Abstract
Silencing of the paternal allele of three imprinted genes (Igf2r, Slc22a2 and Slc22a3) requires cis expression of the Air RNA that overlaps the promoter of one of them (Igf2r). Air is a non-coding RNA whose mode of action is unknown. We tested the role of the Igf2r promoter and the role of transcriptional overlap between Igf2r and Air in silencing in this cluster. We analyzed imprinted expression in mice in which the Igf2r promoter is replaced by a thymidine kinase promoter that preserves a transcription overlap with Air, and in mice with a deleted Igf2r promoter that lack any transcriptional overlap with Air. Imprinted silencing of Air, Slc22a2 and Slc22a3 is maintained by the replacement promoter and also in the absence of transcriptional overlap with Air. These results exclude a role for the Igf2r promoter and for transcriptional overlap between Igf2r and Air in silencing Air, Slc22a2 and Slc22a3. Although these results do not completely exclude a role for a double-stranded RNA silencing mechanism, they do allow the possibility that the Air RNA has intrinsic cis silencing properties.
Keywords: genomic imprinting/Igf2r/non-coding RNA/Slc22a2/Slc22a3
Introduction
Mammalian genomic imprinting is an epigenetic gene regulatory mechanism that results in parental-specific gene expression of a small number of genes in diploid somatic cells (Beechey et al., 2001; Reik and Walter, 2001; Li, 2002; Sleutels and Barlow, 2002). Several features of the imprinting mechanism have been identified; however, it is not yet clear whether imprinting is regulated by a unique process or whether it is part of the general epigenetic apparatus used to regulate mammalian gene expression. Clustering and coordinate regulation is one feature imprinted genes share with non-imprinted genes (Engemann et al., 2000; Onyango et al., 2000), and it is now clear that many imprinted genes are functionally grouped such that imprinted expression of several genes is regulated by one long-range imprint control element (Thorvaldsen et al., 1998; Horike et al., 2000; Zwart et al., 2001; Fitzpatrick et al., 2002). The frequent occurrence of an imprinted non-coding RNA within an imprinted gene cluster has also been observed (Sleutels and Barlow, 2002); however, non-coding RNAs have also been identified in several ‘normal’ bi-allelic expressed genes such as N-myc, BFGF, Hox11, RPS14 and Kelch-like1 (Krystal et al., 1990; Tasheva and Roufa, 1995; Li et al., 1996; Potter and Branford, 1998; Benzow and Koob, 2002), and, in the case of the human β-globin cluster, a non-coding RNA has been linked to chromatin remodeling (Gribnau et al., 2000). Approximately one-quarter of imprinted transcripts are non-coding, and the majority is expressed from the parental chromosome that carries the silent allele of an imprinted protein-coding gene (Beechey et al., 2001; Sleutels and Barlow, 2002). This reciprocal parental expression pattern has been proposed as either the cause or the consequence of imprinted expression, and evidence exists to support both these proposals.
An example of where reciprocal expression of a non-coding RNA is proposed to occur as a consequence of imprinting is shown by the imprinted cluster on mouse chromosome 7 that contains two paternally expressed protein-coding genes (Ins2 and Igf2) located 80 kb upstream of a maternally expressed non-coding RNA (H19). A methylation-sensitive insulator element known as the ‘H19 DMR’ is located between Ins2/Igf2 and H19 and controls their access to a common enhancer, located downstream of all three genes, that activates either H19 or Ins2/Igf2 (Hark et al., 2000; Szabo et al., 2000; Bell et al., 2001). On the paternal chromosome, the H19 DMR inherits a methylation imprint that inactivates its insulator function and allows the access of Ins2/Igf2 to the enhancer, thus permitting their expression on this allele. On the maternal chromosome, the H19 DMR insulator is unmethylated and active, which blocks the access of Ins2/Igf2 to the downstream enhancer. H19 gains access by default and thus shows maternal-specific expression (Schmidt et al., 1999; Thorvaldsen and Bartolomei, 2000). Thus, the maternal-specific expression of the non-coding H19 RNA is a consequence of the imprinted expression of Ins2/Igf2.
In contrast, a direct silencing function for a non-coding RNA has been demonstrated in an imprinted gene cluster on mouse chromosome 17 that contains three maternally expressed protein-coding genes (Igf2r, Slc22a2 and Slc22a3) and one paternally expressed non-coding RNA (Air). The Air promoter lies within intron 2 of Igf2r and expresses, on the paternal chromosome only, a non-coding RNA that overlaps the Igf2r promoter in an antisense orientation and extends 79 kb upstream (Lyle et al., 2000). The Air promoter is methylated and silent on the maternal chromosome. Thus, maternal expression of Igf2r, Slc22a2 and Slc22a3 correlates with methylation and silencing of the Air promoter in cis. Igf2r expression has previously been shown to require DNA methylation (Li et al., 1993). Deletion of the Air promoter from the paternal chromosome demonstrated its action as a cis-acting bidirectional silencer on the upstream Igf2r promoter and on the downstream Slc22a2 and Slc22a3 (Zwart et al., 2001). Recent experiments that truncated Air to within 3 kb of its promoter have identified the Air RNA itself or its active transcription as the cause of Igf2r, Slc22a2 and Slc22a3 silencing (Rougeulle and Heard, 2002; Sleutels et al., 2002).
The Air RNA originates from an antisense-orientated promoter within intron 2 of Igf2r and overlaps by 29 kb the 5′ part of Igf2r including the promoter (Figure 1A). The Slc22a2 and Slc22a3 promoters lie, respectively, 170 and 215 kb upstream of the Air promoter and thus are not overlapped by the Air RNA (Zwart et al., 2001). Since the Air RNA silences Slc22a2 and Slc22a3 without overlapping them, this may indicate that Air or its active transcription has a direct action on its susceptible target genes. However, it is also possible that silencing of this gene cluster acts in a two-step manner that is initially dependent on the transcript overlap between Air and Igf2r (Rougeulle and Heard, 2002). In a two-step model, silencing of the non-overlapped Slc22a2 and Slc22a3 would depend on the initial silencing of the Igf2r promoter by the overlapping Air RNA.
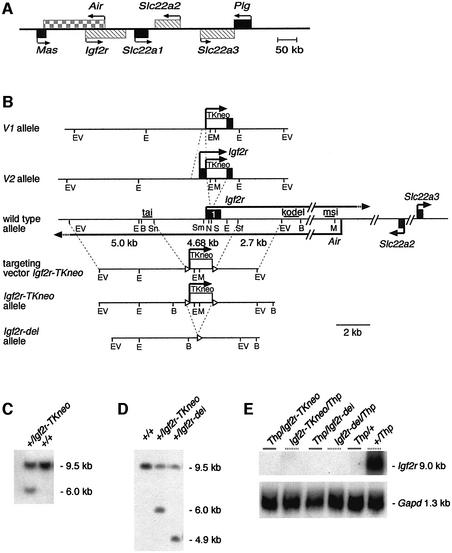
Fig. 1. Four Igf2r promoter replacement/deletion alleles. (A) A map of the Igf2r, Air, Slc22a2 and Slc22a3 imprinted cluster with arrows marking transcriptional orientation. Black box, genes with bi-allelic expression; gray cross-hatched box, imprinted protein-coding genes with maternal-specific expression; gray checked box, imprinted non-coding Air RNA with paternal-specific expression. (B) The V1 and V2 alleles in which, respectively, 444 and 29 bp of the Igf2r allele were replaced by a thymidine kinase promoter–neomycin (TKneo) cassette (Ludwig et al., 1996) are drawn above the wild-type allele. The targeting construct for the Igf2r-TKneo allele is shown below and contains a 12.4 kb EcoRV fragment (bp 89 965–102 345; AJ249895) from the Igf2r locus, from which a 4682 bp SnaBI–SfuI fragment including the entire Igf2r CpG-island promoter and exon 1 was replaced by a 1200 bp loxP (open triangles) flanked cassette containing a TKneo resistance gene and polyadenylation signal (box labeled TKneo). Homologous recombination in embryonic stem (ES) cells yielded the Igf2r-TKneo allele, and in vivo Cre-mediated deletion of the TKneo cassette generated the Igf2r-del allele. Fragments: EcoRV (EV); EcoRI (E); BglII (B); SnaBI (Sn); SmaI (Sm); NotI (N); SalI (S); SfuI (Sf); and MluI (M). The probes (tai, kodel and msi) used for the methylation analyses are shown as black bars above the wild-type allele. (C) Correctly targeted Igf2r-TKneo ES clones were identified by DNA blot of BglII-digested DNA and probe kodel, yielding a 6 kb fragment instead of the 9.5 kb wild-type fragment. (D) The Igf2r-del allele was generated by crossing Igf2r-TKneo mice with mice carrying a CMV-Cre transgene (Schwenk et al., 1995), identified by DNA blot of BglII-digested DNA and probe kodel. Cre-mediated deletion of the TKneo cassette changes the 6.0 kb fragment from the Igf2r-TKneo allele to 4.9 kb, generating the Igf2r-del allele. (E) Absence of Igf2r mRNA from the Igf2r-TKneo and Igf2r-del alleles. RNA blot of 11.5 d.p.c. embryo RNA hybridized with cDNA probes detecting Igf2r exons 3–6 or Gapd used as loading control.
In this study, we tested the role of the Igf2r CpG-island promoter itself and the role of transcriptional overlap between the Air and Igf2r transcripts in regulating imprinted silencing of Air on the maternal chromosome and of Slc22a2 and Slc22a3 on the paternal chromosome. Three different alleles in which part or all of the Igf2r promoter were replaced by a thymidine kinase (TK) promoter–neomycin cassette were analyzed, and each showed normal imprinted expression of Air, Slc22a2 and Slc22a3 when the Igf2r promoter was replaced by a foreign promoter. In addition, a fourth allele, deleted for the entire Igf2r promoter, also preserved normal imprinted expression of Air, Slc22a2 and Slc22a3. These results show that paternal-specific silencing of two coding genes plus maternal-specific silencing of one non-coding RNA do not require the Igf2r promoter, a silent promoter at the same position or transcriptional overlap between Igf2r and Air. Although these results do not exclude a role for a double-stranded RNA silencing mechanism, the data presented here, in combination with the recent demonstration of the direct involvement of the Air RNA in gene silencing (Sleutels et al., 2002), allow the possibility that Air has intrinsic cis regulatory properties and can act directly to silence autosomal genes.
Results
Replacement of the Igf2r promoter: three alleles
The replacement V1 and V2 alleles have, respectively, a 444 and 29 bp fragment from within the Igf2r CpG-island promoter region replaced by a 1100 bp fragment containing a herpes simplex TK promoter linked to neomycin/poly(A) (TKneo). The V1 444 bp deletion removes 330 bp of upstream promoter sequences, including up to codon 38 of Igf2r exon 1, and the smaller V2 allele deletion is contained within exon 1 and removes codons 28–38. These alleles do not express Igf2r from the targeted allele due to the poly(A) signal included in the neomycin gene and show maternal-specific embryonic lethality (Ludwig et al., 1996). The Igf2r-TKneo allele generated for this work contains a 4682 bp deletion spanning the complete Igf2r CpG-island promoter and exon 1 replaced by a 1200 bp TKneo cassette flanked by loxP sites. Figure 1B–E shows the relationship of the V1 and V2 alleles to the wild-type locus and the derivation of the Igf2r-TKneo allele in embryonic stem (ES) cells and mice.
In order to examine Igf2r expression from the targeted allele, heterozygous +/Igf2r-TKneo mice were mated to mice carrying the Thp chromosome. The Thp chromosome contains a 3 cM deletion that includes the Igf2r, Air, Slc22a2 and Slc22a3 region, and these crosses allow examination of parental-specific expression in the absence of the second parental allele. Paternal transmission of the Thp allele in wild-type laboratory mouse strains (+/Thp; note that the maternal allele is written on the left side) has no effect on viability, but maternal transmission (Thp/+) is lethal between 15.5 and 17.5 days post-coitum (d.p.c.) because the absence of Igf2r leads to a lethal excess of Igf2 (Wutz et al., 2001). RNA blots of 11.5 d.p.c. embryos show that a maternally transmitted Igf2r-TKneo allele lacks Igf2r expression, in contrast to a wild-type maternal allele (Figure 1E; compare lanes Igf2r-TKneo/Thp and +/Thp), and that Igf2r is not expressed from a paternal Igf2r-TKneo allele (Thp/Igf2r-TKneo) or a paternal wild-type allele (Thp/+). These results demonstrate that, although the targeting event abolished maternal Igf2r expression downstream of the inserted TKneo cassette, it did not affect silencing on the paternal allele. The absence of maternal Igf2r expression explains the absence of viable offspring with a maternally inherited Igf2r-TKneo allele (data not shown), as has been reported for other Igf2r loss-of-function alleles (Lau et al., 1994; Wang et al., 1994; Ludwig et al., 1996).
Parental-specific expression in the Igf2r promoter replacement alleles
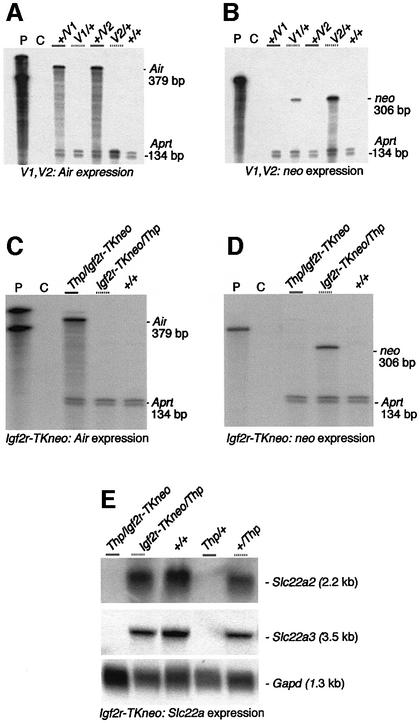
Parental-specific expression of the replacement TK promoter and the downstream imprinted promoters (Air, Slc22a2 and Slc22a3) was tested in the Igf2r promoter replacement/deletion alleles in embryonic and placental tissue. Both the V1 and V2 alleles showed complete imprinted expression in 16.5 d.p.c. embryos of both Air and TKneo, such that Air is expressed only from the paternal allele and TKneo is expressed only from the maternal allele (Figure 2A and B; note that the Airneo probe does not detect endogenous Air RNA). Maternal expression of TKneo from the V1 allele is reduced, compared with the V2 allele (Figure 2B). The Igf2r-TKneo replacement allele similarly showed paternal-specific Air expression and maternal-specific TKneo expression in 11.5 d.p.c. embryos (Figure 2C and D). In addition, imprinted expression of the downstream Slc22a2 and Slc22a3 was also fully maintained and unchanged from the wild-type state in 11.5 d.p.c. placenta on the Igf2r-TKneo allele (Figure 2E; note that imprinted expression of Slc22a2 and Slc22a3 is restricted to the embryonic placenta; Zwart et al., 2001). In summary, analysis of parental-specific expression in these three alleles, with a TK promoter inserted into or replacing the Igf2r promoter, shows that the Igf2r promoter itself is not required for imprinting the other genes in this cluster and that an exogenous promoter that fully replaces the Igf2r promoter can acquire full imprinted expression.
Fig. 2. Parental-specific expression in three Igf2r promoter replacement alleles. (A and B) The V1 and V2 alleles have imprinted paternal expression of Air (gray solid bar) and imprinted maternal expression of neomycin (neo, gray dotted bar). RNase protection analysis (RPA) on 16.5 d.p.c. embryonic RNA with probe Airneo that detects the Air RNA at the position of the thymidine kinase promoter–neomycin (TKneo) cassette (A) or with probe tkneo that detects the neomycin RNA produced from the TK promoter (B). The probes Airneo and tkneo do not recognize endogenous Air RNA. Aprt exon 3: RNA loading control. Lane P, input probes; lane C, tRNA hybridization control. (C and D) The Igf2r-neo allele has imprinted paternal expression of Air (gray solid bar) and imprinted maternal expression of neomycin (neo, gray dotted bar). RPA on 11.5 d.p.c. embryo RNA from Thp/Igf2r-neo reciprocal heterozygous crosses with probe Airneo that detects the Air RNA at the position of the TKneo cassette (C) or with probe tkneo that detects the neomycin RNA produced from the TK promoter (D). Controls as above. (E) The Igf2r-neo allele shows imprinted maternal expression of Slc22a2 and Slc22a3. RNA blot of 11.5 d.p.c. placenta RNA from Thp/Igf2r-neo reciprocal heterozygous crosses hybridized with a cDNA probes detecting Slc22a2 (top), Slc22a3 (middle) or Gapd (bottom) used as loading control.
Parental-specific methylation of the Igf2r promoter replacement alleles
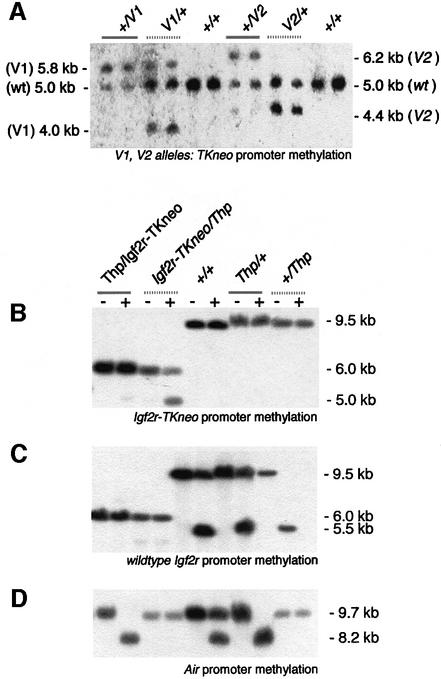
Parental-specific methylation of the TK promoter in the replacement alleles was tested in embryos. Figure 3A shows that maternally inherited V1 and V2 alleles have either reduced or an absent methylation, compared with paternally inherited alleles, in 16.5 d.p.c. embryos. Maternal methylation (as quantified by PhosphorImager; data not shown) was present to ∼40% of the paternal level on the V1 but completely absent on the V2. Paternal methylation was complete for V1 but incomplete for V2 in 16.5 d.p.c. embryos (Figure 3A; lanes V1/+ or V2/+ show the presence of smaller fragments not found in lanes +/V1 and seen as faint bands in +/V2; note that methylation of the paternal allele is incomplete on the wild-type chromosome in embryos; Figure 3C, lane Thp/+; Stoeger et al., 1993). Identical results were obtained for 16.5 d.p.c. embryonic membrane and for the MluI site in the TK promoter and the SmaI sites that flank the replacement (data not shown). The V2 methylation pattern is the same as that on wild-type Igf2r promoter, which is methylated over a 1500 bp region in late development; complete methylation is seen only after birth, and the maternal wild-type Igf2r promoter is always unmethylated (Stoeger et al., 1993). The wild-type Air promoter that lies 29 kb downstream of the Igf2r promoter is normally only methylated on the maternal allele (Stoeger et al., 1993), and this methylation (as determined by analysis of an MluI site inside the Air promoter) remains maternal specific in both the V1 and V2 alleles (data not shown). Slc22a2 and Slc22a3 lack parental-specific methylation (Zwart et al., 2001).
Fig. 3. Parental-specific methylation of three Igf2r promoter replacement alleles. (A) Thymidine kinase promoter–neomycin (TKneo) methylation analysis in V1 and V2 alleles. Heterozygous 16.5 d.p.c. embryonic DNA carrying either paternally derived (gray solid bars) or maternally derived (gray dotted bars) V1 and V2 replacement alleles was digested with EcoRI and hybridized with probe tai (Figure 1B). Note that EcoRI is methyl sensitive when G flanks the GAATTC recognition site and the CG dinucleotide is methylated (Ludwig et al., 1996). The V1 allele generates 4.0 kb when the EcoRI is unmethylated and 5.8 kb for a methylated site. V1 paternal transmission (gray solid bar) shows full methylation at this site, and V1 maternal transmission (gray dotted bar) shows partial (∼40%) methylation at this site. The V2 allele yields 4.4 or 6.2 kb for an unmethylated and methylated sites, respectively. V2 paternal transmission (gray solid bar) shows partial (∼80%) methylation at this site, and V2 maternal transmission (gray dotted bar) shows no methylation at this site. The Igf2r wild-type (wt) promoter fragment is 5.0 kb and does not contain a methyl-sensitive EcoRI site. (B) TKneo promoter methylation analysis in the Igf2r-TKneo allele. Heterozygous 13.5 d.p.c. embryonic DNA carrying either a paternally derived (gray solid bar) or maternally derived (gray dotted bar) Igf2r-TKneo replacement allele was digested with BglII (–) or BglII and MluI (+) and hybridized with probe kodel (Figure 1B). Note that, in crosses with the Thp allele, only bands from the opposite allele are seen. The Igf2r-TKneo allele yields 5.0 or 6.0 kb for an unmethylated and methylated sites, respectively. Igf2r-TKneo paternal transmission (gray solid bar) shows almost complete methylation at this site (a faint 5.0 kb unmethylated can be seen on the original image), and Igf2r-TKneo maternal transmission (gray dotted bar) shows partial (∼50%) methylation at this site. The wild-type Igf2r promoter fragment is 9.5 kb and does not contain an MluI site. (C) Igf2r promoter (NotI) methylation analysis in the wild-type Igf2r promoter. Thp/+ heterozygous 13.5 d.p.c. embryonic DNA carrying either a paternally derived (gray solid bar) or maternally derived (gray dotted bar) wild-type Igf2r promoter was digested with BglII (–) or BglII and NotI (+) and hybridized with probe kodel (Figure 1B). The wild-type Igf2r promoter yields 5.5 or 9.5 kb for an unmethylated and methylated sites, respectively. Igf2r wild-type promoter paternal transmission (gray solid bar) shows partial (∼50%) methylation at this site, and Igf2r wild-type promoter maternal transmission (gray dotted bar) shows no methylation at this site. The Igf2r-TKneo promoter fragment is 6.0 kb and lacks this NotI site (it was deleted in the targeting event). (D) Air promoter (MluI) methylation analysis in the Igf2r-TKneo and wild-type Igf2r alleles. Heterozygous 13.5 d.p.c. embryonic DNA carrying either a paternally derived (gray solid bar) or maternally derived (gray dotted bar) Igf2r-TKneo or a wild-type Igf2r allele was digested with BglII (–) or BglII and MluI (+) and hybridized with probe msi (Figure 1B). The Air promoter yields 8.2 or 9.7 kb for an unmethylated and methylated sites, respectively. Both the Igf2r-TKneo and wild-type Igf2r alleles show no methylation at this site on paternal transmission (gray solid bar) and full methylation on maternal transmission (gray dotted bar).
Parental-specific methylation in the Igf2r-TKneo allele was tested in a similar manner by restriction enzyme digest and DNA blot hybridization in 13.5 d.p.c. embryos in mice heterozygous for the Thp deletion (Figure 3B, C and D). Methylation of the MluI site present in the TKneo promoter is almost complete on paternal transmission (Figure 3B, lane Thp/Igf2r-TKneo); however; this methylation is only reduced by 50% on maternal transmission (lane Igf2r-TKneo/Thp) as quantified by PhosphorImager (data not shown). This result is similar to that shown by the V1 allele (Figure 3A) and contrasts to the strictly paternal-specific methylation that is incomplete in 13.5 d.p.c. embryo tissue (Stoeger et al., 1993) seen in the wild-type Igf2r promoter at the SalI site (data not shown) and the NotI site (Figure 3C; compare lanes Thp/+ and +/Thp). Methylation of the downstream Air promoter at the SfuI site (data not shown) and the MluI site was restricted to the maternal allele in both the Igf2r-TKneo and wild-type alleles in 13.5 d.p.c. embryos (Figure 3D, lanes Igf2r-TKneo/Thp and +/Thp).
In summary, the methylation studies of the three TKneo alleles show that paternal methylation is retained after targeted insertion/replacement of the Igf2r promoter by a TK promoter. However, the absence of maternal methylation that is a feature of the wild-type Igf2r promoter is only seen in the V2 allele that is deleted for 29 bp but retains the Igf2r promoter. The V1 and Igf2r-TKneo alleles (which lack, respectively, 444 and 4682 bp) gain methylation on maternal transmission, albeit at a reduced level compared with paternal transmission. The gain of maternal methylation on the TK promoter in the V1 allele, compared with the V2 allele, likely explains the reduced TKneo expression from the V1 allele (Figure 2B). The methylation status of the linked Air promoter remained unchanged in all three replacement alleles.
Deletion without replacement of the Igf2r promoter
An Igf2r-del allele that replaced the 4682 bp Igf2r promoter fragment with a single loxP site was generated by mating +/Igf2r-TKneo males with female mice containing a Cre transgene driven by a CMV promoter (Schwenk et al., 1995). Heterozygous offspring in which the TKneo cassette was deleted to produce the Igf2r-del allele (Figure 1D) were obtained and crossed with Thp mice to generate embryos and placentas with a germline-transmitted Igf2r-del allele that could be analyzed for allele-specific expression. RNA blot analysis of 11.5 d.p.c. embryos showed that removal of this 4682 bp fragment abolished all transcription through the Igf2r locus as detected by a cDNA probe spanning exons 3–6 (Figure 1A and E). Imprinted expression of Slc22a2 and Slc22a3 on the Igf2r-del allele was maintained, as in wild-type mice, in 13.5 d.p.c. placentas (data not shown) and in 11.5 d.p.c. placentas (Figure 4A; compare lanes Thp/Igf2r-del and Igf2r-del/Thp). Expression from the Air promoter was analyzed in 13.5 d.p.c. embryos (data not shown) and in 11.5 d.p.c. embryos (Figure 4B). These results show that the Air promoter on the Igf2r-del allele is expressed only after paternal transmission and thus retains its normal wild-type parental-specific expression pattern (Figure 4B; compare lanes Thp/Igf2r-del and Igf2r-del/Thp; protected multiple fragments are paternal specific and represent the multiple transcription starts mapped in the Air promoter; Sleutels et al., 2002). Analysis of imprinted methylation was only possible at the Air promoter (since the Igf2r promoter is deleted in this allele and since Slc22a2 and Slc22a3 lack parental-specific methylation), and this analysis in 13.5 d.p.c. embryos showed that SfuI (data not shown) and MluI retained their wild-type pattern and were methylated only on the maternal chromosome (Figure 4C; compare lanes Igf2r-del/Thp and Thp/Igf2r-del; the unmethylated 8.2 kb fragment is paternal specific).
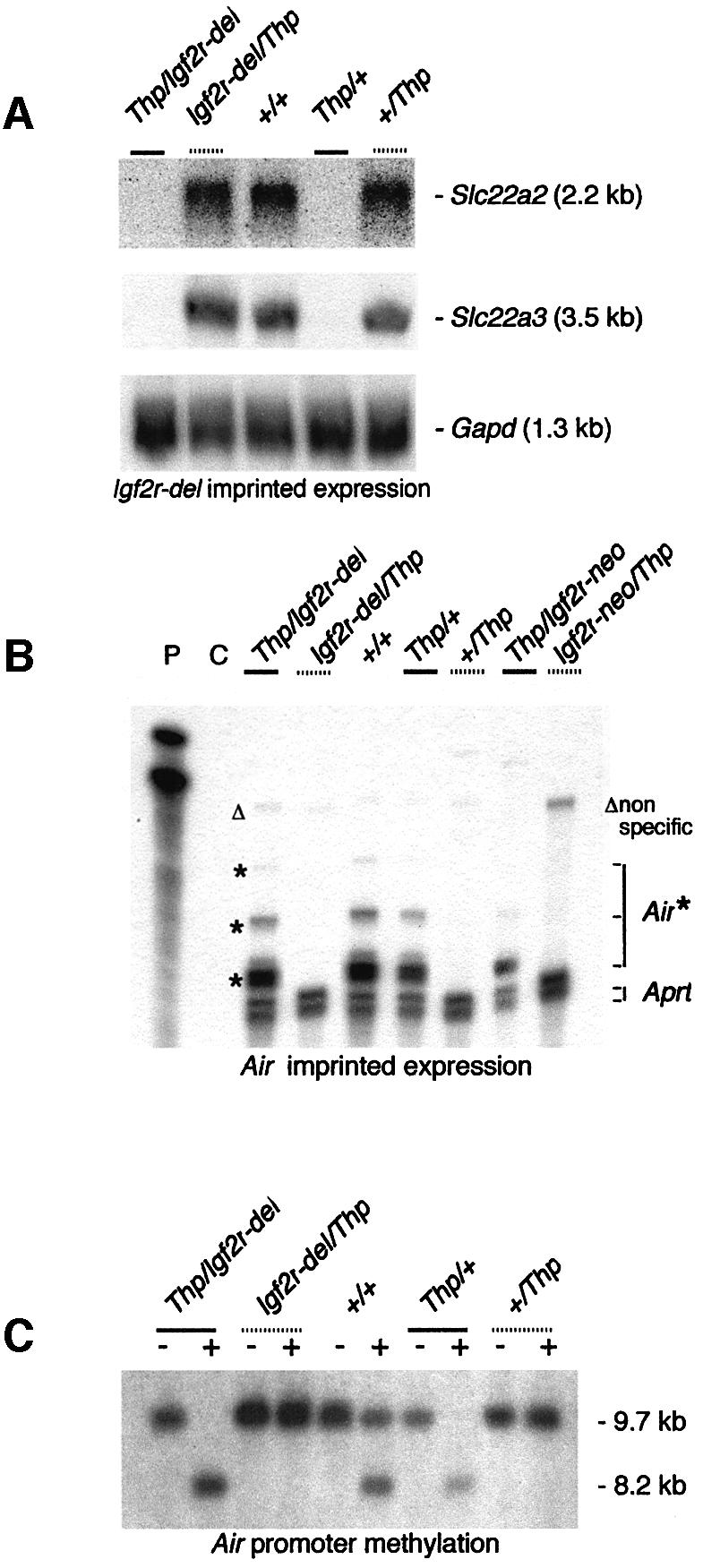
Fig. 4. Imprinted expression and methylation in the Igf2r-del allele. (A) Slc22a2 and Slc22a3 imprinted expression from the Igf2r-del allele is maternal specific (gray dotted bars), identical to the wild-type situation. RNA blot of 11.5 d.p.c. placenta RNA from Thp/Igf2r-del and Thp/wild-type reciprocal crosses hybridized with cDNA probes detecting Slc22a2, Slc22a3 or Gapd used as loading control. Note that imprinted expression of Slc22a2 and Slc22a3 is restricted to the embryonic placenta (Zwart et al., 2001). (B) Air RNA imprinted expression from the Igf2r-del allele is paternal specific (gray solid bars), identical to the wild-type situation. RNase protection analysis using probe MlMs1 on 11.5 d.p.c. embryo RNA from Thp/Igf2r-del and from Thp/wild-type reciprocal crosses. This probe detects multiple fragments (asterisk) as it overlaps multiple Air transcription start sites. The open triangle indicates non-specific protected bands. Controls as in Figure 3. (C) Air promoter methylation on the paternal (gray solid bar) and maternal (gray dotted bar) Igf2r-del allele is identical to the wild-type Air promoter. Heterozygous 13.5 d.p.c. embryonic DNA from Thp/Igf2r-del and from Thp/wild-type reciprocal crosses was cut with BglII (–) or BglII and MluI (+) and hybridized with probe msi (Figure 1B).The unmethylated and methylated alleles are 8.2 and 9.7 kb, respectively.
In summary, analysis of parental-specific expression in an allele completely deleted for the Igf2r promoter confirms that the Igf2r promoter has no role in a silent or an active form in imprinting genes in this cluster. These results also show that transcriptional overlap between the Air and Igf2r RNAs is not needed for silencing Air on the maternal chromosome and Slc22a2 and Slc22a3 on the paternal chromosome.
Discussion
Paternal expression of the Air RNA is required for silencing Igf2r, Slc22a2 and Slc22a3 on the same chromosome but has no effect on maternal expression of these three genes; thus, imprinted silencing is strictly a cis-acting mechanism (Sleutels et al., 2002). On the paternal chromosome, Air expression from an antisense-orientated promoter lying within intron 2 of Igf2r creates a potential transcriptional overlap of 29 kb with the 5′ end of that gene. A transcriptional overlap allows the possibility that Air could silence the overlapped gene by a mechanism different from those that silence non-overlapped genes. For example, Igf2r could be silenced by promoter occlusion (Villemure et al., 2001) or by the formation of a double-stranded RNA intermediate that could induce an RNA interference (RNAi) post-transcriptional silencing response (Hannon, 2002). RNAi was initially excluded from involvement in imprinting, since it was described as trans-acting and imprinting requires a cis-acting mechanism. However, the identification of a putative cis-acting form of RNAi in the nucleus of Schizosaccharomyces pombe involved in centromeric heterochromatin (Volpe et al., 2002) allows the possibility of a similar mechanism in mammals. If Igf2r was repressed because of transcriptional overlap with Air on the same chromosome, then silencing of the upstream Slc22a2 and Slc22a3 may be a secondary event due to spreading of repressive chromatin from the silenced Igf2r allele. The experiments described here were thus designed to test the role of the Igf2r promoter, the role of transcriptional overlap in cis between Igf2r and Air and, finally, whether the silencing of Slc22a2 and Slc22a3 is secondary to the action of Air on Igf2r.
The relevance of the Igf2r promoter in the imprinting mechanism
All three replacement alleles contained the same 1100 bp TKneo cassette combined with different deletions of the endogenous Igf2r promoter, and together they allowed the analysis of the ‘foreign’ TK promoter in different ‘host’ environments. The V1 and V2 alleles share the same distal deletion endpoint (codon 38 of exon 1). The V1 deletion extends 330 bp upstream to exon 1, whereas the V2 deletion extends to codon 28 of exon 1. The deletion in the Igf2r-TKneo allele extends 2733 bp upstream and 1560 bp downstream to exon 1 and thus removes all of the Igf2r promoter. All three alleles showed imprinted maternal-specific expression of the neomycin gene expressed by the foreign TK promoter and also maintained wild-type imprinted expression of Air, Slc22a2 and Slc22a3. Thus, imprinting of the foreign TK promoter resembled that of the endogenous Igf2r promoter in the presence or the absence of this promoter. In addition, a role for the endogenous Igf2r promoter in the imprinting mechanism acting on this gene cluster is excluded. The TK and the Igf2r promoters are very different. Igf2r has a CpG-island-type promoter with a 1 kb CG-rich core (Stoeger et al., 1993). The TK promoter is composed of a 141 bp CpG-poor polyomavirus late-region fragment and a 129 bp CpG-rich herpes simplex promoter fragment (see Materials and methods). The common imprinting of these different promoters indicates that Air may silence in a non-specific, promoter-independent manner that supports arguments that Air has intrinsic silencing properties. A comparable promoter-independent mode of silencing has been observed for the Xist non-coding RNA that is responsible for X-chromosome inactivation (Wutz and Jaenisch, 2000). Although the presence or the absence of the Igf2r promoter had no influence on the imprinted expression of the replacement TK promoter, the absence of methylation on the maternal allele was affected. The wild-type Igf2r promoter is normally free of methylation on maternal transmission but becomes partially methylated following paternal transmission in late embryogenesis (full paternal methylation is only found in postnatal stages; Stoeger et al., 1993). The TK promoter showed ‘wild-type’ methylation behavior only in the V2 allele that also contains the full Igf2r promoter. The V1 and Igf2r-TKneo alleles that are deleted for part or all of the Igf2r promoter were methylated on maternal transmission to a level of 40–50% of that seen following paternal transmission. This indicates that the Igf2r promoter contains sequences that act to prevent maternal methylation that are not present in the TK promoter.
Imprinted silencing of Slc22a2 and Slc22a3 is not a two-step mechanism
The Igf2r-del allele substituted a loxP site for a 4682 bp fragment containing the complete Igf2r promoter and exon 1 and lacked any transcription downstream from the deleted promoter (Figures 1E and 4B). This allele was used to test the role of transcriptional overlap in cis between Igf2r and Air in silencing this imprinted gene cluster. The results show that imprinted expression of the remaining genes (Air, Slc22a2 and Slc22a3) was unaffected by the removal of the Igf2r promoter. All three genes maintained their wild-type pattern of imprinted expression and, in addition, the methylation imprint on the maternal Air promoter was unchanged. Thus, transcriptional overlap between Air and Igf2r is not needed for paternal silencing of Slc22a2 and Slc22a3 or maternal silencing of Air. Since the absence of the Igf2r promoter precluded the existence of a ‘silenced’ promoter, these results also exclude models in which silencing of a promoter in cis is necessary for subsequent silencing of the neighboring Slc22a2 and Slc22a3 (Zwart et al., 2001). Models based on maternal Igf2r transcription playing a role in maintaining maternal methylation on the Air promoter are also now excluded. In addition, this results question the general applicability of a multiple CpG-island requirement for an imprinting mechanism (Onyango et al., 2000).
Slc22a2 and Slc22a3 are paternally silenced by Air but have no transcription overlap with it (Zwart et al., 2001). The demonstration here that this silencing is independent of Igf2r indicates that Air has a direct action on these genes, supporting arguments that Air has intrinsic silencing properties. Despite their common regulation by Air, imprinted silencing of Slc22a2 and Slc22a3 is different in quality from that of Igf2r. The molecular basis of this difference is unknown. One correlation can be made: the degree of imprinted silencing on the paternal allele correlates with distance from the Air promoter. The Igf2r promoter is closest (29 kb) and is silenced in embryo and adult, Slc22a2 is 170 kb distant and is silenced in embryonic placenta and partly silenced in adult kidney (expression is limited to these tissues) and Slc22a3 is 215 kb distant and is silenced in 11.5 but not 15.5 d.p.c. placentas and has widespread bi-allelic expression in adult tissues (Zwart et al., 2001). Thus, Air may have reduced ability to affect promoters with distance. Currently, we have no explanation of why two non-CpG-island promoters in this region (Mas and Slc22a1) escape imprinted silencing despite close proximity to the Air promoter.
Homology and imprinted gene silencing
The physical arrangement of the Air non-coding RNA in this imprinted gene cluster (Figure 1A), as well as the demonstration that other non-coding RNAs are expressed from antisense-orientated promoters within introns of imprinted protein-coding genes (Rougeulle et al., 1998; Smilinich et al., 1999; Lee et al., 2000; Wroe et al., 2000), presents a strong argument that homology and double-stranded RNA formation between non-coding RNAs and target gene transcripts may be involved in imprinted gene silencing. The results presented here, however, show that transcription overlap of 29 kb between Igf2r and Air is not needed for paternal silencing of Air or maternal silencing of the upstream Slc22a2 and Slc22a3. Although these data argue against a role for homology in silencing these specific genes, other possibilities exist whereby homology could play a role in the silencing mechanism at this locus. One possibility is that homology exists between the interspersed repeats present in the mature Air RNA and in the unspliced precursor Slc22a2 and Slc22a3 RNAs. Another possibility is that transcripts in the opposite orientation to Air could exist in the region upstream of the Igf2r promoter and contribute to gene silencing in the Igf2r-del allele described here. At this time, however, an analysis of all expressed sequence tags (ESTs) mapped to this interval (mouse chromosome 17; 12.168–12.288 Mbp; http://www.ensembl.org/Mus_musculus/contigview) shows an abundance of antisense ESTs corresponding to Air but no significant sense transcription in this region. Since silencing of Igf2r cannot be tested in the Igf2r-del allele, it also remains a possibility that Air has two independent silencing modes that act in parallel to silence genes in this cluster: one based on transcriptional overlap, acting on Igf2r (and on foreign promoters replacing Igf2r); and one independent of transcriptional overlap, acting on Slc22a2 and Slc22a3.
The Air RNA may have intrinsic cis silencing properties
In summary, the results presented here show that neither prior silencing of Igf2r nor the transcriptional overlap between Air and Igf2r are necessary for Slc22a2 and Slc22a3 silencing. Thus, models based on this 29 kb transcriptional overlap and those based on a two-step mechanism for silencing Slc22a2 and Slc22a3 are excluded from operating at this imprinted gene cluster. The absence of a two-step mechanism for silencing Slc22a2 and Slc22a3, combined with the lack of specificity in promoters susceptible to Air, allows the possibility that the Air RNA has intrinsic promoter-independent cis silencing properties. However, the existence of nearby genes that are not silenced by Air also indicates that not all promoters are equally susceptible to silencing. Based on these findings, the silencing model we propose for the Air RNA is analogous but different to the Xist model for X-chromosome inactivation in mammals (Avner and Heard, 2001). This model proposes that the Air RNA would generate a cis silencing effect that can repress susceptible gene promoters within a specific region whose boundaries are not yet known. Although the limitation of silencing to a small subchromosomal region marks a major difference between Air and Xist, the suggestion that X-chromosome inactivation evolved from a localized form of imprinting that initially affected only a small region of the X chromosome supports this model (Graves, 1996; Lyon, 1999; Lee, 2003).
Materials and methods
Generation of the Igf2r-TKneo allele
The targeting vector contained a 12.4 kb EcoRV fragment (bp 89 965–102 345; AJ249895) isolated from BAC 18p11 (Research Genetics). A 4682 kb SnaBI–SfuI fragment (bp 95 005–99 687; AJ249895) that includes the entire Igf2r CpG-island promoter and exon 1 was replaced by a loxP-flanked cassette of 1.2 kb containing a herpes simplex TK promoter, neomycin resistance gene and polyadenylation signal obtained from pMC1neo-poly(A) (bp 455–1597; U43612; Stratagene). This construct left 5.0 and 2.7 kb for recombination at the 5′ and 3′ ends, respectively. E14 ES cells (15 × 106) were electroporated with 0.02 mg of NotI linearized targeting construct and selected with 0.2 mg/ml G418; the targeting efficiency was 2%. Correctly targeted ES cells were identified by DNA blot, and chimeric mice were subsequently generated by injecting heterozygous Igf2r-neo ES cells into C57/Bl6 blastocysts (Hogan et al., 1994). All mice, except the V1 and V2 mice, were maintained on an FVB/N background and identified by DNA blot analyses. The V1 and V2 mice were maintained on a C57/Bl6 background. Embryos and placentas (including membranes) were collected after timed mating where the vaginal plug counts as 0.5 d.p.c.
DNA and methylation analyses
Genomic DNA preparation and DNA blots were performed according to standard procedures. Digestion of methyl-sensitive enzymes was monitored by hybridization with mitochondrial DNA (Walsh et al., 1998). The following methylation analyses probes were used (Figure 1B): kodel, a 325 bp fragment (bp 102 813–103 137; AJ249895); msi, an SfuI–MluI fragment (bp 124 992–126 086; AJ249895); and tai, an EcoRI–HindIII fragment (bp 94 104–94 986; AJ249895).
RNA analyses
Total RNA was isolated with Tri Reagent (Molecular Research Center). RNase protection analysis (RPA) was performed with the RPAIII kit (Ambion). The probes for RPA were tkneo and Airneo: a 685 bp PstI fragment (bp 923–1554; U43611) from the neomycin resistance gene taken from plasmid pMC1neo-poly(A) (Stratagene) and cloned into pBluescriptII (Stratagene) cut with NcoI to generate the Airneo and the tkneo template. The Airneo probe (T3) is 446 bp and protects 379 bp of Air RNA. The tkneo probe (T7) is 365 bp and protects 305 bp from the neomycin gene. The Aprt probe is a 252 bp XhoI–XbaI fragment (bp 2165–2417; M11310) that protects 134 bp of Aprt exon 3. The MlMs1 probe is an MluI–MseI fragment (bp 126 086–126 293; AJ249895) that overlaps multiple Air transcription starts and detects 207, 171 and 148 bp for Air RNA (Sleutels et al., 2002). For RNA blot analyses, the following probes were used: Igf2r, exons 3–6 cDNA fragment; Slc22a2 (bp 989–1605; AJ006036); Slc22a3 (bp 1–2766; AF078750); and Gapd (bp 1–1066; NM_008084).
Acknowledgments
Acknowledgements
We thank Karin van Veen, Karin van het Wout and Paul Krimpenfort for help with the ES cells and generating the chimeric mice and Cre transgenic mice; Nell Bosnie for taking care of the mice; Anton Berns for help and encouragement throughout this project; and Laura Spahn for reading the manuscript. The Dutch Cancer Society (KWF) supported this research. D.P.B. is supported by the Austrian Academy of Science.
References
- Avner P. and Heard,E. (2001) X-chromosome inactivation: counting, choice and initiation. Nat. Rev. Genet., 2, 59–67. [DOI] [PubMed] [Google Scholar]
- Beechey C.V., Cattanach,B.M. and Blake,A. (2001) MRC Mammalian Genetics Unit, Harwell, Oxfordshire. World Wide Web Site—Mouse Imprinting Data and References (http://www.mgu.har.mrc.ac.uk/imprinting/imprinting.html).
- Bell A.C., West,A.G. and Felsenfeld,G. (2001) Insulators and boundaries: versatile regulatory elements in the eukaryotic genome. Science, 291, 447–450. [DOI] [PubMed] [Google Scholar]
- Benzow K.A. and Koob,M.D. (2002) The KLHL1-antisense transcript (KLHL1AS) is evolutionarily conserved. Mamm. Genome, 13, 134–141. [DOI] [PubMed] [Google Scholar]
- Engemann S., Strodicke,M., Paulsen,M., Franck,O., Reinhardt,R., Lane,N., Reik,W. and Walter,J. (2000) Sequence and functional comparison in the Beckwith–Wiedemann region: implications for a novel imprinting centre and extended imprinting. Hum. Mol. Genet., 9, 2691–2706. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick G.V., Soloway,P.D. and Higgins,M.J. (2002) Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet., 32, 426–431. [DOI] [PubMed] [Google Scholar]
- Graves J.A. (1996) Mammals that break the rules: genetics of marsupials and monotremes. Annu. Rev. Genet., 30, 233–260. [DOI] [PubMed] [Google Scholar]
- Gribnau J., Diderich,K., Pruzina,S., Calzolari,R. and Fraser,P. (2000) Intergenic transcription and developmental remodeling of chromatin subdomains in the human β-globin locus. Mol. Cell, 5, 377–386. [DOI] [PubMed] [Google Scholar]
- Hannon G.J. (2002) RNA interference. Nature, 418, 244–251. [DOI] [PubMed] [Google Scholar]
- Hark A.T., Schoenherr,C.J., Katz,D.J., Ingram,R.S., Levorse,J.M. and Tilghman,S.M. (2000) CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature, 405, 486–489. [DOI] [PubMed] [Google Scholar]
- Hogan B.L.M., Beddington,R.S.P., Costantini,F. and Lacy,E. (1994) Manipulating the Mouse Embryo. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Horike S., Mitsuya,K., Meguro,M., Kotobuki,N., Kashiwagi,A., Notsu,T., Schulz,T.C., Shirayoshi,Y. and Oshimura,M. (2000) Targeted disruption of the human LIT1 locus defines a putative imprinting control element playing an essential role in Beckwith–Wiedemann syndrome. Hum. Mol. Genet., 9, 2075–2083. [DOI] [PubMed] [Google Scholar]
- Krystal G.W., Armstrong,B.C. and Battey,J.F. (1990) N-myc mRNA forms an RNA–RNA duplex with endogenous antisense transcripts. Mol. Cell. Biol., 10, 4180–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau M.M., Stewart,C.E., Liu,Z., Bhatt,H., Rotwein,P. and Stewart,C.L. (1994) Loss of the imprinted IGF2/cation-independent mannose 6-phosphate receptor results in fetal overgrowth and perinatal lethality. Genes Dev., 8, 2953–2963. [DOI] [PubMed] [Google Scholar]
- Lee J.T. (2003) Molecular links between X-inactivation and autosomal imprinting: X-inactivation as a driving force for the evolution of imprinting? Curr. Biol., 13, R242–R254. [DOI] [PubMed] [Google Scholar]
- Lee Y.J., Park,C.W., Hahn,Y., Park,J., Lee,J., Yun,J.H., Hyun,B. and Chung,J.H. (2000) Mit1/Lb9 and Copg2, new members of mouse imprinted genes closely linked to Peg1/Mest(1). FEBS Lett., 472, 230–234. [DOI] [PubMed] [Google Scholar]
- Li E. (2002) Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet., 3, 662–673. [DOI] [PubMed] [Google Scholar]
- Li E., Beard,C. and Jaenisch,R. (1993) Role for DNA methylation in genomic imprinting. Nature, 366, 362–365. [DOI] [PubMed] [Google Scholar]
- Li A.W., Seyoum,G., Shiu,R.P. and Murphy,P.R. (1996) Expression of the rat BFGF antisense RNA transcript is tissue-specific and developmentally regulated. Mol. Cell. Endocrinol., 118, 113–123. [DOI] [PubMed] [Google Scholar]
- Ludwig T., Eggenschwiler,J., Fisher,P., D’Ercole,A.J., Davenport,M.L. and Efstratiadis,A. (1996) Mouse mutants lacking the type 2 IGF receptor (IGF2R) are rescued from perinatal lethality in Igf2 and Igf1r null backgrounds. Dev. Biol., 177, 517–535. [DOI] [PubMed] [Google Scholar]
- Lyle R. et al. (2000) The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat. Genet., 25, 19–21. [DOI] [PubMed] [Google Scholar]
- Lyon M.F. (1999) Imprinting and X-chromosome inactivation. Results Probl. Cell Differ., 25, 73–90. [DOI] [PubMed] [Google Scholar]
- Onyango P., Miller,W., Lehoczky,J., Leung,C.T., Birren,B., Wheelan,S., Dewar,K. and Feinberg,A.P. (2000) Sequence and comparative analysis of the mouse 1-megabase region orthologous to the human 11p15 imprinted domain. Genome Res., 10, 1697–1710. [DOI] [PubMed] [Google Scholar]
- Potter S.S. and Branford,W.W. (1998) Evolutionary conservation and tissue-specific processing of Hoxa 11 antisense transcripts. Mamm. Genome, 9, 799–806. [DOI] [PubMed] [Google Scholar]
- Reik W. and Walter,J. (2001) Genomic imprinting: parental influence on the genome. Nat. Rev. Genet., 2, 21–32. [DOI] [PubMed] [Google Scholar]
- Rougeulle C. and Heard,E. (2002) Antisense RNA in imprinting: spreading silence through Air. Trends Genet., 18, 434–437. [DOI] [PubMed] [Google Scholar]
- Rougeulle C., Cardoso,C., Fontes,M., Colleaux,L. and Lalande,M. (1998) An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat. Genet., 19, 15–16. [DOI] [PubMed] [Google Scholar]
- Schmidt J.V., Levorse,J.M. and Tilghman,S.M. (1999) Enhancer competition between H19 and Igf2 does not mediate their imprinting. Proc. Natl Acad. Sci. USA, 96, 9733–9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk F., Baron,U. and Rajewsky,K. (1995) A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res., 23, 5080–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleutels F. and Barlow,D.P. (2002) The origins of genomic imprinting in mammals. Adv. Genet., 46, 119–163. [DOI] [PubMed] [Google Scholar]
- Sleutels F., Zwart,R. and Barlow,D.P. (2002) The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature, 415, 810–813. [DOI] [PubMed] [Google Scholar]
- Smilinich N.J. et al. (1999) A maternally methylated CpG island in KvLQT1 is associated with an antisense paternal transcript and loss of imprinting in Beckwith–Wiedemann syndrome. Proc. Natl Acad. Sci. USA, 96, 8064–8069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeger R., Kubicka,P., Liu,C.G., Kafri,T., Razin,A., Cedar,H. and Barlow,D.P. (1993) Maternal-specific methylation of the imprinted mouse Igf2r locus identifies the expressed locus as carrying the imprinting signal. Cell, 73, 61–71. [DOI] [PubMed] [Google Scholar]
- Szabo P., Tang,S.H., Rentsendorj,A., Pfeifer,G.P. and Mann,J.R. (2000) Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr. Biol., 10, 607–610. [DOI] [PubMed] [Google Scholar]
- Tasheva E.S. and Roufa,D.J. (1995) Regulation of human RPS14 transcription by intronic antisense RNAs and ribosomal protein S14. Genes Dev., 9, 304–316. [DOI] [PubMed] [Google Scholar]
- Thorvaldsen J.L. and Bartolomei,M.S. (2000) Molecular biology. Mothers setting boundaries. Science, 288, 2145–2146. [DOI] [PubMed] [Google Scholar]
- Thorvaldsen J.L., Duran,K.L. and Bartolomei,M.S. (1998) Deletion of the H19 differentially methylated domain results in loss of imprinted expression of H19 and Igf2. Genes Dev., 12, 3693–3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villemure J.F., Savard,N. and Belmaaza,A. (2001) Promoter suppression in cultured mammalian cells can be blocked by the chicken β-globin chromatin insulator 5′HS4 and matrix/scaffold attachment regions. J. Mol. Biol., 312, 963–974. [DOI] [PubMed] [Google Scholar]
- Volpe T.A., Kidner,C., Hall,I.M., Teng,G., Grewal,S.I. and Martienssen,R.A. (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science, 297, 1833–1837. [DOI] [PubMed] [Google Scholar]
- Walsh C.P., Chaillet,J.R. and Bestor,T.H. (1998) Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat. Genet., 20, 116–117. [DOI] [PubMed] [Google Scholar]
- Wang Z.Q., Fung,M.R., Barlow,D.P. and Wagner,E.F. (1994) Regulation of embryonic growth and lysosomal targeting by the imprinted Igf2/Mpr gene. Nature, 372, 464–467. [DOI] [PubMed] [Google Scholar]
- Wroe S.F., Kelsey,G., Skinner,J.A., Bodle,D., Ball,S.T., Beechey,C.V., Peters,J. and Williamson,C.M. (2000) An imprinted transcript, antisense to Nesp, adds complexity to the cluster of imprinted genes at the mouse Gnas locus. Proc. Natl Acad. Sci. USA, 97, 3342–3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A. and Jaenisch,R. (2000) A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol. Cell, 5, 695–705. [DOI] [PubMed] [Google Scholar]
- Wutz A., Theussl,H.C., Dausman,J., Jaenisch,R., Barlow,D.P. and Wagner,E.F. (2001) Non-imprinted Igf2r expression decreases growth and rescues the Tme mutation in mice. Development, 128, 1881–1887. [DOI] [PubMed] [Google Scholar]
- Zwart R., Sleutels,F., Wutz,A., Schinkel,A.H. and Barlow,D.P. (2001) Bidirectional action of the Igf2r imprint control element on upstream and downstream imprinted genes. Genes Dev., 15, 2361–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]