). EPM1 patients also develop other neurological symptoms such as ataxia, incoordination and dysarthria (Koskiniemi et al., 1974a; Norio and Koskiniemi, 1979). On histopathological examination of the brain, widespread non-specific degenerative changes, and loss of Purkinje cells, but no intracellular inclusions have been observed (Haltia et al., 1969; Koskiniemi et al., 1974a; Eldridge et al., 1983).
Molecular genetic basis of EPM1
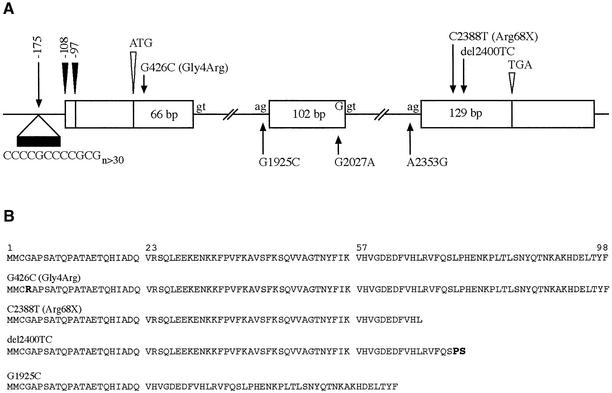
The mutated gene on chromosome 21q22.3 responsible for EPM1 was identified using positional cloning (Lehesjoki et al., 1991, 1993; Pennacchio et al., 1996; Stone et al., 1996; Virtaneva et al., 1996). It encodes a previously described and characterized, but unmapped protein, cystatin B (CSTB), a cysteine protease inhibitor (Järvinen and Rinne, 1982; Ritonja et al., 1985). The CSTB gene is ubiquitously expressed with a transcript of ∼0.8 kb in northern blot analysis (Pennacchio et al., 1996). Lalioti et al. (1997a) used an RNase protection assay to determine the transcription start site of CSTB, and found two sites, 97 bp and 108 bp upstream of the translation initiation codon and downstream of a dodecamer repeat unit (Figure 1). To date, seven CSTB gene mutations underlying EPM1 have been described (Pennacchio et al., 1996; Lafreniere et al., 1997; Lalioti et al., 1997a,b; Virtaneva et al., 1997; Kagitani-Shimono et al., 2002; Figure 1).
Fig. 1. Schematic overview of the CSTB gene structure, mutations and predicted amino acid sequences of mutant proteins. (A) The 98 amino acid CSTB protein is transcribed from three exons depicted as white boxes, with the corresponding lengths of coding sequences in base pairs. White arrowheads indicate the translation start (ATG) and stop (TGA) sites. Black arrowheads indicate the positions of the two transcription start sites. The positions of the EPM1-associated mutations are shown with arrows. (B) Predicted sequences of CSTB proteins representing EPM1 mutations. The major mutation underlying EPM1, the dodecamer repeat expansion in the promoter region, results in production of reduced amounts of CSTB with normal sequence, shown on top. The del2400TC mutation creates a frameshift with two out-of-frame amino acids before an early stop codon. Of the three mutations affecting exon–intron boundaries, the G1925C mutation has been shown to result in skipping of exon 2 (Bespalova et al., 1997b), with consequent in-frame deletion of 34 amino acids from the polypeptide. The consequences of the other two splice site mutations have not been studied.
An unstable expansion of a dodecamer (12 bp) or minisatellite repeat unit of 5′-ccccgccccgcg-3′ located 175 bp upstream from the translation initiation codon in the putative promoter region of CSTB (Figure 1) is the most common EPM1-associated mutation and accounts for ∼90% of disease alleles (Lafreniere et al., 1997; Lalioti et al., 1997a; Virtaneva et al., 1997). The CSTB minisatellite repeat is normally polymorphic, with 2–3 copies, but EPM1-associated expanded alleles contain at least 30 repeat copies (Lalioti et al., 1998). The majority of EPM1 patients are homozygous for the minisatellite expansion. No correlation between the repeat size and the age of onset or the severity of the clinical phenotype has been observed (Lafreniere et al., 1997; Lalioti et al., 1997a, 1998; Virtaneva et al., 1997). Expanded, pathogenic alleles of the EPM1 minisatellite show a high (47%) mutation rate, with contractions or expansions of the minisatellite typically by a single repeat unit (Larson et al., 1999). The EPM1-associated dodecamer repeat has been shown to form stable secondary structures under physiological conditions, which may be, at least in part, responsible for the expansion (Pataskar et al., 2001; Saha and Ushdin, 2001).
The six further EPM1-associated mutations occur within the transcription unit of CSTB (Pennacchio et al., 1996; Lalioti et al., 1997b; Kagitani-Shimono et al., 2002; Figure 1). Three mutations, G1925C, G2027A and A2353G (GenBank acccession No. U46692), affect conserved splice site sequences and predict severe splicing defects, while two mutations in exon 3 (C2388T and del2400TC; GenBank accession No. U46692) predict a truncated protein through either creating a nonsense codon or causing a frameshift. The sixth mutation is a G426C (GenBank accession No. U46692) transversion in exon 1 that results in the substitution of a highly conserved glycine by an arginine at amino acid position 4 (Gly4Arg). This is, so far, the only missense mutation reported in EPM1 patients.
Cystatin B, a cysteine protease inhibitor
CSTB belongs to family 1 of a large superfamily of protease inhibitors, the cystatins, which are known to inhibit in vitro several papain-family cysteine proteases, cathepsins, by tight and reversible binding (Barrett, 1986; Barrett et al., 1986; Rawlings and Barrett, 1990; Turk and Bode, 1991). Cystatins of family 1 are small molecular weight proteins comprising a single polypeptide chain with no disulfide bonds or carbohydrates. Human CSTB, also known as neutral cysteine protease inhibitor and stefin 1, was first identified and characterized from lymphatic tissue (Rinne et al., 1981; Järvinen and Rinne, 1982) and later from liver (Ritonja et al., 1985). It is a ubiquitously expressed 98 amino acid protein and has a mol. wt of 11 kDa (Järvinen and Rinne, 1982; Ritonja et al., 1985; Jerala et al., 1988). CSTB has isoelectric variants and it often forms dimers by intermolecular disulfide bond formation. CSTB binds in vitro tightly to cathepsins H, L and S, and less tightly to cathepsin B (Green et al., 1984; Abrahamson et al., 1986; Brömme et al., 1991; Machleidt et al., 1991; Lenarcic et al., 1996).
The crystal structures of chicken cystatin B (Bode et al., 1988) and of recombinant human CSTB–papain complex (Stubbs et al., 1990; PDB entry 1STF) have been determined. Both the chicken and human CSTB consist of a five-stranded β-sheet wrapped around a five-turn α-helix. In addition, the human CSTB contains a C-terminal strand that runs along the convex side of the sheet. CSTB interacts with papain through a tripartite wedge formed by conserved residues at the most N-terminal part of CSTB, the first hairpin loop containing the highly conserved QVVAG sequence, and the second hairpin loop (Stubbs et al., 1990). The extended C-terminus of human CSTB provides an additional binding site relative to chicken CSTB (Stubbs et al., 1990). Although CSTB has been characterized in detail in vitro, its physiological function is unknown.
Consequences of EPM1 mutations on CSTB mRNA and protein
CSTB mRNA expression in EPM1 patients has been studied both by northern analysis and by RNase protection assay, with somewhat controversial results. In northern analysis of lymphoblastoid cell mRNA, dramatically reduced levels of CSTB mRNA were observed in patients either homozygous or compound heterozygous for the minisatellite expansion mutation (Pennacchio et al., 1996; Lalioti et al., 1997b; Bespalova et al., 1997a,b; Lafreniere et al., 1997; Alakurtti et al., 2000). Contrary to the northern data, Lalioti et al. (1997a) showed with an RNase protection assay that the level of CSTB mRNA was markedly reduced in blood leukocytes, but was either normal or only slightly reduced in fibroblasts and lymphoblastoid cell lines of patients. These data suggested a cell-specific reduction in CSTB gene expression and/or modulation of the expression of CSTB by the repeat expansion in some cell types.
Consistent with the results in northern analyses of CSTB expression, Rinne et al. (2002) showed that the papain inhibitory (cystatin) activity was significantly decreased or absent in lymphoblastoid cells of EPM1 patients. This reduction correlated with a significant increase in general cathepsin activity, and in particular of cathepsin B, L and S activities. Taking these data together with the in vitro kinetic data, which have shown very strong binding of CSTB to cathepsin S, intermediate binding to cathepsins H and L, and relatively weak binding to cathepsin B (Green et al., 1984; Abrahamson et al., 1986; Brömme et al., 1991; Machleidt et al., 1991; Lenarcic et al., 1996), it was suggested that the consequences of decreased CSTB activity in EPM1 pathogenesis may be, at least in part, due to increased activity of cathepsins S and L.
The consequences of the only EPM1-associated missense mutation (Gly4Arg) have not been experimentally tested. However, Gly4 is known to be highly conserved in cystatins from various species (Rawlings and Barrett, 1990; Turk and Bode, 1991; Lalioti et al., 1997b). Based on the known crystal structure of the CSTB–papain complex (Stubbs et al., 1990), the Gly4Arg substitution that is located in the N-terminal interacting part is likely to modify the papain-binding pocket. Three-dimensional modelling suggests that the Gly4Arg mutation, by bringing in an amino acid with a long and charged side chain, which causes major steric hindrance, is likely to strongly affect the papain-binding capacity of CSTB (Lalioti et al., 1997b).
Characteristics of the CSTB gene promoter
As the expansion mutation is located upstream from the transcription initiation sites of CSTB in the putative promoter region, two studies have aimed to characterize the CSTB promoter (Lalioti et al., 1999; Alakurtti et al., 2000). Alakurtti et al. (2000) limited the promoter analysis to downstream of an Alu-rich repeat region starting at ∼0.7 kb upstream from translation initiation and used different promoter–luciferase reporter constructs in transient transfection experiments in COS-1 cells to map the promoter within 670 bp from the translation initiation codon. Sixteen extra copies of the dodecamer in the 670 bp promoter construct resulted in 10-fold reduced luciferase expression, indicating that the repeat expansion downregulates transcription in vitro, compatible with northern analysis of lymphoblastoid cell RNA of patients (Alakurtti et al., 2000). Using electrophoretic mobility shift assays, active binding to five Sp1 and four AP1 sites as well as weak binding to an androgen response element half-site were demonstrated (Alakurtti et al., 2000).
Lalioti et al. (1999) used a 3.2 kb fragment upstream of the translation initiation codon in characterization of the promoter. They showed that a 600 bp repeat expansion in this promoter fragment reduced luciferase activity 2- to 4-fold compared with an identical fragment with a normal sized repeat. This reduction was only seen in some cell types (SK-N-BE neuroblastoma and HeLa), whereas in others (CHP neuroblastoma and COS-7) either a small decrease or an increase in luciferase activity was observed (Lalioti et al., 1999). These data were compatible with the observations on the cell type-specific reduction in mRNA expression in EPM1 patients using the RNase protection assay (Lalioti et al., 1997a). Introduction of heterologous DNA fragments of 730 and 1000 bp into the normal promoter instead of the repeat expansion showed similarly reduced activity, suggesting that altered spacing of promoter elements due to the dodecamer repeat expansion contributed to reduced CSTB gene expression (Lalioti et al., 1999).
In addition to altered spacing of promoter elements, the possibility of abnormal hypermethylation of the promoter region in the presence of the expansion mutation as a cause of reduced transcription has been considered. However, restriction analysis of the CSTB promoter region with methylation-sensitive enzymes has revealed no significant hypermethylation (Lalioti et al., 1997b; own unpublished findings), although the methylation status of the dodecamer repeat itself has not been determined. It is possible that the stable secondary structures that the dodecamer repeat forms under physiological conditions (Pataskar et al., 2001; Saha and Usdin, 2001) affect transcription or even translation, which has been shown to occur, for example, in association with trinucleotide repeat expansions (Feng et al., 1995; Parsons et al., 1998).
Cellular localization and molecular interactions of CSTB
Two studies have reported the results of immunofluorescence analysis of endogenous CSTB in a variety of cultured cell types (Calkins et al., 1998; Riccio et al., 2001), with somewhat controversial results. Calkins et al. (1998) used a monoclonal anti-CSTB antibody in confocal immunofluorescence analysis of CSTB and cathepsin B in two liver cell lines. CSTB distribution was distinct from that of cathepsin B, and it was found diffusely throughout the cytoplasm with apparent concentration at membranes of vesicular structures. More recent data have also indicated nuclear localization for CSTB (Riccio et al., 2001). Using several different cell types and polyclonal anti-CSTB antibodies, Riccio et al. (2001) demonstrated that CSTB was mainly localized in the nucleus in proliferating cells and both in the nucleus and the cyto plasm in differentiated cells. By comparison, cathepsin B was shown to be essentially cytoplasmic, and CSTB and cathepsin B were shown to co-localize only partially (Riccio et al., 2001). The separate compartmentalization of CSTB and cathepsin B as well as the localization of CSTB in both nucleus and cytoplasm could suggest that CSTB has functions in the cell other than cathepsin inhibition.
In order to identify proteins interacting with CSTB, Di Giaimo et al. (2002) used the yeast two-hybrid technique with rat CSTB cDNA as a bait in screening a cDNA library from developing rat cerebella. They identified five proteins interacting with CSTB. Three were known proteins: rat neurofilament light polypeptide (NF-L), rat activated protein kinase C receptor (RACK-1) and rat brain β-spectrin. One protein was related to human myotubu larin, while the fifth was a novel protein of unknown function. Interestingly, although present in the cDNA library, no interaction of CSTB was detected with cathepsins B, H or L. The three previously known proteins identified in the screen were shown to interact with CSTB in GST pull-down assays as well as to partially co-localize with CSTB in immunofluorescence analysis of differentiated cultured primary cerebellar granule cells (Di Giaimo et al., 2002). Moreover, all three proteins were shown to co-immunoprecipitate in a specific manner with CSTB from rat cerebellar cell extracts, but not from cerebral hemispheres, suggesting in vivo interaction of the four proteins in the cerebellum. Confocal immunofluorescence analysis indicated that all four proteins were mostly expressed in the granule cells of developing rat cerebellum, and in Purkinje cells of adult rat cerebellum. Based on these data, the authors concluded that CSTB participates in the formation of a multiprotein complex that has a specific cerebellar function, possibly involved in cell growth and differentiation (Di Giaimo et al., 2002).
Cystatin B-deficient mouse model for Unverricht–Lundborg disease
Mice deficient for CSTB have been produced by targeted disruption of the mouse Cstb gene (Pennacchio et al., 1998). The mice develop a phenotype that resembles the human phenotype, with progressive ataxia and myoclonic seizures. However, in contrast to the human phenotype, no tonic–clonic seizures, photosensitivity or spike–wave complexes in EEGs, typical in human patients, have been observed in the mice. Initial analysis of the mice revealed apoptotic death of cerebellar granule cells, indicating that CSTB has a role in preventing cerebellar apoptosis and suggesting that EPM1 should be classified as a primary neurodegenerative disorder that selectively targets specific mammalian cells (Pennacchio et al., 1998). Further evidence supporting the hypothesis of an endogenous neuroprotective role for CSTB arises from studies in a rat kindling model of epilepsy (D’Amato et al., 2000), in which seizure activity has been shown to induce marked and widespread upregulation of CSTB mRNA and protein in rat forebrain neurons.
Later, additional neuropathology in Cstb-knockout mice was reported (Shannon et al., 2002). In addition to cerebellar granule cell apoptosis, less marked neuronal apoptosis was detected in the hippocampal formation and entorhinal cortex in 3- to 4-month-old mice. In older mice, gliosis was present in the hippocampal formation, entorhinal cortex, neocortex and striatum. Widespread gliosis was also present in the white matter, which may be a secondary phenomenon (Shannon et al., 2002). In addition to neuronal death by apoptosis, the superficial neurons of the prosubiculum in the cerebral cortex displayed prominent cellular atrophy. The observed neuropathology was similar in seizure-prone and seizure-resistant genetic backgrounds. The data indicate that neuronal atrophy is an important consequence of CSTB deficiency independent of seizure events, suggesting that CSTB is important in maintaining normal neuronal architecture and size and that in addition to cellular death, cellular dysfunction may also be important in producing the phenotype (Shannon et al., 2002). Interestingly, the neuropathological changes were distributed unevenly in the mouse brain, suggesting differential neuronal sensitivity to CSTB deficiency.
Consistent with the observed pathology in the mice, mRNA expression profiling of CSTB-deficient mice shows increased expression of seven genes involved in proteolysis, apoptosis and glial activation, among them that for cathepsin S (Lieuallen et al., 2001).
Conclusion
Recent advances in the molecular understanding of PMEs have important implications. Pinpointing the molecular defects in individual diseases allows an aetiological diagnosis, which has already resulted in improved clinical practice and classification of these disorders. Moreover, identification of the primary defects underlying PMEs has provided a starting point towards understanding the molecular pathogenesis of these disorders. Towards this goal, important tools including animal and cellular models have been created. However, the physiological function of the PME proteins and the disease mechanisms are still largely unknown. Further research utilizing both classical methods and the modern tools provided by the genome project is needed to dissect the diseases at a molecular and cellular level and to compile this information into an understanding of the basic mechanisms underlying PMEs. This knowledge will be the basis for the development of rational methods for the prevention and treatment of these devastating disorders. Moreover, the research is expected to provide insights into molecular mechanisms involved in neuronal function and survival as well as seizure generation, thus providing insights into more common disorders that share underlying mechanisms.
Acknowledgments
Acknowledgements
I thank the past and present members of my laboratory for their commitment and dedication. Special thanks to Kirsi Alakurtti for help in designing Figure 1. Work related to EPM1 in my laboratory has been supported by the Academy of Finland (projects 43029, 50011 and 44870), the EC (project QLG3-CT-2000-01405), the Folkhälsan Research Foundation and the Sigrid Juselius Foundation.
References
- Abrahamson M., Barrett,A.J., Salvesen,G. and Grubb,A. (1986) Isolation of six cysteine proteinase inhibitors from human urine. Their physicochemical and enzyme kinetic properties and concentrations in biological fluids. J. Biol. Chem., 261, 11282–11289. [PubMed] [Google Scholar]
- Alakurtti K., Virtaneva,K., Joensuu,T., Palvimo,J.J. and Lehesjoki,A.-E. (2000) Characterization of the cystatin B gene promoter harboring the dodecamer repeat expanded in progressive myoclonus epilepsy, EPM1. Gene, 242, 65–73. [DOI] [PubMed] [Google Scholar]
- Barrett A.J. (1986) The cystatins, a diverse superfamily of cysteine peptidase inhibitors. Biomed. Biochim. Acta, 45, 1363–1373. [PubMed] [Google Scholar]
- Barrett A.J. et al. (1986) Nomenclature and classification of the proteins homologous with the cysteine-proteinase inhibitor chicken cystatin. Biochem. J., 236, 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkovic S.F., Andermann,F., Carpenter,S. and Wolfe,L. (1986) Progressive myoclonus epilepsies: specific causes and diagnosis. N. Engl. J. Med., 315, 296–304. [DOI] [PubMed] [Google Scholar]
- Bespalova I.N., Adkins,S., Pranzatelli,M. and Burmeister,M. (1997a) Novel cystatin B mutation and diagnostic PCR assay in an Unverricht–Lundborg progressive myoclonus epilepsy patient. Am. J. Med. Genet., 74, 467–471. [DOI] [PubMed] [Google Scholar]
- Bespalova I.N., Pranzatelli,M. and Burmeister,M. (1997b) G to C transversion at a splice acceptor site causes exon skipping in the cystatin B gene. Mutat. Res. Gen., 382, 67–74. [DOI] [PubMed] [Google Scholar]
- Bode W., Engh,R., Musil,D., Thiele,U., Huber,R., Karshikov,A., Brzin,J., Kos,J. and Turk,V. (1988) The 2.0 Å X-ray crystal structure of chicken egg white cystatin and its possible mode of interaction with cysteine proteinases. EMBO J., 7, 2593–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonten E., van der Spoel,A., Fornerod,M., Grosveld,G. and d’Azzo,A. (1996) Characterization of human lysosomal neuraminidase defines the molecular basis of the metabolic storage disorder sialidosis. Genes Dev., 10, 3156–3169. [DOI] [PubMed] [Google Scholar]
- Brömme D., Rinne,R. and Kirschke,H. (1991) Tight-binding inhibition of cathepsin S by cystatins. Biomed. Biochim. Acta, 50, 631–635. [PubMed] [Google Scholar]
- Calkins C.C., Sameni,M., Koblinski,J., Sloane,B.F. and Moin,K. (1998) Differential localization of cysteine protease inhibitors and a target cysteine protease, cathepsin B, by immuno-confocal microscopy. J. Histochem. Cytochem., 46, 745–751. [DOI] [PubMed] [Google Scholar]
- D’Amato E., Kokaia,Z., Nanobashvili,A., Reeben,M., Lehesjoki,A.-E., Saarma,M. and Linvall,O. (2000) Seizures induce widespread upregulation of cystatin B, the gene mutated in progressive myoclonus epilepsy, in rat forebrain neurons. Eur. J. Neurosci., 12, 1687–1695. [DOI] [PubMed] [Google Scholar]
- Di Giaimo R., Riccio,M., Santi,S., Galeotti,C., Ambrosetti,D.C. and Melli,M. (2002) New insights into the molecular basis of progressive myoclonus epilepsy: a multiprotein complex with cystatin B. Hum. Mol. Genet., 11, 2941–2950. [DOI] [PubMed] [Google Scholar]
- Eldridge R., Iivanainen,M., Stern,R., Koerber,T. and Wilder,B.J. (1983) ‘Baltic’ myoclonus epilepsy: hereditary disorder of childhood made worse by phenytoin. Lancet, 2, 838–842. [DOI] [PubMed] [Google Scholar]
- Feng Y., Zhang,F., Lokey,L.K., Chastain,J.L., Lakkis,L., Eberhart,D. and Warren,S.T. (1995) Translational suppression by trinucleotide repeat expansion at FMR1. Science, 268, 731–734. [DOI] [PubMed] [Google Scholar]
- Ganesh S. et al. (2000) Laforin, defective in the progressive myoclonus epilepsy of Lafora type, is a dual-specificity phosphatase associated with polyribosomes. Hum. Mol. Genet., 9, 2251–2261. [DOI] [PubMed] [Google Scholar]
- Gao H. et al. (2002) Mutations in a novel CLN6-encoded transmembrane protein cause variant neuronal ceroid lipofuscinosis in man and mouse. Am. J. Hum. Genet., 70, 324–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green G.D., Kembhavi,A.A., Davies,M.E. and Barret,A.J. (1984) Cystatin-like cysteine proteinase inhibitors from human liver. Biochem. J., 218, 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltia M., Kristensson,K. and Sourander,P. (1969) Neuropathological studies in three Scandinavian cases of progressive myoclonus epilepsy. Acta Neurol. Scand., 45, 63–77. [DOI] [PubMed] [Google Scholar]
- International Batten Disease Consortium (1995) Isolation of a novel gene underlying Batten Disease, CLN3. Cell, 82, 949–957. [DOI] [PubMed] [Google Scholar]
- Järvinen M. and Rinne,A. (1982) Human spleen cysteine protease inhibitor. Purification, fractionation into isoelectric variants and some properties of the variants. Biochim. Biophys Acta, 708, 210–217. [PubMed] [Google Scholar]
- Jerala R., Trstenjak,M., Lenarcic,B. and Turk,V. (1988) Cloning a synthetic gene for human stefin B and its expression in E.coli. FEBS Lett., 239, 41–44. [DOI] [PubMed] [Google Scholar]
- Kagitani-Shimono K., Imai,K., Okamoto,N., Ono,J. and Okada,S. (2002) Unverricht–Lundborg disease with cystatin B gene abnormalities. Pediatr. Neurol., 26, 55–60. [DOI] [PubMed] [Google Scholar]
- Koide R. et al. (1994) Unstable expansion of CAG repeat in hereditary dentatorubral–pallidoluysian atrophy (DRPLA). Nat. Genet., 6, 9–13. [DOI] [PubMed] [Google Scholar]
- Koskiniemi M., Donner,M., Majuri,H., Haltia,M. and Norio,R. (1974a) Progressive myoclonus epilepsy: a clinical and histopathologic study. Acta Neurol. Scand., 50, 307–332. [PubMed] [Google Scholar]
- Koskiniemi M., Toivakka,E. and Donner,M. (1974b) Progressive myoclonus epilepsy: electroencephalographical findings. Acta Neurol. Scand., 30, 333–359. [PubMed] [Google Scholar]
- Lafreniere R.G. et al. (1997) Unstable insertion in the 5′-flanking region of the cystatin B gene is the most common mutation in progressive myoclonus epilepsy type 1, EPM1. Nat. Genet., 15, 298–302. [DOI] [PubMed] [Google Scholar]
- Lalioti M.D., Scott,H.S., Buresi,C., Rossier,C., Bottani,A., Morris,M.A., Malafosse,A. and Antonarakis,S.E. (1997a) Dodecamer repeat expansion in cystatin B gene in progressive myoclonus epilepsy. Nature, 386, 847–851. [DOI] [PubMed] [Google Scholar]
- Lalioti M.D. et al. (1997b) Identification of mutations in cystatin B, the gene responsible for the Unverricht–Lundborg type of progressive myoclonus epilepsy (EPM1). Am. J. Hum. Genet., 60, 342–351. [PMC free article] [PubMed] [Google Scholar]
- Lalioti M.D. et al. (1998) A PCR amplification method reveals instability of the dodecamer repeat in progressive myoclonus epilepsy (EPM1) and no correlation between the size of the repeat and age at onset. Am. J. Hum. Genet., 62, 842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalioti M.D., Scott,H.S. and Antonarakis,S.E. (1999) Altered spacing of promoter elements due to the dodecamer repeat expansion contributes to reduced expression of the cystatin B gene in EPM1. Hum. Mol. Genet., 8, 1791–1798. [DOI] [PubMed] [Google Scholar]
- Larson G.P., Ding,S., Lafrenière,R.G., Rouleau,G.A. and Krontiris,T.G. (1999) Instability of the EPM1 minisatellite. Hum. Mol. Genet., 8, 1985–1988. [DOI] [PubMed] [Google Scholar]
- Lehesjoki A.-E., Koskiniemi,M., Sistonen,P., Miao,J., Hästbacka,J., Norio,R. and de la Chapelle,A. (1991) Localization of a gene for progressive myoclonus epilepsy to chromosome 21q22. Proc. Natl Acad. Sci. USA, 883, 3696–3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehesjoki A.-E., Koskiniemi,M., Norio,R., Tirrito,S., Sistonen,P., Lander,E. and de la Chapelle,A. (1993) Localization of the EPM1 gene for progressive myoclonus epilepsy on chromosome 21: linkage disequilibrium allows high resolution mapping. Hum. Mol. Genet., 2, 1229–1234. [DOI] [PubMed] [Google Scholar]
- Lenarcic B., Krizaj,I., Zunec,P. and Turk,V. (1996) Differences in specificity for the interactions of stefins A, B and D with cysteine proteinases. FEBS Lett., 395, 113–118. [DOI] [PubMed] [Google Scholar]
- Lieuallen K., Pennacchio,L.A., Park,M., Myers,R.M. and Lennon,G.G. (2001) Cystatin B-deficient mice have increased expression of apoptosis and glial activation genes. Hum. Mol. Genet., 10, 1867–1871. [DOI] [PubMed] [Google Scholar]
- Machleidt W., Thiele,U., Assfalg-Machleidt,I., Forger,D. and Auerswald,E.A. (1991) Molecular mechanism of inhibition of cysteine proteinases by their protein inhibitors: kinetic studies with natural and recombinant variants of cystatins and stefins. Biomed. Biochim. Acta, 50, 613–620. [PubMed] [Google Scholar]
- Marseille Consensus Group (1990) Classification of progressive myoclonus epilepsies and related disorders. Ann. Neurol., 28, 113–116. [DOI] [PubMed] [Google Scholar]
- Minassian B.A. et al. (1998) Mutations in a gene encoding a novel protein tyrosine phosphatase cause progressive myoclonus epilepsy. Nat. Genet., 20, 171–174. [DOI] [PubMed] [Google Scholar]
- Nagafuchi S. et al. (1994) Dentatorubral and pallidoluysian atrophy expansion of an unstable CAG trinucleotide on chromosome 12p. Nat. Genet., 6, 14–18. [DOI] [PubMed] [Google Scholar]
- Norio R. and Koskiniemi,M. (1979) Progressive myoclonus epilepsy; genetic and nosological aspects with special reference to 107 Finnish patients. Clin. Genet., 15, 382–398. [DOI] [PubMed] [Google Scholar]
- Parsons M.A., Sinden,R.R. and Izban,M.G. (1998) Transcriptional properties of RNA polymerase II within triplet repeat-containing DNA from the human myotonic dystrophy and fragile X loci. J. Biol. Chem., 273, 26998–27008. [DOI] [PubMed] [Google Scholar]
- Pataskar S.S., Dash,D. and Brahmachari,S.K. (2001) Progressive myoclonus epilepsy [EPM1] repeat d(CCCCGCCCCGCG)n forms folded hairpin structures at physiological pH. J. Biomol. Struct. Dyn., 19, 293–305. [DOI] [PubMed] [Google Scholar]
- Pennacchio L.A. et al. (1996) Mutations in the gene encoding cystatin B in progressive myoclonus epilepsy (EPM1). Science, 271, 1731–1734. [DOI] [PubMed] [Google Scholar]
- Pennacchio L.A., Bouley,D.M., Higgins,K.M., Scott,M.P., Noebels,J.L. and Myers,R.M. (1998) Progressive ataxia, myoclonic epilepsy and cerebellar apoptosis in cystatin B-deficient mice. Nat. Genet., 20, 251–258. [DOI] [PubMed] [Google Scholar]
- Pshezhetsky A.V. et al. (1997) Cloning, expression and chromosomal mapping of human lysosomal sialidase and characterization of mutations in sialidosis. Nat. Genet., 15, 316–320. [DOI] [PubMed] [Google Scholar]
- Ranta S. et al. (1999) The neuronal ceroid lipofuscinosis in human EPMR and mnd mutant mice are associated with mutations in CLN8. Nat. Genet., 23, 233–236. [DOI] [PubMed] [Google Scholar]
- Rawlings N.D. and Barret,A.J. (1990) Evolution of proteins of the cystatin superfamily. J. Mol. Evol., 30, 60–71. [DOI] [PubMed] [Google Scholar]
- Rawlings N.D. and Barrett,A.J. (1999) Tripeptidyl-peptidase I is apparently the CLN2 protein absent in classical late-infantile neuronal ceroid lipofuscinosis. Biochim. Biophys Acta, 1429, 496–500. [DOI] [PubMed] [Google Scholar]
- Riccio M., Di Giaimo,R., Pianetti,S, Palmieri,P.P., Melli,M. and Santi,S. (2001) Nuclear localization of cystatin B, the cathepsin inhibitor implicated in myoclonus epilepsy (EPM1). Exp. Cell Res., 262, 84–94. [DOI] [PubMed] [Google Scholar]
- Rinne A., Järvinen,M., Martikainen,J., Alavaikko,M. and Räsänen,O. (1981) Über das Vorkommen des epidermalen SH-Protease-Inhibitors im lymphatischen Gewebe. Verb. Anat. Ges., 75, 573–574. [Google Scholar]
- Rinne R., Saukko,P., Järvinen,M. and Lehesjoki,A.-E. (2002) Reduced cystatin B activity correlates with enhanced cathepsin activity in progressive myoclonus epilepsy. Ann. Med., 34, 380–385. [DOI] [PubMed] [Google Scholar]
- Ritonja A., Machleidt,W. and Barrett,A.J. (1985) Amino acid sequence of the intracellular cysteine proteinase inhibitor cystatin B from human liver. Biochem. Biophys. Res. Commun., 131, 1187–1192. [DOI] [PubMed] [Google Scholar]
- Saha T. and Ushdin,K. (2001) Tetraplex formation by the progressive myoclonus epilepsy type-1 repeat: implications for instability in the repeat expansion diseases. FEBS Lett. 491, 184–187. [DOI] [PubMed] [Google Scholar]
- Savukoski M., Klockars,T., Holmberg,V., Santavuori,P., Lander,E.S. and Peltonen,L. (1998) CLN5, a novel gene encoding a putative transmembrane protein mutated in Finnish variant late infantile neuronal ceroid lipofuscinosis. Nat. Genet., 19, 286–288. [DOI] [PubMed] [Google Scholar]
- Serratosa J.M. et al. (1999) A novel protein tyrosine phosphatase gene is mutated in progressive myoclonus epilepsy of the Lafora type (EPM2). Hum. Mol. Genet., 8, 345–352. [DOI] [PubMed] [Google Scholar]
- Shannon P., Pennacchio,L.A., Houseweart,M.K., Minassian,B.A. and Myers,R.M. (2002) Neuropathological changes in a mouse model of progressive myoclonus epilepsy: cystatin B deficiency and Unverricht–Lundborg disease. J. Neuropathol. Exp. Neurol., 61, 1085–1091. [DOI] [PubMed] [Google Scholar]
- Shoffner J.M., Lott,M.T., Lezza,A.M., Seibel,P., Ballinger,S.W. and Wallace,D.C. (1990) Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell, 61, 931–937. [DOI] [PubMed] [Google Scholar]
- Sleat D.E., Donnelly,R.J., Lackland,H., Liu,C.G., Sohar,I., Pullarkat,R.K. and Lobel,P. (1997) Association of mutations in a lysosomal protein with classical late-infantile neuronal ceroid lipofuscinosis. Science, 277, 1802–1805. [DOI] [PubMed] [Google Scholar]
- Stone N.E. et al. (1996) Construction of a 750 kb bacterial clone contig and restriction map in the region of human chromosome 21 containing the progressive myoclonus epilepsy (EPM1) gene. Genome Res., 6, 218–225. [DOI] [PubMed] [Google Scholar]
- Stubbs M.T., Laber,B., Bode,W., Huber,R., Jerala,R., Lenarcic,B. and Turk,V. (1990) The refined 2.4 Å X-ray crystal structure of recombinant human stefin B in complex with the cysteine proteinase papain: a novel type of proteinase inhibitor interaction. EMBO J., 9, 1939–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk V. and Bode,W. (1991) The cystatins: protein inhibitors of cysteine proteinases. FEBS Lett., 285, 213–219. [DOI] [PubMed] [Google Scholar]
- Vesa J., Hellsten,E., Verkruyse,L.A., Camp,L.A., Rapola,J., Santavuori,P., Hoffmann,S.L. and Peltonen,L. (1995) Mutations in the palmitoyl protein thioesterase gene causing infantile neuronal ceroid lipofuscinosis. Nature, 376, 584–587. [DOI] [PubMed] [Google Scholar]
- Vines D.J. and Warburton,M.J. (1999) Classical late infantile neuronal ceroid lipofuscinosis fibroblasts are deficient in lysosomal tripeptidyl peptidase I. FEBS Lett., 443, 131–135. [DOI] [PubMed] [Google Scholar]
- Virtaneva K. et al. (1996) Progressive myoclonus epilepsy EPM1 locus maps to a 175 kb interval in distal 21q. Am. J. Hum. Genet., 58, 1247–1253. [PMC free article] [PubMed] [Google Scholar]
- Virtaneva K. et al. (1997) Unstable minisatellite expansion causing recessively inherited myoclonus epilepsy, EPM1. Nat. Genet., 15, 393–396. [DOI] [PubMed] [Google Scholar]
- Wheeler R.B., Sharp,J.D., Schultz,R.A., Joslin,J.M., Williams,R.E. and Mole,S.E. (2002) The gene mutated in variant late-infantile neuronal ceroid lipofuscinosis (CLN6) and in nclf mutant mice encodes a novel predicted transmembrane protein Am. J. Hum. Genet., 70, 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda M., Tanno,Y., Horai,S., Ozawa,T., Miyatake,T. and Tsuji,S. (1990) A common mitochondrial DNA mutation in the tRNA-lys of patients with myoclonus epilepsy associated with ragged-red fibers. Biochem. Int., 21, 789–796. [PubMed] [Google Scholar]