Abstract
SNAREs on transport vesicles and target membranes are required for vesicle targeting and fusion. Here we describe a novel yeast protein with a typical SNARE motif but with low overall amino acid homologies to other SNAREs. The protein localized to the endoplasmic reticulum (ER) and was therefore named Use1p (unconventional SNARE in the ER). A temperature-sensitive use1 mutant was generated. use1 mutant cells accumulated the ER forms of carboxypeptidase Y and invertase. More specific assays revealed that use1 mutant cells were defective in retrograde traffic to the ER. This was supported by strong genetic interactions between USE1 and the genes encoding SNAREs in retrograde traffic to the ER. Antibodies directed against Use1p co-immunoprecipitated the SNAREs Ufe1p, myc-Sec20p and Sec22p, which form a SNARE complex required for retrograde traffic from the Golgi to the ER, but neither Bos1p nor Bet1p (members of the SNARE complex in anterograde traffic to the Golgi). Therefore, we conclude that Use1p is a novel SNARE protein that functions in retrograde traffic from the Golgi to the ER.
Keywords: ER/membrane traffic/Saccharomyces cerevisiae/SNARE proteins/Use1p
Introduction
Transport between different organelles and maintenance of organelle identity require fusion between membranes. SNARE proteins on both membranes are an important part of the protein machinery required for recognition and fusion between membranes (Jahn and Südhof, 1999). Different sets of SNAREs are required for each kind of membrane fusion event. The constituents of these protein complexes are conserved from yeast to man. At least one SNARE present on each of the two membranes must be anchored by a transmembrane domain in order to transmit the force generated by formation of the SNARE complex (Grote et al., 2000). Accordingly, most SNAREs possess a C-terminal transmembrane domain. Next to it is the conserved SNARE motif of 58 amino acid residues. Within the SNARE complexes, the SNARE motifs form an extended parallel four-helix bundle. The crystal structures of two SNARE complexes have been determined (Sutton et al., 1998; Antonin et al., 2002). The structures are very similar even though the sequence homologies between the proteins involved are quite low. Amino acid side chains from the four proteins pointing into the middle of the bundle interact in 16 different layers. Most layers consist of hydrophobic amino acids, which are well conserved. However, the most conserved layer, the so-called 0 layer in the center of the bundle, consists of one arginine and three glutamine residues. SNAREs with an arginine in the 0 layer are called R-SNAREs. Q-SNAREs contain a glutamine residue in this position (Fasshauer et al., 1998). Q-SNAREs can be divided into three groups according to their sequence similarities: Qa (related to syntaxin), Qb (similar to SNAP-25 N-terminal helix) and Qc (SNAP-25 C-terminal helix; Bock et al., 2001). SNARE complexes consist of one helix from each of the groups.
So far, 23 different SNAREs have been identified in the yeast Saccharomyces cerevisiae, among them five R-SNAREs, seven Qa-SNAREs, six Qb-SNARE helices and seven Qc-SNARE helices (Pelham, 2001). Since the whole genome is known, it is expected that membrane trafficking in yeast requires not more than these 23 SNAREs.
Additional SNARE-encoding genes may have been overlooked since some Qb- and Qc-SNAREs show little sequence similarity. In database searches, we found an uncharacterized open reading frame (ORF) whose product shares characteristics with SNAREs. Here we show that this protein is indeed a SNARE. It was localized to the endoplasmic reticulum (ER), was part of a SNARE complex and required for retrograde traffic from the Golgi to the ER.
Results
Identification of a SNARE-related sequence
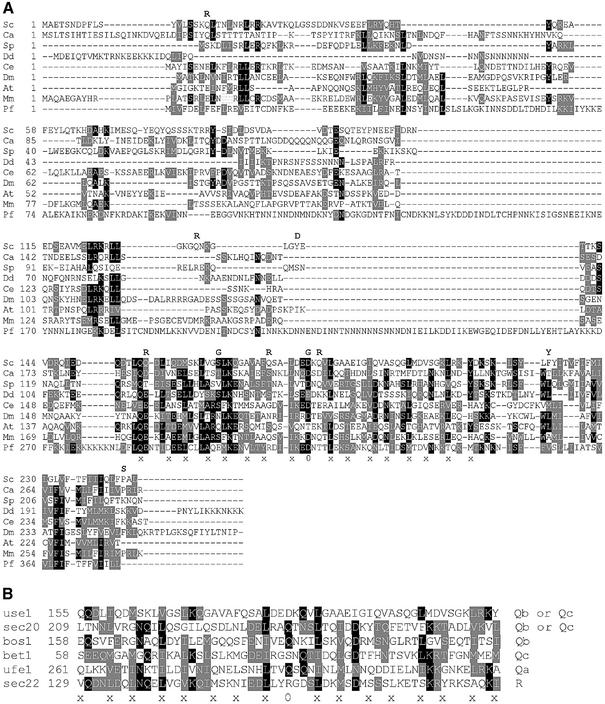
We performed database searches to identify unknown members of the SNARE protein family in yeast. Among different SNAREs, the highest degree of conservation is found in the amino acid residues forming layers of interacting side chains in SNARE complexes within SNARE motifs (x in Figure 1B). Consecutive PSI-Blasts were performed against the S.cerevisiae genome database using multiple yeast SNAREs as starting sequences. This approach seemed to be valid since it yielded Sec20p, the most divergent SNARE. Candidate sequences were evaluated for their potential to be SNAREs. The uncharacterized ORF YGL098w emerged as the only strong candidate. YGL098w encodes an essential protein (Giaever et al., 2002) predicted to have 245 amino acid residues (Figure 1A). This protein has a single C-terminal transmembrane domain, like most SNAREs. A putative SNARE motif is directly in front of this transmembrane domain, as expected. An aspartate residue is found at the position for the 0 layer instead of a glutamine found in most Q-SNAREs. Bulky hydrophobic amino acids form the –1 and +1 layers of SNAREs. Ygl098wp has leucine residues in both positions. An alanine residue forms the –3 layer of Ygl098wp, which is a typical small amino acid side chain found in this position in Qb- and Qc-SNAREs. Searches revealed that genes related to YGL098w may exist in Candida albicans, Schizosaccharomyces pombe, Dictyostelium discoideum, Caenorhabditis elegans, Drosophila, Arabidopsis, mouse and Plasmodium falciparum (Figure 1A). As expected for a SNARE, the highest degree of amino acid conservation was found in the SNARE motif, especially in positions predicted to form the layers in a SNARE complex. As shown in Figure 1B, many of these layer residues are conserved between ER and Golgi SNAREs belonging to different groups.
Fig. 1. Amino acid sequence of Ygl098wp (Use1p) has features of SNARE proteins. (A) Related proteins were found in Saccharomyces cerevisiae (S.c.), Candida albicans (C.a. gnl SDSTC 5476), Schizosaccharomyces pombe (S.p. CAB16218), Dictyostelium discoideum (D.d. IIAFP1D36343), C.elegans (C.e. AAF60428), Drosophila (D.m. AC013969, nucleotides 4505–4792, 4887–5339), Arabidopsis (A.t. AAD25784), mouse (M.m. AK008573) and Plasmodium falciparum (P.f. gi 23612490). Use1 proteins had a C-terminal transmembrane domain. Next was a sequence related to SNARE motifs with amino acid residues that could form layers in a SNARE complex (–8 to +8) and an aspartic acid residue in the putative 0 layer. The ClustalW program was used. Black boxes indicate identical residues, gray boxes conserved exchanges. Amino acid exchanges found in use1-10AA are shown. (B) Comparison of SNARE motifs of Use1p with ER and Golgi SNAREs. 0, 0-layer; x, other layers.
Use1p localizes to the ER
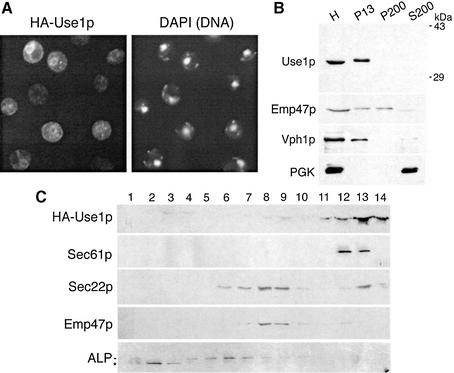
To prove that Ygl098wp protein functions as a SNARE, we first determined the localization of this protein. A segment encoding an HA epitope was introduced after the start codon of YGL098w and cells expressing this protein were inspected by immunofluorescence microscopy. The tagged protein was fully functional (data not shown). Overexpressed tagged protein was localized to ring-like structures around the nucleus identified by the DNA dye 4′,6-diamidino-2-phenylindole (DAPI) (Figure 2A). These structures are typical for yeast ER. Therefore, YGL098w was termed USE1 (unconventional SNARE in the ER). To exclude potential mislocalization due to overexpression, an antiserum against Use1p was raised. This antiserum recognized a single band at ∼35 kDa (Figure 2B). Wild-type yeast cells were spheroplasted, osmotically lysed and subjected to differential centrifugation. The immunoblot analysis showed that the Golgi protein Emp47p (Schröder et al., 1995) fractionated to both the 13 000 and 200 000 g pellets (P13 and P200). Endogenous Use1p was found exclusively in P13, which contained vacuolar (identified by Vph1p) and ER membranes (Figure 2B). Organelles were separated by sucrose density gradient centrifugation. Endogenous Use1p (data not shown) as well as HA-Use1p expressed at about wild-type levels migrated to fractions 12 and 13 close to the bottom of the gradient together with the ER protein Sec61p (Figure 2C). Low amounts of HA-Use1p were seen in fractions 8 and 9. The Golgi protein Emp47p was concentrated in fractions 8 and 9. Sec22p was present in Golgi and ER fractions. By contrast, the vacuolar protein alkaline phosphatase (ALP) was most abundant in fractions 4–7. Taken together, these data demonstrate that Use1p is localized predominantly to the ER with low amounts in the Golgi.
Fig. 2. Use1p is localized to the ER. (A) use1Δ cells expressing HA-Use1p from a 2µ vector were processed for immunofluorescence. HA-Use1p was found in ring-like structures around the nucleus (identified by DNA staining using DAPI) which are typical for yeast ER. (B) Wild-type yeast homogenate (H) was fractionated into a 13 000 g pellet (P13), a 200 000 g pellet (P200) and a 200 000 g supernatant (S200) by differential centrifugation. Fractions were analyzed by immunoblotting using antisera against Use1p, Emp47p, Vph1p and PGK. Endogenous Use1p was found in P13, which contains ER and vacuoles. Vph1p is the 100 kDa subunit of the vacuolar ATPase. Emp47p is localized to the Golgi, PGK is a soluble protein. (C) use1Δ cells expressing HA-Use1p from a centromeric vector were used. Cleared yeast homogenate was fractionated on a 19–42% sucrose gradient and analyzed by immunoblotting. HA-Use1p co-fractionated with the ER protein Sec61p. Sec22p was present in ER as well as Golgi fractions. The vacuolar protein ALP was found in fractions 4–7. A soluble degradation product of ALP (*) was present in the load fractions 1–3.
Generation of use1 mutant cells
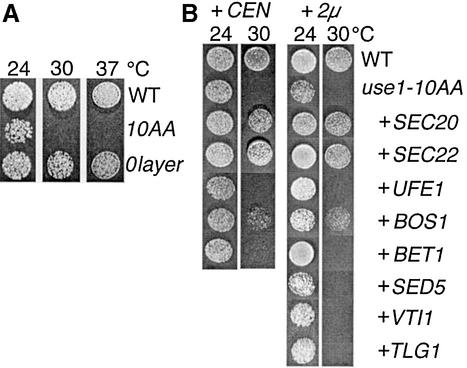
We confirmed that USE1 is an essential gene. To analyze the function of Use1p, a temperature-sensitive mutation was obtained after random mutagenesis of USE1, plasmid shuffling and screening for growth defects. The protein encoded by this mutant allele carried 10 amino acid replacements, as indicated in Figure 1A, and was called use1-10AA. Five amino acid replacements were found in the SNARE motif, including the mutation D183G in the 0 layer. To determine the importance of the 0 layer, the allele use1-0layer with the mutation D183G was created. use1-10AA cells grew more slowly at 24°C than wild-type cells and did not grow at 30 or 37°C (Figure 3A). use1-0layer cells did not display a growth defect.
Fig. 3. Overexpression of ER and Golgi SNAREs partially suppresses the growth defect of use1-10AA cells. (A) Dilutions of wild-type, use1-0layer and use1-10AA cells were grown at 24, 30 or 37°C on plates with rich medium. use1-10AA cells grew slower at 24°C and did not grow at 30°C; use1-0layer cells did not display a growth defect. (B) Dilutions of wild-type (WT), use1-10AA and use1-10AA cells overexpressing different SNAREs from CEN or 2µ plasmids were incubated at 24 or 30°C. Overexpression of SEC20, a SNARE in retrograde traffic from the Golgi to the ER, allowed for growth at 30°C. The growth defect was also suppressed by SEC22, which is required for both anterograde and retrograde traffic, and by the Qb SNARE BOS1. Prolonged incubation at 30°C revealed that overexpression of BET1 improved growth slightly; UFE1 and SED5 had small effects.
Genetic interactions between use1-10AA and SNAREs of the ER and Golgi
Genetic interactions provide a method to identify components of a common pathway. Genetic interactions with SNARE-encoding genes required for traffic between the ER and the Golgi were used to analyze which SNAREs have the closest functional connections with Use1p. A SNARE complex consisting of Sec22p (R-SNARE), Sed5p (Qa), Bos1p (Qb) and Bet1p (Qc) is required for anterograde traffic from the ER to the Golgi (Newman et al., 1990; Hardwick and Pelham, 1992; Søgaard et al., 1994). Sec22p is also involved in retrograde traffic from the Golgi to the ER (Spang and Schekman, 1998). Sec22p forms a complex with Ufe1p (Qa) and Sec20p (Qb). Functional data indicate that Bet1p is required for retrograde traffic to the ER (Spang and Schekman, 1998), but it was not found in a complex with Ufe1p (Lewis et al., 1997).
We tested whether overexpression of different SNAREs suppressed growth defects. use1-10AA cells were transformed with plasmids carrying SNARE-encoding genes. CEN plasmids (low copy) or 2µ plasmids (high copy) were used to achieve different levels of overexpression. Growth of the resulting strains was tested at 24 and 30°C (Figure 3B). Overexpression of SEC20 or SEC22 from either a CEN or a 2µ plasmid suppressed the growth defect at 30°C. Overexpression of the anterograde Qb-SNARE Bos1p allowed for slow growth at 30°C. Overexpression of these SNAREs did not influence growth of wild-type cells (data not shown). The growth defect of use1-10AA cells was not suppressed by 2µ plasmids encoding the SNAREs Tlg2p (Qa in TGN fusion), Tlg1p (Qc in TGN fusion) or Vti1p (Qb in Golgi, endosomal and vacuolar fusion).
A functional relationship of gene products is likely if the combination of mutations aggravates the growth defects of single defects. To observe such synthetic effects, heterozygous diploids were obtained by mating use1-10AA cells and cells carrying mutations in SNAREs. Tetrads were dissected and the resulting spores analyzed. Spores carrying both a use1-10AA and a sec20-1, sec22-3 or ufe1-1 double mutation were not found, indicating that these mutations were synthetically lethal (Table I). Combinations of use1-10AA with sec32-1/bos1-1 or sed5-1 mutations were viable but resulted in a reduction of the restrictive temperatures to 27°C. use1-10AA bet1-1 cells did not display a synthetic growth defect.
Table I. Phenotypes of double mutants of use1-10AA and an ER or Golgi SNARE.
| Double mutant with | Phenotype |
|---|---|
| sec20-1 | Inviable |
| sec22-3 | Inviable |
| ufe1-1 | Inviable |
| sec32-1(bos1-1) | RT reduced to 27°C |
| bet1-1 | RT unchanged at 30°C |
| sed5-1 | RT reduced to 27°C |
use1-10AA cells were mated with strains carrying mutations in ER or Golgi SNAREs, diploids were sporulated, tetrads dissected and spores analyzed. Mutations in SNAREs required for retrograde traffic from the Golgi to the ER were synthetic lethal with use1-10AA.
RT, restrictive temperature.
These data indicate that USE1 exhibits stronger genetic interactions with retrograde than with anterograde SNAREs.
use1 mutant cells are defective in traffic between the ER and the Golgi
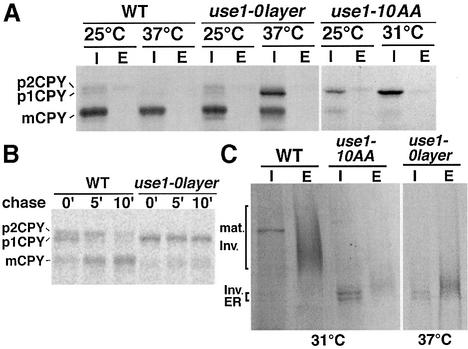
Next we analyzed traffic out of the ER in use1 mutant cells. The processing of the vacuolar protein carboxypeptidase Y (CPY) can serve as an indicator of ER to Golgi traffic. The ER form of CPY, p1CPY, is further glycosylated to p2CPY in the Golgi. Proteolytic cleavage in the vacuole yields the mature mCPY form. CPY maturation was studied by pulse–chase labeling after a 15 min pre-incubation at the indicated temperature, followed by immunoprecipitation of CPY. Little CPY was transported to the vacuole in use1-10AA cells at 25°C, as indicated by reduced amounts of mCPY (Figure 4A). Instead, p1CPY accumulated in the ER. This defect was more pronounced at 31°C. CPY transport was not affected in use1-0layer cells at 25°C. An accumulation of p1CPY was observed at 37°C, demonstrating the importance of the putative 0 layer for function. A total of 25–50% of CPY reached the vacuole, pointing towards a partial defect. Since no p2CPY was observed, transport from the Golgi to the vacuole appeared to be normal. This indicates that Use1p was not required for later steps in Golgi or vacuolar transport. To determine the onset of the transport defect, wild-type and use1-0layer cells were pulsed at 37°C for 10 min without pre-incubation at 37°C and chased for different time periods (Figure 4B). Significant amounts of p2CPY and mCPY were present in wild-type cells at 0′ chase but absent in use1-0layer cells, indicating that transport was already defective during the pulse. Secretion of invertase served as a second marker for anterograde transport. use1-10AA cells accumulated the ER form of invertase (Figure 4C, I) and secreted only small amounts of partially glycosylated invertase (E) at 31°C. This was confirmed by quantifying invertase activity (not shown). The ER form of invertase was also observed in use1-0layer cells at non-permissive temperature. However, most of the newly synthesized invertase was secreted as partially glycoslyated form. Defects in genes required for cargo sorting at the ER (EMP24), anterograde traffic to the Golgi (SEC22, YPT1), retrograde traffic to the Golgi (VTI1) or retrograde traffic to the ER (COPI subunits, DSL1, SEC22) result in secretion of partially glycosylated invertase. After derepression and shift to 37°C for 30 min, 35% of the total invertase activity was found intracellularly in use1-0layer cells (wild type 15%, three experiments). After 60 min, the difference was not significant (use1-0layer cells 19%, wild type 14%), suggesting that invertase secretion was slower in use1-0layer cells. These data indicate that Use1p is required for traffic between the ER and the Golgi.
Fig. 4. use1 mutant cells are defective in traffic between the ER and the Golgi. (A) Wild-type, use1-0layer and use1-10AA cells were grown at 24°C. CPY was immunoprecipitated from cellular extracts (I) and the medium (E) after a 15 min pre-incubation and pulse–chase labeling at the indicated temperatures. CPY accumulated in the ER as p1CPY in use1-10AA cells at 25 and at 31°C. use1-0layer cells transported CPY to the vacuole at 25°C but accumulated p1CPY in the ER at 37°C. p1CPY, ER proCPY (carboxypeptidase Y); p2CPY, Golgi proCPY; mCPY, vacuolar mature CPY. (B) The CPY sorting defect had a fast onset in use1-0layer cells. Wild-type and use1-0layer cells were pulsed for 10 min at 37°C without pre-incubation and chased for the indicated time periods. CPY was immunoprecipitated from cellular extracts. (C) Invertase accumulated in the ER in use1-10AA; a partially glycosylated form was secreted in use1-0layer cells. Cells were grown at 24°C. Invertase was immunoprecipitated from cellular extracts (I) and periplasm (E) after a 15 min pre-incubation and pulse–chase labeling at the indicated temperatures. Mat. Inv., mature invertase.
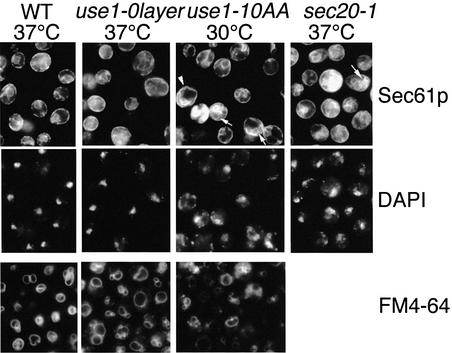
The morphology of the ER at non-permissive temperature was studied by immunofluorescence using antiserum against the ER membrane protein Sec61p (Figure 5). Sec61p localizes to the cortical ER below the plasma membrane and the nuclear envelope in wild-type cells (Stirling et al., 1992). Invaginations of the cortical ER (arrowhead) and additional Sec61p-containing membranes (arrows) were observed in use1-10AA cells, indicating that ER membranes accumulated. This was paralleled by a fragmentation of the nucleus visualized by DAPI staining. A similar ER accumulation and nuclear fragmentation were observed in many sec20-1 cells at 37°C. use1-10AA cells and some use1-0layer cells at non-permissive temperature were bigger than wild-type cells. Sec61p staining was almost normal in use1-0layer cells. Endocytosis and vacuolar morphology were investigated using FM4-64 uptake (Figure 5, bottom row). FM4-64 stained vacuolar structures in use1 mutant cells. Vacuoles were slightly more fragmented in some use1-10AA cells.
Fig. 5. use1-10AA cells accumulate ER membranes. Wild-type, use1-0layer, use1-10AA and sec20-1 cells were grown at 24°C, shifted to 30 or 37°C for 1.75 h and ER membranes detected by immunofluorescence using antisera against Sec61p. DNA was stained with DAPI. use1-10AA cells were larger, accumulated ER (arrows and arrowhead) and nuclei appeared fragmented in some cells. Cells grown at 24°C were shifted to 30 or 37°C for 15 min and incubated with FM4-64 for an additional 15 min at the same temperature to stain vacuolar membranes. Vacuoles had wild-type morphology in most use1 mutant cells, but were slightly more fragmented in some of them.
use1 mutant cells are defective in retrograde traffic to the ER
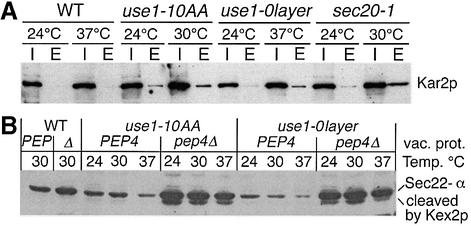
Anterograde and retrograde traffic between the ER and the Golgi is closely coupled. A block in retrograde transport has an indirect but rapid effect on anterograde transport (Lewis and Pelham, 1996). Several assays have been developed to distinguish between these transport steps. Kar2p/BiP is a soluble ER protein, which is retrieved from the Golgi (Semenza et al., 1990). Kar2p is secreted into the medium if retrograde traffic to the ER is blocked. However, some mutants display this phenotype but are not solely involved in retrograde traffic. We determined the amount of Kar2p secreted in a time period of 2 h at permissive and restrictive temperatures. use1-10AA cells secreted some Kar2p at 24°C and slightly larger amounts at 30°C (Figure 6A). A shift to 37°C increased Kar2p secretion from use1-0layer cells to levels clearly above that from wild-type cells. use1-0layer cells secreted less Kar2p at non-permissive temperature than use1-10AA cells. sec20-1 cells were used for comparison since this defect primarily affects the retrograde traffic to the ER. Wild-type cells secreted 0.37% (SD 0.2), sec20-1 cells 3.7% (SD 0.9) and use1-10AA cells 4.9% (SD 3.4) of the intracellular Kar2p within 3 h at 30°C in four independent experiments; use1-0layer cells secreted 0.8% in 3 h at 37°C.
Fig. 6. Kar2p/BiP secretion as well as Sec22-α cleavage and vacuolar degradation indicate that retrograde Golgi to ER traffic is defective in use1 mutant cells. (A) Wild-type, use1-0layer, use1-10AA and sec20-1 cells were grown at 24°C, placed in fresh medium and incubated at the indicated temperatures for 2 h. Proteins precipitated from the medium (E) and cellular extracts (I) were analyzed by immunoblotting with antiserum against Kar2p. use1-10AA and use1-0layer cells secreted more Kar2p than wild-type cells. (B) A fusion protein containing Sec22p, a luminal myc tag, Kex2p-cleavage site and α-factor (Sec22-α) was used to monitor defects in ER retrieval. Cells expressing Sec22-α were grown at 24°C and shifted to the indicated temperatures for 30 min. Strains marked Δ or pep4Δ did not contain active vacuolar proteases due to a pep4Δ mutation. Homogenates were analyzed by SDS–PAGE and immunoblotting with antiserum against the myc tag. Reduced levels of Sec22-α were seen in use1 mutant cells, especially at 37°C. Sec22-α was degraded in the vacuole as the intact Sec22-α and the Kex2p cleavage product were stabilized in use1 pep4Δ mutant cells.
In addition, we used a fusion protein that monitors the failure to recycle Sec22p to the ER (Ballensiefen et al., 1998). A luminal myc epitope, a Kex2p-cleavage site and α-factor were added to the C-terminus of this SNARE (Sec22-α). The α-factor moiety is cleaved off by the Kex2p protease, which is localized to the late Golgi. This can be monitored by the reduction in size in immunoblots probed for the myc tag. Reduced amounts of Sec22-α were detected in use1-10AA and use1-0layer cells (Figure 6B, PEP and PEP4 lanes). This decrease was stronger at 37°C. Cells defective in vacuolar proteases due to deletion of PEP4 were used to analyze whether Sec22-α was degraded in vacuoles of use1 mutant cells. Sec22-α was stabilized in use1-10AA and use1-0layer cells without active vacuolar proteases (Figure 6B, pep4Δ lanes). In addition, the Kex2p cleavage product of Sec22-α was observed, which lacked the α-factor moiety as it was not detected by antiserum against α-factor (data not shown). These data indicate that retrograde transport to the ER was defective in use1-10AA and use1-0layer cells, resulting in transport of Sec22-α to the vacuole and vacuolar degradation.
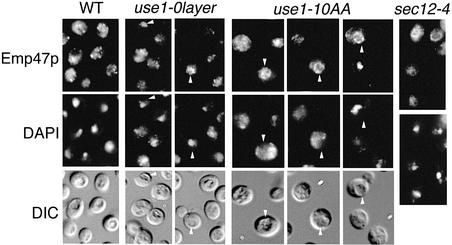
As a third assay, the localization of Emp47p was analyzed. Emp47p is localized to the Golgi at steady state and cycles continuously between the Golgi and the ER (Schröder et al., 1995). Emp47p accumulates in the ER if forward traffic from the ER to the Golgi is defective, as for example in sec12-4 cells. Emp47p remains in the Golgi or is transported slowly to the vacuole after a block in retrograde traffic from the Golgi to the ER. Localization of Emp47p was determined in wild-type, use1-0layer and use1-10AA cells lacking active vacuolar proteases and in sec12-4 cells by immunofluorescence microscopy (Figure 7). In wild-type cells, Emp47p was found in punctate structures typical for yeast Golgi. Emp47p localized to the Golgi in use1-10AA cells at 24°C (data not shown). After a shift to 30°C for 1.5 h, Emp47p remained in the Golgi or was seen in vacuoles (arrowheads) of some use1-10AA cells. The yeast vacuole is a ring-like structure distinct from the nucleus and is visible in differential interference contrast (DIC) microscopy. Emp47p redistributed to the vacuole in cells with a fragmented or intact nucleus, indicating that the redistribution was not due to general defects in abnormal cells. Emp47p did not accumulate in the ER in use1-10AA cells, as seen in mutants with a defect in anterograde transport. Only a few use1-0layer cells showed mislocalization of Emp47p to the vacuole (arrowheads). These data point towards a defect in retrograde traffic in use1-10AA cells at restrictive temperature. In summary, the data obtained with the use1-10AA mutant cells confirm the results of the two previous assays, which suggested that Use1p is involved in retrograde traffic from the Golgi to the ER.
Fig. 7. Golgi and vacuolar localization of Emp47p indicated that ER to Golgi traffic is not selectively affected in use1 mutant cells. Wild-type, use1- 0layer and use1-10AA cells with a pep4Δ mutation and sec12-4 cells were grown at 24°C, shifted to 37°C (WT, use1-0layer), 30°C (use1-10AA) or 35°C (sec12-4) for 1.5 h and Emp47p detected by immunofluorescence. Emp47p is predominantly localized to the Golgi (punctate structures) in wild-type cells. Emp47p accumulates in the ER upon block of ER to Golgi traffic in sec12-4 cells. Emp47p was found in the Golgi and in some use1-0layer and use1-10AA cells in vacuoles (arrowheads) but not in the ER at non-permissive temperature. DIC, differential interference contrast.
Use1p is part of the SNARE complex required for retrograde traffic to the ER
A defining feature of SNAREs is the formation of SNARE complexes. We tested whether Use1p was bound to different ER and Golgi SNAREs. Membrane extracts were prepared from sec18-1 cells expressing myc-Sec20p. Antiserum against Use1p co-immunoprecipitated Ufe1p, myc-Sec20p as well as Sec22p (Figure 8). About 3% of total myc-Sec20p and 4% of Sec22p were found in a complex with Use1p. Quantification was not possible for Ufe1p due to low signals in the starting material. However, when we repeated the experiment using cells producing myc-Ufe1p, the results indicated that the fraction of myc-Ufe1p co-immunoprecipitated with Use1p was not higher than that of myc-Sec20p or Sec22p (data not shown). By contrast, Bos1p and Bet1p did not co-immunoprecipitate with Use1p (Figure 8). These data show that Use1p is part of the SNARE complex consisting of Ufe1p, Sec20p and Sec22p, which is required for retrograde traffic to the ER. By contrast, Use1p does not participate in the SNARE complex in traffic to the Golgi, which is formed by Sed5p, Bos1p, Bet1p and Sec22p.
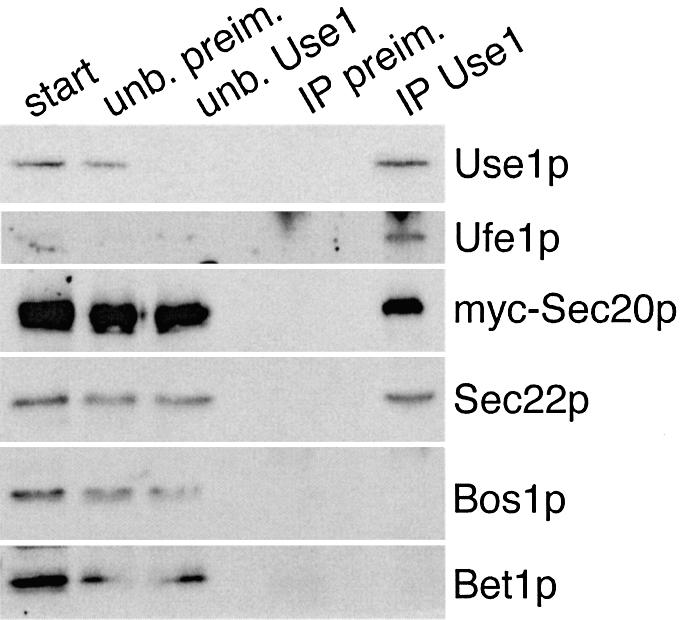
Fig. 8. Antibodies directed against Use1p co-immunoprecipitate SNAREs required for retrograde traffic from the Golgi to the ER. Detergent extracts from sec18-1 cells expressing myc-Sec20p grown at 24°C (start) were incubated with antiserum against Use1p or pre- immune serum coupled to protein A–Sepharose. Unbound fractions (unb. preim., unb. Use1) were separated from the immunoprecipitates (IP preim., IP Use1). Fractions were analyzed by immunoblotting using antisera against Use1p, Ufe1p, myc tag, Sec22p, Bos1p and Bet1p. Starting samples correspond to 50% of the precipitated material for Use1p and to 4.4% for the other SNAREs. Antiserum against Use1p co-immunoprecipitated Ufe1p, myc-Sec20p and Sec22p. No unspecific binding to pre-immune serum was observed.
In summary, multicopy suppression, synthetic lethality, defects in three assays for retrograde transport to the ER and SNARE complex formation showed that Use1p is a SNARE required for retrograde traffic to the ER.
Discussion
In this work we present evidence that the ER SNARE complex from yeast is made up of four different subunits. We add a new SNARE protein, Use1p, to the list of three subunits that have been characterized (Ufe1p, Sec22p and Sec20p). Our findings suggest that Use1p is specifically required for Golgi–ER retrograde transport.
Features of the amino acid sequence of Use1p
Use1p from S.cerevisiae and related sequences from other organisms contain a domain similar to the conserved SNARE motif. An unusual feature of Use1p is an aspartic acid residue in the putative 0 layer instead of a glutamine found in nearly all Q-SNAREs. 0 layers with aspartate instead of glutamine residues are present in yeast Sft1p (Qc) and mammalian vti1a proteins (Qb). By contrast, glutamine residues are found in vti1a proteins from Drosophila and C.elegans (Bock et al., 2001). This means that related proteins can function with either a glutamine or an aspartic acid residue in this position. An aspartate side chain should fit into the structure of a SNARE complex. The Use1p-related sequences in Arabidopsis and other plants (maize, wheat and rice) contain a threonine residue in the 0 layer. A similar divergence was noted previously: Bet1 proteins (Qc) from fungi have a serine or threonine residue in the 0 layer (Gupta and Heath, 2002), while a glutamine is found in fly and mammalian Bet1 homologs (Bock et al., 2001). Use1p is not a Qa-SNARE because it lacks a large amino acid residue in the –3 layer (Fasshauer et al., 1998). The alanine residue in the –3 layer of Use1p is typical for a Qb- or Qc-SNARE. However, the SNARE motif of Use1p is so divergent that it is not possible to assign it to either of these two groups. Its SNARE partner Sec20p has been assigned as a Qb-SNARE or Bos1p-related SNARE on functional grounds, even though it shows little sequence homology (Pelham, 2001). Interestingly, the amino acid sequence of Ufe1p as the Qa-SNARE of this complex is also very divergent. Therefore, all three Q-SNAREs of the SNARE complex in retrograde traffic to the ER are only distantly related to other Q-SNAREs.
SNARE complex in retrograde traffic to the ER
An antiserum against Use1p co-immunoprecipitated Ufe1p, myc-Sec20p and Sec22p. It has been shown that Sec22p, Ufe1p and Sec20p form a complex that is required for retrograde traffic from the Golgi to the ER (Lewis et al., 1997). We found ∼3% of myc-Sec20p and 4% of Sec22p in a complex with Use1p. The fraction of SNAREs found in this retrograde SNARE complex is in agreement with earlier studies. Antiserum against Ufe1p precipitates ∼5–10% of myc-Sec20p and Sec22p. An additional 5% of Sec22p is present in the anterograde SNARE complex isolated with antiserum against Sed5p (Lewis et al., 1997). Less than 1% of Sec22p co-precipitated with antisera against Sed5p or Bos1p in a different study (Parlati et al., 2002). Neither Bos1p nor Bet1p co-precipitated with Ufe1p (Lewis et al., 1997) or with Use1p as shown here. According to Spang and Schekman (1998), intact Bet1p is required for anterograde and retrograde traffic. Anterograde and retrograde transport were reconstituted with artifical cargo using membranes from wild-type and mutant cells. Bet1p may act as a chaperone for the retrograde SNAREs in the transport vesicles or may be required to package retrograde SNAREs or other components of the transport machinery. An additional SNARE-associated protein is Tip20p. Tip20p binds Sec20p and co-precipitates with the SNARE complex (Lewis et al., 1997). However, Tip20p does not contain a recognizable SNARE motif and may function in regulating the SNARE complex.
Phenotypes of use1 mutant cells
USE1 is an essential gene, like almost all genes required for transport between the ER and the Golgi. Two mutant alleles were generated. use1-10AA mutant proteins carry five amino acid exchanges in the SNARE motif, including replacements of aspartate in the 0 layer by glycine and of three glutamine by arginine residues. use1-10AA cells showed severe defects even at 24°C and did not grow at 30°C. use1-0layer cells contained only the 0 layer mutation D183G. use1-0layer cells had partial defects in several transport assays but no growth defect at 37°C. This indicates that the 0 layer was important, but that the mutant protein retained partial function. use1-10AA and use1- 0-layer cells had defects in transport of CPY from the ER to the Golgi. The onset of the CPY sorting defect was rapid in use1-0layer cells. However, rapid effects on secretion have also been observed for other mutants defective in retrograde traffic: for ufe1-1 and sec20-1, as well as for sec21-1 encoding defective γ-COPI (Lewis and Pelham, 1996), indicating that direct and indirect effects cannot be distinguished this way. COPI mutants have cargo-selective defects in anterograde traffic as CPY transport is blocked, while invertase is secreted normally even though it is partially glycosylated (Gaynor and Emr, 1997). This cargo selectivity may be due to different cargo receptors, which vary in the type of machinery involved in recycling or in recycling rates required. sec20-2 cells secrete HPS150 (Gaynor and Emr, 1997), while sec20-1 (Novick et al., 1980), ufe1-1 (Lewis and Pelham, 1996) as well as use1-10AA cells are defective in invertase secretion. As these SNAREs form a complex, differences may be due to allele-specific variations in residual traffic. use1-10AA cells accumulated ER membranes, which is a phenotype shared by cells defective in either anterograde or retrograde traffic between the ER and the Golgi. use1-10AA cells were larger than wild-type cells both at 24 and at 30°C. Wild-type cells strongly overexpressing SED5 increase in size, secrete Kar2p, and accumulate p1CPY and ER (Hardwick and Pelham, 1992). Therefore, the increase in size may be due to defects in transport between the ER and the Golgi. We used three different assays to distinguish between these transport steps: secretion of Kar2p, failure to recycle Sec22-α and mislocalization of Emp47p. Defects of use1-10AA cells pointed towards involvement in retrograde transport in all three assays.
Genetic interactions with USE1
The use1-10AA allele showed synthetic lethality with mutant alleles of all three SNAREs found in the complex required for retrograde traffic to the ER. All of these effects are due to amino acid exchanges in positions predicted to form layers of interacting amino acid side chains in the SNARE complex. sec22-3 carries the mutation R157G in the 0 layer (Sacher et al., 1997). The amino acid exchange L234S is found in the –1 layer of sec20-1 (Lewis et al., 1997). ufe1-1 has the mutations S282N in the –2 layer and L295P (Lewis et al., 1997). Synthetic lethality was described for a combination of ufe1-1 and sec20-1 (Frigerio, 1998), but not for sec20-1 and sec22-3 (Kaiser and Schekman, 1990). Mutations in SNAREs required for anterograde ER to Golgi traffic are not synthetically lethal with use1-10AA even though they carry mutations in layer residues. sec32-1/bos1-1 has the amino acid exchange L190S in the +1 layer, bet1-1 has L72F in the –4 layer and sed5-1 has R255G in the –8 layer (Banfield et al., 1995; Stone et al., 1997). Synthetic lethal interactions are found between members of the anterograde SNARE complex (Newman et al., 1990; Sacher et al., 1997). These data indicate that amino acid exchanges in the SNARE motifs of two SNAREs of either anterograde or retrograde SNARE complex often result in complete loss of function, suggesting that Use1p is a retrograde SNARE. Further confirmation was obtained from overexpression data. Overproduction of Sec20p or Sec22p suppressed growth defects in use1-10AA cells. In addition, overexpression of SEC20 or SEC22 partially suppressed the CPY sorting defect and normalized nuclear morphology in use1-10AA cells shifted to 30°C (data not shown). Larger amounts of SNARE partners may allow for more efficient usage of the residual activity in mutant Use1p. Overproduction of Ufe1p was without effect in use1-10AA cells. However, lack of multicopy suppression is not informative as suppression can be restricted to certain alleles or may be absent even if the encoded proteins interact physically. For example, overexpression of SED5 does not suppress defects in sec32-1/bos1-1 cells (Wuestehube et al., 1996). Overproduction of the anterograde Bos1p suppressed growth defects in use1-10AA cells somewhat. Overexpression of BOS1 does not have a general effect on retrograde SNAREs as ufe1-1 is not suppressed (Lewis et al., 1997). Deletion of one SNARE can result in the recruitment of a different SNARE of the same group into a SNARE complex, which does not form in wild-type cells. The endosomal Pep12p and the vacuolar Vam3p can mutually replace each other (Darsow et al., 1997; Götte and Gallwitz, 1997). Deletion of SEC22 is not lethal because it is replaced by the R-SNARE Ykt6p (Liu and Barlowe, 2002). Therefore, overproduced Bos1p may replace defective Use1p in the retrograde SNARE complex. This suggests that Use1p is a Qb-SNARE like Bos1p, while Sec20p may represent a Qc-SNARE like Bet1p. By analogy, Sec20p may function as a v-SNARE, since Bet1p was shown to act on one liposome with Bos1p, Sec22p and Sed5p present together on the other liposome (Parlati et al., 2002). In a more physiological assay, Sed5p was required on the Golgi, Bet1p together with Bos1p on the vesicles (Cao and Barlowe, 2000). Sec22p is also needed (Liu and Barlowe, 2002). Therefore, it is possible that Sec22p, Use1p and Sec20p act together on vesicles. Sec22p is present on these vesicles because it cycles between ER and Golgi (Ballensiefen et al., 1998).
We conclude that Sec22p, Ufe1p, Sec20p and Use1p form a SNARE complex which is required for retrograde traffic from the Golgi to the ER.
Materials and methods
Methods
Reagents were used from the following sources: enzymes for DNA manipulation from New England Biolabs (Beverly, MA), [35S]methionine and protein A–sepharose from Amersham Pharmacia (Braunschweig, Germany), fixed Staphylococcus aureus cells (Pansorbin) from Calbiochem (San Diego, CA), zymolyase from Seikagaku (Tokyo, Japan) and Ni-NTA–Sepharose from Qiagen (Hilden, Germany). All other reagents were purchased from Sigma (St Louis, MO). Antibodies against CPY, invertase, ALP, Vph1p and PGK were provided by T.H.Stevens (University of Oregon, Eugene, OR). Antiserum against Ufe1p was a gift from M.Lewis (MRC, Cambridge, UK). Antisera against Bet1p, Bos1p, Sec22p, Emp47p and BiP/Kar2p were provided by H.D.Schmitt. The antiserum against Sec61p has been described (Panzner et al., 1995). The c-myc tag was detected with monoclonal antibody 9E10. Cy2-conjugated secondary antibody was purchased from Dianova (Hamburg, Germany).
Plasmid manipulations were performed in the Escherichia coli strain XL1Blue. Yeast strains (Table II) were grown in rich media (YEPD) or standard minimal medium (SD) with appropriate supplements.
Table II. Yeast strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| SEY6210 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 mel- | Robinson et al. (1988) |
| SEY6211 | MATa leu2-3,112 ura3-52 his3-Δ200 ade2-101 trp1-Δ901 suc2-Δ9 mel- | Robinson et al. (1988) |
| BKY3 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 mel- use1Δ::TRP1 use1-plasmid | This study |
| BKY4 | MATa leu2-3,112 ura3-52 his3-Δ200 ade2-101 trp1-Δ901 suc2-Δ9 mel- use1Δ::TRP1 use1-plasmid | This study |
| BKY10 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 mel- pep4Δ::URA3 use1Δ ::TRP1 + use1-plasmid | This study |
| BKY12 | MATα leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 lys2-801 suc2-Δ9 mel- pep4Δ::URA3 | This study |
| HMSF226 | MATa mal mel gal2 sec12-4 | R.Schekman |
| BSH-7C | MATα ura3 trp1 his3 suc2-Δ9 bet1-1 | H.D.Schmitt |
| S20P4/3-9A | MATα ura3 leu2 lys2 pep4Δ::HIS3 sec20-1 | H.D.Schmitt |
| RH236-3A | MATα ura3 leu2 lys2 sec20-1 | H.Riezman |
| SHC22-12A | MATα ura3 his3 lys2 suc2-Δ9 sec22-3 | Andag et al. (2001) |
| S32G-8A | MATα ura3 leu2 his3 sec32-1/bos1 | Andag et al. (2001) |
| TNY51 | MATα ura3 leu2 his3 trp1 lys2 ade2 sed5-1 | Andag et al. (2001) |
| MLY-101 | MATα ura3 ade2 trp1 ufe1Δ::TRP1, containing pUT1 (ufe1-1) | Lewis and Pelham (1996) |
| MLY201 | MATα sec18-1 URA3::SEC20-(myc)3 his4-619 | Lewis et al. (1997) |
Plasmids and strains
Precise deletions of the USE1 ORF were generated by PCR in SEY6210×SEY6211 diploid cells. After transformation with a USE1 plasmid, diploids were sporulated and haploid cells carrying the deletion identified (BKY3, BKY4; Table II). A SEY6210 strain with a pep4Δ mutation was mated with BKY4, and tetrads dissected to generate wild-type and use1Δ strains defective in vacuolar proteases (BKY12 and BKY10). An N-terminal His6-tagged version of the cytoplasmic domain of Use1p (amino acids 1–215) was generated by PCR amplification and cloning into pET28b via NdeI–EcoRI (pMD19; Table III). A 1 kb fragment coding for USE1 was PCR amplified from genomic yeast DNA and cloned into pBluescript KS via endogenous HindIII–EcoRI sites (HindIII 100 nucleotides upstream of ATG; EcoRI 150 nucleotides downstream of stop). This USE1 fragment was subcloned into pRS315 (CEN6-LEU2) via BamHI–XhoI sites to yield pBK85. To introduce a triple HA tag, a BglII site was generated after the start codon by PCR-based site-directed mutagenesis and a 126 bp BglII fragment encoding three HA epitopes inserted (pBK55). The insert of pBK55 was moved to YEp352 via HindIII–SacI digestions (pBK64, 2µ-URA3). PCR random mutagenesis was performed as described (Muhlrad et al., 1992) using USE1 in pBluescript KS as template. The PCR reaction contained 0.25 mM MnCl2 and 25 µM dATP, 250 µM dGTP, dCTP, dTTP or 25 µM dGTP, 250 µM dATP, dCTP, dTTP. The PCR product was co-transformed with PstI–HindIII-digested pRS315 into use1Δ cells with a USE1-URA3 plasmid. Transformants were grown on 5-fluoroorotic acid to remove the USE1-URA3 plasmid, and screened for growth defects at 37°C resulting in the identification of pBK83. The mutations D183G and K184T together with a KpnI site were introduced into USE1 in pBluescript KS by PCR-based site-directed mutagenesis using the primers ACCCAAGTTCTTGGAGCTGCAG and ACCTTCATCAAGTGCTGATTGAAA (use1-2AA). use1-2AA was used for mutagenesis to remove the KpnI site, leaving the 0 layer mutation D183G, with the primers AGGCAAGTTCTTGGAGCTGCAG and ACCTTCATCAAGTGCTGATTGAAA.
Table III. Plasmids used in this study.
| Plasmid | Description | Reference |
|---|---|---|
| pBK55 | 1.0-kb USE1 (chr.VII, 317239–318228) with N-terminal 3×HA tag, F242S in pRS315 (CEN6-LEU2) | This study |
| pBK64 | 1.0-kb USE1 (chr.VII, 317239–318228) with N-terminal 3×HA tag, F242S in YEp352 (2µ-URA3) | This study |
| pBK85 | 1.0-kb USE1 (chr.VII, 317239–318228) in pRS315, F242S | This study |
| pBK83 | 1.0-kb use1-10AA in pRS315 with mutations Q18R, Q132R, E139D, Q156R, S168G, Q177R, D183G, Q185R, F220Y, F242S | This study |
| pMD16 | 1.0-kb use1-2AA in pRS315 with KpnI, D183G, K184T, F242S | This study |
| pMD19 | 6His-Use1p (aa 1–215) in pET-28b (E.coli expression vector) | This study |
| pMD25 | 1.0-kb use1-0 layer in pRS315 with mutations D183G, F242S | This study |
| pMD26 | 1.0-kb use1-10AA in pRS313 | This study |
| pFvM104 | SUC2 in pRS316 (CEN6-URA3) | This laboratory |
| pWB-Acycα | CYC1-SEC22-myc-α (CEN6-URA3) | Boehm et al. (1997) |
| pUA20 | pRS315-SEC22 (CEN6-LEU2) | Andag et al. (2001) |
| 2µ-SEC22 | SEC22 in YEp351 (2µ-LEU2) | H.D.Schmitt |
| pUA37 | pRS315-BOS1 (CEN6-LEU2) | Andag et al. (2001) |
| 2µ-BOS1 | pRS323-BOS1 (2µ-HIS3) | H.D.Schmitt |
| pUA39 | pRS315-UFE1 (CEN6-LEU2) | Andag et al. (2001) |
| 2µ-UFE1 | pRS323-UFE1 (2µ-HIS3) | H.D.Schmitt |
| CEN-BET1 | BET1 in pRS316 (CEN6-URA3) | H.D.Schmitt |
| 2µ-BET1 | BET1 in YEp351 (2µ-LEU2) | H.D.Schmitt |
| pMD20 | SED5 in YEp352 (2µ-URA3) | This study |
| pFvM32 | VTI1 in YEp352 (2µ-URA3) | This laboratory |
| pTlg1 | TPI-TLG1 in JS209 (2µ-URA3) | Holtius et al. (1998) |
| pBK124 | SEC20 in pRS315 (CEN6-LEU2) | This study |
| 2µ-SEC20 | 1.5-kb SEC20 XhoI-blunt in pRS323 (2µ-HIS3) | H.D.Schmitt |
Generation of antiserum against Use1p
A fusion protein containing an N-terminal His6 tag and amino acid residues 1–215 of Use1p was expressed in E.coli and purified from inclusion bodies after solubilization with 7.2 M urea in PBS using Ni-NTA–Sepharose. The purified protein was dialyzed against PBS and used to immunize a rabbit.
Subcellular fractionation
Subcellular fractionation was performed by differential centrifugation (Paravicini et al., 1992). SEY6211 cells were spheroplasted, osmotically lysed and centrifuged at 500 g to remove debris (homogenate H). The homogenate of SEY6211 and use1Δ cells expressing HA-USE1 from a CEN6 plasmid was separated by sucrose density gradient centrifugation (Becherer et al., 1996). The gradient consisted of the following steps: 0.5 ml 60%, 1 ml 42%, 1 ml 37%, 1.5 ml 34%, 2 ml 32%, 2 ml 29%, 1 ml 27%, 1.5 ml 22%, 1 ml 19% (w/w) sucrose in 10 mM HEPES–NaOH pH 7.6, with centrifugation for 16 h at 38 000 r.p.m. in a SW40 rotor. Fractions were separated by SDS–PAGE and immunoblotted using HRP-conjugated secondary antibodies and ECL.
Immunoprecipitations of 35S-labeled CPY and invertase
Cells were grown in log phase at 24°C and 0.5 OD were preshifted for 15 min to the labeling temperature unless indicated otherwise. Proteins were labeled for 10 min with [35S]methionine (100 µCi/0.5 OD), chased for 30 min and CPY immunoprecipitated from cellular extracts (I) and medium (E) as described earlier (Vater et al., 1992). Invertase was derepressed in minimal medium containing 0.1% glucose for 30 min at 24°C and for 15 min at restrictive temperature in cells expressing SUC2 from a CEN plasmid (Fischer von Mollard et al., 1997). Proteins were labeled for 10 min with [35S]methionine and chased for 30 min. Invertase was immunoprecipitated from lysed spheroplasts (I) and the combined periplasmic and medium fraction (E). Immunoprecipitates were analyzed by SDS–PAGE and autoradiography. A BAS1000 (Fuji) was used for quantification.
Secretion of Kar2p/BiP
Experiments were performed according to Boehm et al. (1994). Cells were grown in YEPD at 24°C to an OD of 0.3–0.5, harvested, resuspended in fresh medium and incubated for 2 h at the indicated temperatures. Proteins were precipitated from supernatants by addition of 10% TCA. The pellet was washed with acetone, resuspended in 8.3 µl of 2× sample buffer per 1 OD of cells and neutralized with 1 M Tris. To extract proteins from cell pellets, glass beads and 40 µl of Thorner buffer (8 M urea, 5% SDS, 50 mM Tris–HCl pH 6.8, 5% β-mercaptoethanol, protease inhibitors) per 1 OD were added. Equivalents of 0.19 OD for the pellet fractions and 3.6 OD for the external fractions were subjected to SDS–PAGE, transferred to a nitrocellulose membrane and analyzed by immunoblotting using antiserum against Kar2p.
Proteolytic processing of α-factor-tagged Sec22p (Sec22-α)
ER retrieval was monitored using a Sec22-α construct as described (Boehm et al., 1997). PEP4-positive and PEP4-deficient yeast strains containing the centromeric Sec22-α plasmid were grown in minimal medium at 24°C to early log phase and shifted for 30 min to 24, 30 or 37°C. Protein extracts were prepared as described above. Equivalents of 0.19 OD were subjected to SDS–PAGE and analyzed by immunoblotting using antibodies against the myc tag.
Indirect immunofluorescence
Indirect immunofluorescence was performed with use1Δ cells expressing HA-Use1p from a 2µ vector as described previously using a monoclonal antibody against the HA tag (Raymond et al., 1992). Sec61p (Panzner et al., 1995) and Emp47p were detected using specific antisera following an established procedure (Schröder et al., 1995). After the antibody incubations, DNA was stained for 10 min with 0.1 µg/ml DAPI in PBS/10% sorbitol. Cells were washed three times with PBS/10% sorbitol and embedded. Cells were viewed in an Olympus IX50 fluorescence microscope with the CCD camera imago (Photonics) or for Emp47p in a Zeiss Axiophot with a VarioCam CCD camera (Phase, Lübeck, Germany).
Vacuolar staining by FM4-64
Cells were grown at 24°C and shifted to restrictive temperature for 15 min. According to published procedures (Vida and Emr, 1995), 65 µM FM4-64 (Molecular Probes, Eugene, OR) was added and the incubation continued for 15 min. Cells were washed once and viewed immediately under a fluorescence microscope.
Immunoprecipitations
Immunoprecipitations of SNAREs were performed similar to described procedures (Søgaard et al., 1994; Lewis et al., 1997). Thirty microliters of antiserum against Use1p as well as pre-immune serum were cross-linked to 200 µl of protein A–Sepharose with dimethylpimelinediimidate. Ten OD of sec18-1 cells with SEC20-(myc)3 integrated at the URA3 locus (Lewis et al., 1997) were harvested, then spheroplasted for 1 h at 24°C in spheroplast buffer (1.2 M sorbitol, 50 mM KPi pH 7.3, 1 mM MgCl2) containing 300 µg/ml zymolyase-20T. Cells were washed, suspended in 10 ml YEPD/1 M sorbitol and incubated for 1 h at 24°C. Cells were suspended in 1 ml of lysis buffer (20 mM HEPES–KOH pH 7.0, 100 mM KCl, 2 mM EDTA, 0.5% Triton X-100, protease inhibitors) and dounced 20 times on ice. The detergent extract was centrifuged for 20 min at 50 000 r.p.m. Four hundred and fifty microliters of supernatant and 48 µl of beads were incubated overnight at 4°C. Beads were washed three times with lysis buffer and suspended in 30 µl of 1× sample buffer (without β-mercaptoethanol). Samples were analyzed by SDS–PAGE and immunoblotting. Starting samples correspond to 50% of the precipitated material for Use1p and to 4.4% for the other SNAREs.
Acknowledgments
Acknowledgements
We are grateful to T.H.Stevens (University of Oregon, Eugene, OR) for the generous gifts of antibodies against CPY, invertase, ALP, Vph1p and PGK, and to Mike Lewis (MRC, Cambridge, UK) for strains and antiserum against Ufe1p. We wish to thank Dieter Gallwitz for his support. This work was supported by grants from the Volkswagen Stiftung (program: Nachwuchsgruppen an Universitäten) to G.F.v.M., from the Deutsche Forschungsgemeinschaft (SFB 523 TP A4) to H.D.S. and from the Fonds der chemischen Industrie to E.H.
Note added in proof
Localization and function of Use1p have been investigated recently by N.Belgareh-Touzé, M.Corral-Debrinski, H.Launhardt, J.M.Galan, T.Munder, S.Le Panse and R.Haguenauer-Tsapis (2003) Yeast functional analysis: identification of two essential genes involved in ER to Golgi trafficking. Traffic, in press.
References
- Andag U., Neumann,T. and Schmitt,H. (2001) The coatomer-interacting protein Dsl1p is required for Golgi-to-endoplasmic reticulum retrieval in yeast. J. Biol. Chem., 276, 39150–39160. [DOI] [PubMed] [Google Scholar]
- Antonin W., Fasshauer,D., Becker,S., Jahn,R. and Schneider,T. (2002) Crystal structure of the endosomal SNARE complex reveals common structural principles of all SNAREs. Nat. Struct. Biol., 9, 107–111. [DOI] [PubMed] [Google Scholar]
- Ballensiefen W., Ossipov,D. and Schmitt,H. (1998) Recycling of the yeast v-SNARE Sec22p involves COPI-proteins and the ER transmembrane proteins Ufe1p and Sec20p. J. Cell Sci., 111, 1507–1520. [DOI] [PubMed] [Google Scholar]
- Banfield D.K., Lewis,M.J. and Pelham,H.R. (1995) A SNARE-like protein required for traffic through the Golgi complex. Nature, 375, 806–809. [DOI] [PubMed] [Google Scholar]
- Becherer K.A., Rieder,S.E., Emr,S.D. and Jones,E.W. (1996) Novel syntaxin homolog, Pep12p, required for the sorting of lumenal hydrolases to the lysosome-like vacuole in yeast. Mol. Biol. Cell, 7, 579–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock J.B., Matern,H.T., Peden,A.A. and Scheller,R.H. (2001) A genomic perspective on membrane compartment organization. Nature, 409, 839–841. [DOI] [PubMed] [Google Scholar]
- Boehm J., Ulrich,H.D., Ossig,R. and Schmitt,H.D. (1994) Kex2-dependent invertase secretion as a tool to study the targeting of transmembrane proteins which are involved in ER→Golgi transport in yeast. EMBO J., 13, 3696–3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm J., Letourneur,F., Ballensiefen,W., Ossipov,D., Demolliere,C. and Schmitt,H. (1997) Sec12p requires Rer1p for sorting to coatomer (COPI)-coated vesicles and retrieval to the ER. J. Cell Sci., 110, 991–1003. [DOI] [PubMed] [Google Scholar]
- Cao X. and Barlowe,C. (2000) Asymmetric requirements for a Rab GTPase and SNARE proteins in fusion of COPII vesicles with acceptor membranes. J. Cell Biol., 149, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darsow T., Rieder,S.E. and Emr,S.D. (1997) A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J. Cell Biol., 138, 517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasshauer D., Sutton,R.B., Brunger,A.T. and Jahn,R. (1998) Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl Acad. Sci. USA, 95, 15781–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer von Mollard G., Nothwehr,S.F. and Stevens,T.H. (1997) The yeast v-SNARE Vti1p mediates two vesicle transport pathways through interactions with the t-SNAREs Sed5p and Pep12p. J. Cell Biol., 137, 1511–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio G. (1998) The Saccharomyces cerevisiae early secretion mutant tip20 is synthetic lethal with mutants in yeast coatomer and the SNARE proteins Sec22p and Ufe1p. Yeast, 14, 633–646. [DOI] [PubMed] [Google Scholar]
- Gaynor E. and Emr,S. (1997) COPI-independent anterograde transport: cargo-selective ER to Golgi protein transport in yeast COPI mutants. J. Cell Biol., 136, 789–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G. et al., (2002) Functional profiling of the Saccharomyces cerevisiae genome. Nature, 418, 387–391. [DOI] [PubMed] [Google Scholar]
- Götte M. and Gallwitz,D. (1997) High expression of the yeast syntaxin-related Vam3 protein suppresses the protein transport defects of a pep12 null mutant. FEBS Lett., 411, 48–52. [DOI] [PubMed] [Google Scholar]
- Grote E., Baba,M., Ohsumi,Y. and Novick,P. (2000) Geranylgeranylated SNAREs are dominant inhibitors of membrane fusion. J. Cell Biol., 151, 453–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta G. and Heath,I. (2002) Predicting the distribution, conservation and function of SNAREs and related proteins in fungi. Fungal Genet. Biol., 36, 1–21. [DOI] [PubMed] [Google Scholar]
- Hardwick K.G. and Pelham,H.R. (1992) SED5 encodes a 39-kD integral membrane protein required for vesicular transport between the ER and the Golgi complex. J. Cell Biol., 119, 513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R. and Südhof,T. (1999) Membrane fusion and exocytosis. Annu. Rev. Biochem., 68, 863–911. [DOI] [PubMed] [Google Scholar]
- Kaiser C.A. and Schekman,R. (1990) Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell, 61, 723–733. [DOI] [PubMed] [Google Scholar]
- Lewis M.J. and Pelham,H.R. (1996) SNARE-mediated retrograde traffic from the Golgi complex to the endoplasmic reticulum. Cell, 85, 205–215. [DOI] [PubMed] [Google Scholar]
- Lewis M.J., Rayner,J.C. and Pelham,H.R.B. (1997) A novel SNARE complex implicated in vesicle fusion with the endoplasmic reticulum. EMBO J., 16, 3017–3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. and Barlowe,C. (2002) Analysis of Sec22p in endoplasmic reticulum/Golgi transport reveals cellular redundancy in SNARE protein function. Mol. Biol. Cell, 13, 3314–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D., Hunter,R. and Parker,R. (1992) A rapid method for localized mutagenesis of yeast genes. Yeast, 8, 79–82. [DOI] [PubMed] [Google Scholar]
- Newman A.P., Shim,J. and Ferro-Novick,S. (1990) BET1, BOS1 and SEC22 are members of a group of interacting yeast genes required for transport from the endoplasmic reticulum to the Golgi complex. Mol. Cell. Biol., 10, 3405–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P., Field,C. and Schekman,R. (1980) Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell, 21, 205–215. [DOI] [PubMed] [Google Scholar]
- Panzner S., Dreier,L., Hartmann,E., Kostka,S. and Rapoport,T.A. (1995) Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell, 81, 561–570. [DOI] [PubMed] [Google Scholar]
- Paravicini G., Horazdovsky,B.F. and Emr,S.D. (1992) Alternative pathways for the sorting of soluble vacuolar proteins in yeast: a vps35 null mutant missorts and secretes only a subset of vacuolar hydrolases. Mol. Biol. Cell, 3, 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parlati F., Varlamov,O., Paz,K., McNew,J., Hurtado,D., Söllner,T. and Rothman,J. (2002) Distinct SNARE complexes mediating membrane fusion in Golgi transport based on combinatorial specificity. Proc. Natl Acad. Sci. USA, 99, 5424–5429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham H.R.B. (2001) SNAREs and the specificity of membrane fusion. Trends Cell Biol., 11, 99–101. [DOI] [PubMed] [Google Scholar]
- Raymond C.K., Howald-Stevenson,I., Vater,C.A. and Stevens,T.H. (1992) Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell, 3, 1389–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M., Stone,S. and Ferro-Novick,S. (1997) The synaptobrevin-related domains of Bos1p and Sec22p bind to the syntaxin-like region of Sed5p. J. Biol. Chem., 272, 17134–17138. [DOI] [PubMed] [Google Scholar]
- Schröder S., Schimmöller,F., Singer-Krüger,B. and Riezman,H. (1995) The Golgi-localization of yeast Emp47p depends on its di-lysine motif but is not affected by the ret1-1 mutation in α-COP. J. Cell Biol., 131, 895–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza J.C., Hardwick,K.G., Dean,N. and Pelham,H.R. (1990) ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell, 61, 1349–1357. [DOI] [PubMed] [Google Scholar]
- Søgaard M., Tani,K., Ye,R.R., Geromanos,S., Tempst,P., Kirchhausen,T., Rothman,J.E. and Sollner,T. (1994) A rab protein is required for the assembly of SNARE complexes in the docking of transport vesicles. Cell, 78, 937–948. [DOI] [PubMed] [Google Scholar]
- Spang A. and Schekman,R. (1998) Reconstitution of retrograde transport from the Golgi to the ER in vitro. J. Cell Biol., 143, 589–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling C.J., Rothblatt,J., Hosobuchi,M., Deshaies,R. and Schekman,R. (1992) Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol. Biol. Cell, 3, 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S., Sacher,M., Mao,Y., Carr,C., Lyons,P., Quinn,A.M. and Ferro-Novick,S. (1997) Bet1p activates the v-SNARE Bos1p. Mol. Biol. Cell, 8, 1175–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton R.B., Fasshauer,D., Jahn,R. and Brunger,A.T. (1998) Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature, 395, 347–353. [DOI] [PubMed] [Google Scholar]
- Vater C.A., Raymond,C.K., Ekena,K., Howald,S.,I. and Stevens,T.H. (1992) The VPS1 protein, a homolog of dynamin required for vacuolar protein sorting in Saccharomyces cerevisiae, is a GTPase with two functionally separable domains. J. Cell Biol., 119, 773–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida T.A. and Emr,S.D. (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol., 128, 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuestehube L., Duden,R., Eun,A., Hamamoto,S., Korn,P., Ram,R. and Schekman,R. (1996) New mutants of Saccharomyces cerevisiae affected in the transport of proteins from the endoplasmic reticulum to the Golgi complex. Genetics, 142, 393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]