Abstract
While membrane insertion of single-spanning membrane proteins into the endoplasmic reticulum (ER) is relatively well understood, it is unclear how multi-spanning proteins integrate. We have investigated the cotranslational ER integration of a double-spanning protein that is derived from leader peptidase. Both transmembrane (TM) segments are inserted into the membrane by the Sec61 channel. While the first, long and hydrophobic TM segment (TM1) inserts into the lipid bilayer on its own, the second, shorter TM anchor (TM2) collaborates with TM1 during its integration. TM1 diffuses away from the Sec61 complex in the absence of TM2, but is close to Sec61 when TM2 arrives inside the channel. These data suggest that the exit of a weak TM segment from the Sec61 channel into the lipid phase can be facilitated by its interaction with a previously integrated strong and stabilizing TM anchor.
Keywords: endoplasmic reticulum membrane/membrane proteins/translocation
Introduction
Both membrane and secretory proteins are transported across the membrane of the endoplasmic reticulum (ER) through the same protein-conducting channel, which is formed by the heterotrimeric Sec61 complex (for review, see Hegde and Lingappa, 1997; Matlack et al., 1998). Secretory proteins are transported completely across the membrane, whereas membrane proteins have some domains translocated to the luminal side of the membrane, while others remain in the cytosol. Transmembrane (TM) segments, which anchor proteins in the membrane, need to be released laterally through the walls of the channel into the lipid phase. Membrane proteins can have many different topologies. The simplest are single-spanning membrane proteins, whose biosynthesis has been studied in some detail (Martoglio et al., 1995; Mothes et al., 1997; Heinrich et al., 2000; Kida et al., 2000). In contrast, very little is known about the integration of multi-spanning membrane proteins.
The cotranslational translocation of both single- and multi-spanning proteins starts in the cytosol when the first hydrophobic domain, a cleavable signal sequence or a TM segment, emerges from the ribosome and is recognized by the signal recognition particle (SRP) (for review, see Walter and Johnson, 1994; Rapoport et al., 1996). The complex of ribosome, nascent polypeptide chain, and SRP is then targeted to the ER membrane, where the ribosome binds to the Sec61 channel and the polypeptide chain inserts into the channel (for review, see Matlack et al., 1998). In the case of a well studied signal–anchor type I protein, which spans the membrane once with an NlumCcyt (cyt, cytosol; lum, luminal) orientation, targeting to the membrane occurs when ∼10 hydrophobic residues of the TM segment have emerged from the ribosome, a length similar to the hydrophobic core of a signal sequence (Heinrich et al., 2000). The second phase in the integration of a signal–anchor type I protein occurs when the entire TM segment has emerged from the ribosome. At this point, the TM domain laterally exits the Sec61 channel and enters the lipid phase. Finally, during the third phase, the TM segment has stopped moving perpendicular to the plane of the membrane. As the cytosolic domain of the membrane protein is extended by translation, the anchor diffuses in the bilayer away from the channel.
Membrane integration of a single-spanning membrane protein can be described as a passive partitioning process, where the Sec61 channel allows the TM segment to equilibrate between the hydrophilic interior of the channel and the hydrophobic core of the lipid bilayer (Heinrich et al., 2000). The equilibrium constant for partitioning into lipid varies according to the hydrophobicity of the TM segment. Long and hydrophobic membrane anchors partition into the lipid bilayer more readily than shorter or less hydrophobic TM segments, which prefer to stay at the amphipathic interphase between lipid and channel for an extended time period (Heinrich et al., 2000).
The molecular details of the integration of multi-spanning proteins are poorly understood. The simplest model is the ‘linear insertion model’ (initially proposed by Blobel, 1980; Sabatini et al., 1982). In this model, TM segments integrate in the order of their synthesis from the N- to the C-terminus. The orientation of the first TM segment defines that of all downstream TM segments. However, a linear insertion model does not explain why downstream TM segments can sometimes alter the topology of upstream TM segments. For example, when two TM sequences with the same preferred orientation are placed adjacent to one another, one may be expelled from the membrane (McGovern et al., 1991; Gafvelin and von Heijne, 1994). An alternative to the ‘linear insertion model’ is the ‘bundling model’, which proposes that the TM segments of a nascent protein remain within the translocation channel until termination of translation and that they are then released as a group into the lipid phase (Borel and Simon, 1996). It is conceivable that for certain proteins both models apply: some hydrophobic anchors may leave the translocation site on their own, while others that are too short or contain too many charges, may need to assemble with other TM segments in the channel before they can stably integrate into the lipid phase as one hydrophobic unit. There is indeed evidence for some model membrane proteins that pairs of TM segments interact with one another before they can be integrated into the lipid phase (Skach and Lingappa, 1993; Lin and Addison, 1995).
To study the simplest case of a multi-spanning protein, we have employed a membrane protein with two TM segments, derived from the bacterial leader peptidase. In the final structure, the first TM segment (TM1), which is long and hydrophobic, interacts with the second, less hydrophobic TM segment (TM2) to form a coiled-coil (Whitley et al., 1993). In a previous study, we used a construct containing only TM1 to investigate in detail the integration of a single-spanning protein (Heinrich et al., 2000). Early stages of membrane integration of the double-spanning protein are therefore identical to those of the protein containing only TM1. Here, we have concentrated on subsequent stages during which TM2 enters the lipid bilayer. Using a variety of methods, we provide evidence that the integration of the weakly hydrophobic TM2 segment requires the presence of TM1. The TM1 segment initially diffuses away from the channel into the lipid phase, but is close to the channel during the integration of TM2.
Results
Model proteins
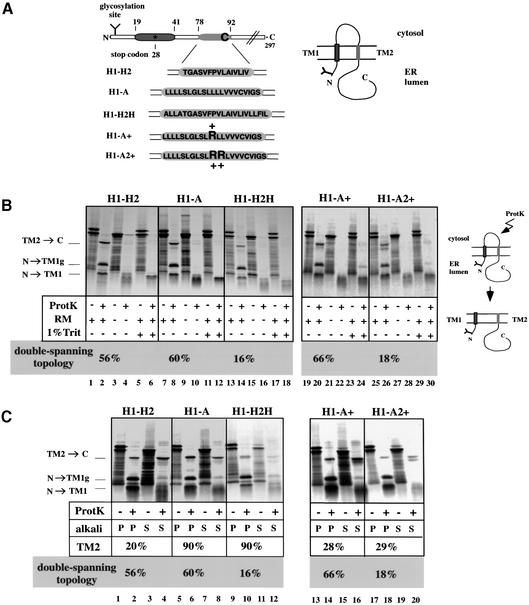
To study the ER integration of the TM segments of a double-spanning protein, we used constructs derived from the bacterial leader peptidase (Figure 1A; von Heijne, 1989). Leader peptidase uses essentially the same integration mechanism in bacteria and mammals, both in vivo and in vitro (Gafvelin et al., 1997). Previous experiments with a single-spanning construct have shown that leader peptidase inserts in a cotranslational and SRP-dependent manner into rough canine pancreatic microsomes (RMs) (Heinrich et al., 2000). For the present study we used a series of double-spanning constructs. The H1–H2 protein contains the two TM segments of leader peptidase (Figure 1A). The fully synthesized and integrated protein has a small N-terminal domain (18 amino acids, with an N-glycosylation site at position 3) in the ER lumen, followed by the first hydrophobic TM segment of 23 amino acids (TM1), a cytosolic loop of 35 residues, a second TM segment of 15 amino acids (TM2), and a luminal domain of 205 residues (Figure 1A, scheme on the right). The TM2 segment of leader peptidase is unusually short, and experiments in Escherichia coli have shown that in the mature protein it interacts with the TM1 segment to form a coiled-coil structure in the membrane (Whitley et al., 1993). To test the membrane integration of TM2, we generated different constructs in which it was replaced by other TM segments. In construct H1–H2H, the TM2 segment was made more hydrophobic by extension at the N-terminus by four and at the C-terminus by five hydrophobic residues. In construct H1–A, TM2 was replaced by the TM anchor of the asialoglycoprotein receptor, a single-spanning type II protein with an NcytClum topology. This TM segment is longer and more hydrophobic than wild-type TM2, and it would not be expected to interact with TM1. Finally, we generated constructs in which one or two positive charges were introduced into the TM2 segment of the H1–A protein (H1–A+ and H1–A2+). In the H1–A+ construct, the second TM segment has about the same overall hydrophobicity as wild-type TM2. For all constructs, the TM1 segment and the connecting loop between TM1 and TM2 are identical.
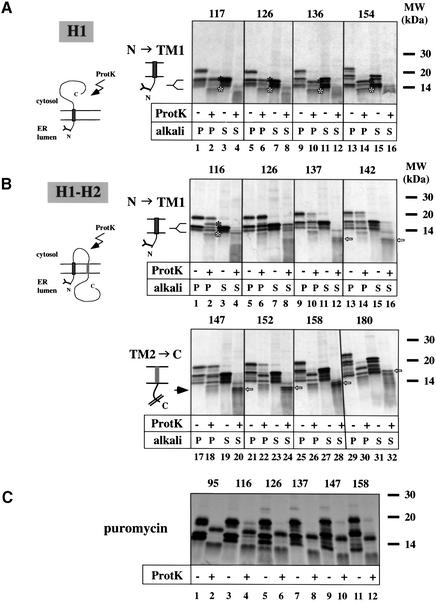
Fig. 1. Membrane topology of double-spanning constructs. (A) Scheme of the double-spanning proteins. The first TM segment (TM1) is shown in dark gray. In photocrosslinking experiments, the probe was introduced at position 28 (star) by suppression of a stop codon. The amino acid sequences of the second TM segments (TM2) of the constructs used in this study are shown in light gray boxes. For crosslinking with a bifunctional reagent, a single cysteine was introduced at position 89 of H1–H2 or at position 90 of H1–A, H1–A+ and H1–A2+. Arginines introduced into TM2 of H1–A are indicated (+). (B) Radiolabeled full-length proteins were synthesized in the presence or absence of RMs. Proteolysis was carried out with proteinase K (ProtK) in the presence or absence of 1% Triton (Trit), as indicated. All samples were analyzed by SDS–PAGE followed by autoradiography. Quantification resulted in estimates of the fraction of proteins with a double-spanning topology. (C) Radiolabeled full-length proteins were synthesized in the presence of RMs and treated with proteinase K as indicated. The membranes were sedimented at alkaline pH, and the pellet (P) and the supernatant (S) fractions were analyzed. The percentage of TM2 in the alkali extracted membrane pellet and the percentage of double-spanning topology are given.
Investigating the integration of the double-spanning constructs by proteolysis
The full-length proteins were synthesized in vitro in the presence of [35S]methionine and RMs, and analyzed by SDS–PAGE. For all proteins two bands were observed (Figure 1B, lanes 1, 7, 13, 19 and 25). The slower migrating band corresponds to the glycosylated, full-length protein since it is not seen in the absence of membranes (lanes 3, 9, 15, 21 and 27) or when a competitor peptide containing an N-glycosylation site was added (data not shown). Glycosylation indicates that the N-terminus has reached the ER lumen.
To determine the membrane topology of the proteins, we used a protease protection assay. Proteolysis of the full-length polypeptides by proteinase K generated three major bands (Figure 1B, lanes 2, 8, 14, 20 and 26). The slowest migrating band corresponds to TM2 and the luminal C-terminus (TM2→C), which are protected from proteolysis by the membrane. The faster migrating bands correspond to the glycosylated and non-glycosylated forms of a fragment comprising the protected luminal N-terminus and TM1 (N→TM1g and N→TM1, respectively). These bands disappeared after proteinase K treatment if the membranes were omitted or solubilized in detergent (Figure 1B, lanes 3–6, 9–12, 15–18, 21–24 and 27–30, respectively). The percentage of double-spanning topology for the different constructs was calculated by dividing the radioactivity in the TM2→C fragment by the total radioactivity in the non-proteolyzed polypeptides, taking into account the loss of methionines during proteolysis. This quantification showed that 56–66% of all H1–H2, H1–A or H1–A+ polypeptide chains span the membrane twice, while only 16–18% of the H1–H2H or H1–A2+ chains adopt a double-spanning topology. Most of the other polypeptide chains adopt a single-spanning topology, with TM1 as the only TM anchor. Some chains are not targeted to the membrane and are thus completely degraded by protease treatment. These data show that the structure of the second TM segment has an influence on the translocation of the C-terminal domain.
To investigate the stability of the second TM segment in the lipid bilayer, full-length proteins were treated with proteinase K to digest the connecting loop between TM1 and TM2. Subsequently, the membranes were sedimented at alkaline pH to extract all but stably integrated TM segments. For all constructs, the full-length, glycosylated proteins and the large majority of N→TM1 and N→TM1g were found in the alkali-extracted membrane fraction (Figure 1C, compare lanes 2 versus 4, 6 versus 8, 10 versus 12, 14 versus 16 and 18 versus 20). Some of the full-length, non-glycosylated proteins remained in the alkali supernatant and probably represent chains that were not targeted to the ER membrane. The cleaved-off TM2 segments of the H1–H2 and H1–A+ proteins were mostly extracted from the membrane (20–28% remained in the alkali pellet; lanes 2 versus 4 and 14 versus 16), indicating that the less hydrophobic TM2 segments are not stably integrated in the membrane on their own. The integration of TM2 of the construct H1–A2+ was very inefficient, but in this case too a significant percentage of TM2 was extractable by alkali (lane 18 versus 20). In contrast, with the constructs H1–A and H1–H2H, the cleaved TM2 remained in the membrane pellet after alkali extraction (∼90% for both H1–A and H1–H2H; lanes 6 and 10, respectively), consistent with the fact that both have a significantly more hydrophobic segment than those in wild-type leader peptidase or the H1–A+ construct. Interestingly, TM2 of H1–H2H was less efficiently integrated than that of the wild-type protein (16% compared with 56%), but was more stable once integrated. This result suggests that the interaction between TM1 and TM2, which may be disturbed in the mutant H1–H2H, facilitates the membrane integration of TM2.
Following membrane integration of TM2 with a bifunctional crosslinker
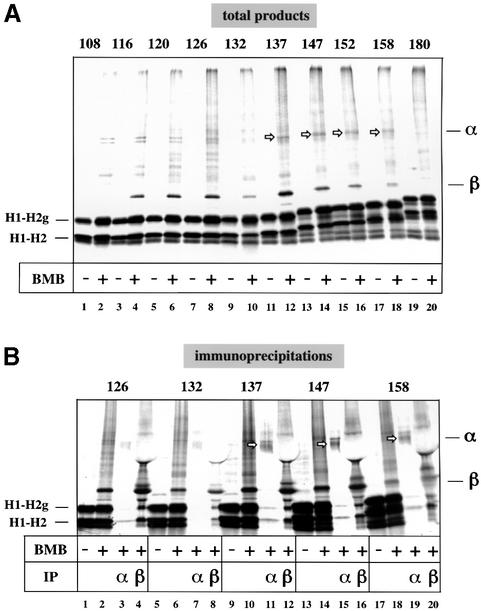
To study the integration of TM2 of the H1–H2 construct in more detail, we generated a series of translocation intermediates and used bismaleimidobutane (BMB) to crosslink a single cysteine in the nascent chain with cysteines in components of the translocation channel. We removed a native cysteine in TM1 at position 35 and introduced a cysteine into TM2 of H1–H2 at position 89 (Cys89) (see Figure 1A). Truncated mRNAs were translated in the presence of [35S]methionine and RMs to generate translocation intermediates of increasing length, each with its C-terminus still associated with the tRNA in the ribosome. To increase the incorporation of radioactivity, the last two amino acids of each translocation intermediate were changed to methionines. One half of each sample was treated with BMB, the other served as a control (Figure 2A). The membranes were then sedimented at alkaline pH and the samples analyzed by SDS–PAGE. To follow the integration of TM2, we used nascent chains that were long enough to allow complete membrane insertion of TM1 (Heinrich et al., 2000). The TM2 segment is expected to emerge from the ribosome with its N-terminus first, but eventually it must turn around and adopt an NcytClum orientation in the membrane.
Fig. 2. Probing the membrane integration of TM2 of H1–H2 by chemical crosslinking. (A) Radiolabeled H1–H2 chains with the only cysteine in TM2 at position 89 were synthesized in the presence of RMs. The chain length is indicated above the gel. The samples were treated with the bifunctional crosslinker BMB, where indicated. Crosslinks to Sec61α (open arrows) and Sec61β are indicated. (B) As in (A), but the samples were also analyzed by immunoprecipitation (IP) with antibodies to Sec61α and Sec61β. Crosslinks to Sec61α are indicated by open arrows.
In the absence of crosslinker, two major bands were seen for all tested chain lengths, corresponding to the glycosylated and non-glycosylated nascent chains (H1–H2g and H1–H2, respectively). After incubation with BMB, additional bands could be detected. With the 108mer or shorter chains, no crosslinks to any channel component were observed (Figure 2A, lane 2 and data not shown; some unidentified bands may represent crosslinks to ribosomal proteins). With a chain of 108 residues, the cysteine at position 89 has not yet emerged from the ribosome, since ∼30 residues at the C-terminus are buried within the ribosome (Malkin and Rich, 1967; Blobel and Sabatini, 1970; Mothes et al., 1994).
With the 116mer, crosslinks to Sec61β appeared (Figure 2A, compare lane 4 with 3 and 2), as confirmed by immunoprecipitation with specific antibodies (data not shown). At this length, more than half of TM2 has emerged from the ribosome (see scheme in Figure 7). All longer chains up to 158 residues also contact Sec61β as shown by immunoprecipitation with antibodies to Sec61β (lanes 4, 8, 12, 16 and 20).
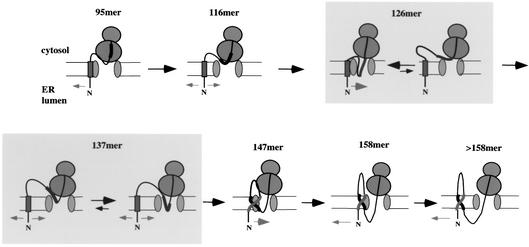
Fig. 7. Model for the integration of TM2. The scheme shows the behavior of the TM segments of H1–H2 chains during the integration of TM2 into the ER membrane, as described in the text. Gray boxes indicate equilibria between two different states, one of which is predominating (unequal arrows).
Crosslinks to Sec61α appeared with a chain length of 137 residues (Figure 2A, lane 12, open arrow), as confirmed by immunoprecipitation with specific antibodies against Sec61α (Figure 2B, lane 11). At this point, the C-terminus of TM2 has emerged from the ribosome by ∼15 residues, presumably allowing the TM segment to flip to its final NcytClum topology, in which Cys89 at the C-terminus of TM2 is near Sec61α at the luminal side of the ER. Interestingly, at a chain length of 126 amino acids very little crosslinking to Sec61α was detected, although the entire TM2 has emerged from the ribosome (Figure 2B, lane 3). These chains are probably too short for TM2 to span the membrane in its final NcytClum orientation (see scheme in Figure 7). Crosslinks to Sec61α were detectable with chains up to 158 amino acids in length, similar to those to Sec61β (Figure 2A, lanes 12, 14, 16 and 18). With longer chains, both Sec61β and Sec61α crosslinks disappeared (Figure 2A, lane 20), consistent with the assumption that TM2 diffuses in the lipid phase away from the channel.
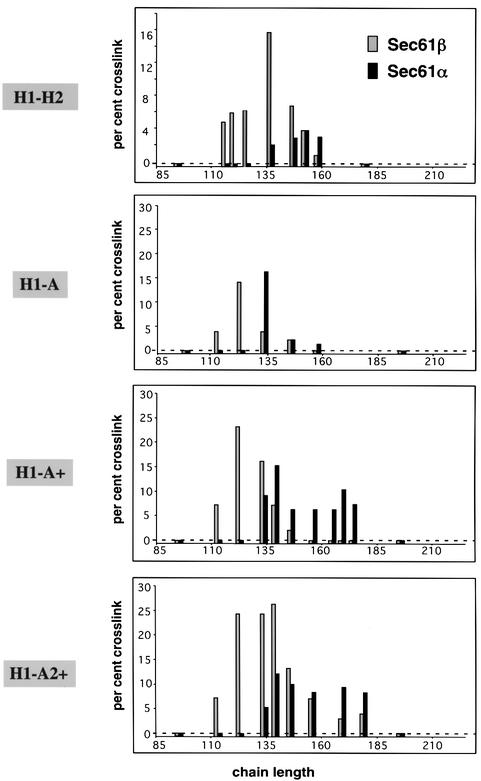
Similar experiments were performed with the constructs H1–A, H1–A+ and H1–A2+, and the crosslinks to Sec61α and Sec61β obtained for the various chain lengths were quantitatively compared (Figure 3). While H1–H2 chains gave strong crosslinks to Sec61β up to 152 residues with a maximum at 137 amino acids (Figure 3, H1–H2), H1–A chains gave most prominent crosslinks with chains of 123 amino acids and almost none beyond 134 residues (Figure 3, H1–A). Thus, although the second TM segment is at a similar position in both constructs (residues 78–92 and 74–94, respectively), TM2 in the H1–H2 protein appears to stay close to the channel over a longer range of chain lengths. A similar conclusion could be drawn from the Sec61α crosslinking results.
Fig. 3. Quantification of Sec61α and Sec61β crosslinks generated by BMB. The graphs show the average of three independent BMB crosslinking experiments with H1–H2, H1–A, H1–A+ and H1–A2+ chains, performed as in Figure 2A. The radioactivity in the crosslinked product is given as a percentage of the total radioactivity in the sample. Light gray bars correspond to Sec61β crosslinks, dark gray bars to Sec61α crosslinks. Bars lower than the dotted line indicate that no crosslinks were observed at the corresponding chain length.
Since TM2 in the H1–H2 protein has a relatively low hydrophobicity, we tested whether this may be the reason for its prolonged residence at the channel, as had been observed previously for single-spanning constructs. Indeed, with the H1–A+ protein crosslinks to Sec61α remained strong for chains between 134 and 174 amino acids, at which the H1–A chains no longer gave crosslinks (Figure 3). In contrast, Sec61β appeared and disappeared at the same chain lengths as in the original H1–A construct. Together these data indicate that TM2 enters the translocation channel at the same chain length in both cases, but it does not diffuse readily into the lipid phase away from the channel if it contains a positive charge. The results with two positive charges in TM2 (H1–A2+) were similar, except that Sec61β crosslinks were also seen for longer chains (Figure 3).
Following membrane insertion by protease protection
To probe more precisely the membrane insertion of the H1–H2 protein as a function of chain length, we performed protease protection experiments (Figure 4). After proteolysis, the membranes were sedimented at alkaline pH, and pellet (P) and supernatant (S) fractions were analyzed by SDS–PAGE. In these experiments we compared proteins containing only TM1 (H1) with those containing both TM1 and TM2 (H1–H2).
Fig. 4. Probing the membrane integration of single-spanning H1 and double-spanning H1–H2 by protease protection. (A) Radiolabeled H1 chains were synthesized in the presence of RMs and treated with proteinase K (ProtK), where indicated. The membranes were sedimented at alkaline pH and both pellet (P) and supernatant (S) fractions were analyzed. Closed and open stars indicate the positions of glycosylated and non-glycosylated fragments comprised of TM1 and the N-terminal segment. The scheme on the left indicates the expected topology of the protein. Molecular weights of marker proteins are indicated on the right. (B) As in (A), but with H1–H2 chains. Open arrows indicate the membrane-protected C-terminal fragment. (C) Radiolabeled H1–H2 chains of different lengths were treated with puromycin before proteinase K digestion, as indicated. The pellet fractions after alkaline extraction were analyzed.
With H1 chains of 117 residues or longer, two major bands were generated, corresponding to glycosylated and non-glycosylated nascent chains (Figure 4A, lanes 1, 5, 9 and 13). Proteolysis produced two fragments that had the same size for all tested chain lengths. These correspond to the protected glycosylated and non-glycosylated polypeptide segments containing the luminal domain and TM1 (lanes 2, 6, 10 and 14, closed and open stars, respectively). As shown before (Heinrich et al., 2000), these fragments are generated because the domain following TM1 emerges into the cytosol and becomes accessible to proteinase K.
With H1–H2 chains of 116 residues, two major bands were also synthesized (Figure 4B, lane 1). After proteolysis, the same bands were generated as for the single-spanning protein, corresponding to the fragment comprising the glycosylated and non-glycosylated N-terminal domain and TM1 (e.g. lane 2, closed and open star, respectively). However, in contrast to H1, some of the nascent chains were not degraded (lane 2; see also quantification in Figure 5A), suggesting that the cytosolic loop between the two TM segments is shielded by the ribosome and the associated Sec61 channel. Protection by the ribosome is indeed indicated by the effect of puromycin, a reagent that releases nascent chains from ribosomes (Figure 4C, lane 4). Upon puromycin treatment, the cytosolic loop between TM1 and TM2 was completely degraded by proteinase K. In contrast, the same loop is not protected in the single-spanning construct (Figure 4A). One interpretation of these results is that a population of TM1 segments in the double-spanning protein is close to the translocation site. With a chain length of 116 residues, more than half of TM2 has emerged from the ribosome, suggesting that the proximity of TM1 to the translocation site may coincide with TM2 entering the channel.
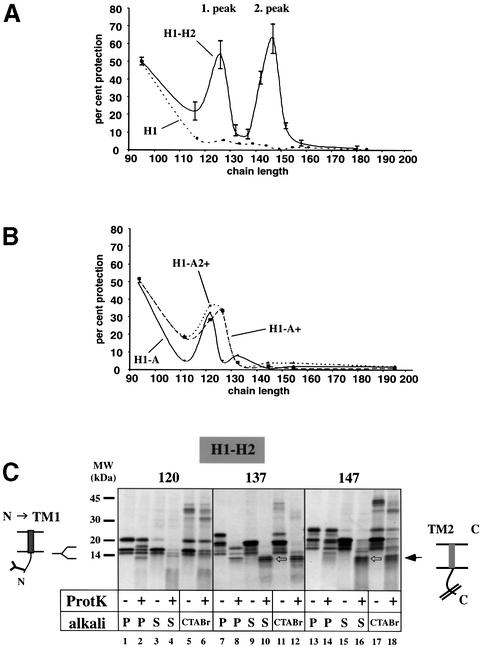
Fig. 5. Probing membrane integration by protease protection. (A) H1 and H1–H2 chains of different length were generated in the presence of membranes and treated with proteinase K. The sum of radioactivity in glycosylated and non-glycosylated chains that remained after proteolysis is given as percentage of the radioactivity in the undigested chains. (B) As in (A), but with H1–A, H1–A+ and H1–A2+ chains. (C) Radiolabeled H1–H2 chains of different lengths were generated in the presence of membranes. The samples were incubated with proteinase K where indicated. One half of the samples was sedimented at alkaline pH and the pellet (P) and supernatant (S) fractions were analyzed by SDS–PAGE. The other half was precipitated with CTABr. The N- and C-terminal (open arrows) fragments protected from proteolysis by the membrane are indicated.
An even more dramatic difference between H1–H2 and H1 was seen with chains of 126 residues. The majority of the H1–H2 chains were protected from proteolysis (Figure 4B, compare lane 6 with 5), while the single-spanning chains were completely degraded. Again, upon puromycin treatment, the H1–H2 chains were accessible to proteolysis (Figure 4C, lane 6). At this chain length, TM2 has completely emerged from the ribosome and is expected to be inside the Sec61 channel.
With the 137mer, the large majority of H1–H2 chains were sensitive to protease digestion (Figure 4B, lane 10 versus 9; see also Figure 5C, lane 8 versus 7). At this chain length, TM2 may have turned around, allowing the loop between TM1 and TM2 to emerge from underneath the ribosome. At a chain length of 142 or 147 amino acids, the cytosolic loop between TM1 and TM2 regains protection from protease digestion (Figure 4B, lanes 14 versus 13 and 18 versus 17, respectively). At this point, TM2 may span the membrane in the right orientation and associate with TM1.
Beginning with the 152mer, the connecting loop between TM1 and TM2 again becomes accessible to protease digestion (Figure 4B, compare lane 22 with 21), likely because TM1 and TM2 together can now diffuse away from the channel.
With H1–H2 chains longer than 137 amino acids, a sharp band appeared in the supernatant fraction after protease digestion and alkali extraction (see open arrows in Figure 4B, lane 12 and Figure 5C, lanes 10 and 16). The size of this fragment increased with chain length. It corresponds to the protease-protected second TM segment and the following C-terminus. As expected from the fact that the C-terminus should be associated with the ribosome as peptidyl-tRNA, the fragment could be precipitated with cetyltrimethylammonium bromide (CTABr) (Figure 5C), whereas it was not precipitable after treatment with puromycin (data not shown). These data show that membrane insertion of TM2 first occurs with a chain length of 137 amino acids, consistent with the assumption that at this chain length, it first adopts its final orientation.
The quantification of three independent protease digestion experiments similar to those in Figure 4A and B is shown in Figure 5A. Single-spanning H1 chains longer than 117 amino acids are completely accessible to protease digestion, whereas double-spanning H1–H2 chains display two peaks of protease protection at 126 and 147 residues. According to our interpretation, the first peak would be indicative of TM2 entering the translocation channel and the second of an interaction between TM1 and TM2. To test this assumption, we performed similar protease protection experiments with the double-spanning constructs H1–A, H1–A+ and H1–A2+. The quantification shows that they gave a first peak at about the same position as the H1–H2 construct (Figure 5B), indicating that in all cases TM2 enters the translocation channel at the same chain length. However, none of the constructs gave a second peak, consistent with the predicted lack of interaction between TM1 and TM2.
Probing the environment of TM1 by photocrosslinking
To test whether TM1 is close to the channel during the integration of TM2, we performed photocrosslinking experiments. A stop codon was introduced at position 28, in the middle of TM1, for both H1–H2 and H1 chains (Figure 1A). It was suppressed in vitro by translation in the presence of a modified phenylalanyl-suppressor tRNA, resulting in the site-specific incorporation of a photoreactive group into TM1 (High et al., 1993). Nascent chains of different lengths were synthesized in the presence of RMs. One half of each sample was irradiated with UV light, while the other served as a control. Irradiated samples were subjected to immunoprecipitation with antibodies to Sec61α (Figure 6A and B).
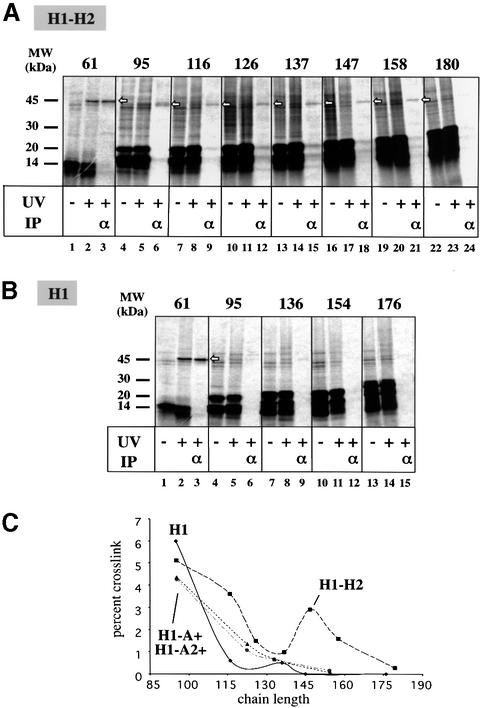
Fig. 6. Probing the environment of TM1 by photocrosslinking. (A) Photocrosslinking experiments were performed in the presence of membranes with H1–H2 chains of different lengths carrying a photoreactive probe in TM1 at position 28. The samples were irradiated with UV light where indicated and sedimented at alkaline pH or subjected to immunoprecipitation (IP) with antibodies to Sec61α. Open arrows indicate crosslinks to Sec61α. (B) As in (A), but with H1 chains. (C) Quantification of photocrosslinking experiments as shown in (A) and (B). Also shown are the results with H1–A+ and H1–A2+ chains. In each case, the radioactivity in the Sec61α crosslinked product was divided by that in the total sample. Given is the crosslinking efficiency relative to that observed for the 61mer (set to 100%).
With the 61mer, strong crosslinks to Sec61α were detected for both H1–H2 and H1 (Figure 6A and B, lanes 3, open arrows). At this chain length, the ribosome-nascent chain complex is targeted to the membrane where TM1 contacts Sec61α. For all chain lengths longer than 61 amino acids strong lipid crosslinks could also be observed (seen in shorter exposures of the autoradiograms; see also Heinrich et al., 2000), indicating that TM1 has entered a lipid environment.
With the 95mer, weak crosslinks to Sec61α could be observed for both H1–H2 and H1 (Figure 6A and B, lanes 6). As shown before (Heinrich et al., 2000), a 95mer of H1 is fully integrated into the membrane, but its diffusion radius is restricted because the C-terminus of the polypeptide chain is tethered to the ribosome.
For all longer H1 chains, crosslinks to Sec61α were extremely weak, indicating that the majority of the TM segments have diffused away from the channel (Figure 6B, lanes 9, 12 and 15). In contrast, TM1 of H1–H2 was in contact with Sec61α for chains up to 158 amino acids in length (Figure 6A, lanes 9, 12, 15, 18 and 21). Quantification showed that the H1–H2 crosslinks had a small peak around 147 residues. The crosslinks disappeared when the chains reached a length of 180 amino acids (Figure 6A, lane 24 and panel C). These results are consistent with a model in which TM1 in H1–H2 is close to the channel during membrane integration of TM2.
Photocrosslinking experiments were also performed to test the behavior of TM1 in the H1–A+ and H1–A2+ constructs (Figure 6C). The crosslinks of TM1 to Sec61α decreased with chain length in a similar way as with the single-spanning H1 construct and did not show a peak with longer chains as observed with the H1–H2 construct. Together, these results indicate that an imperfect TM2 has an extended residence close to the channel, but does not cause TM1 to return to or remain close to the channel; the latter appears to require a specific interaction between TM2 and TM1.
Discussion
We have investigated how double-spanning proteins integrate into the ER membrane. The constructs employed are derived from bacterial leader peptidase, a protein with two interacting TM segments. Since we had previously characterized in detail how the first TM segment (TM1) integrates in the absence of TM2, we could now focus on the integration of the second TM segment and its influence on TM1. We therefore used chain lengths at which TM1 is already integrated into the membrane. Our results lead to a multi-stage model for the integration of TM2. Figure 7 shows the different stages during the integration of wild-type TM2 (in the H1–H2 protein).
The earliest stage is exemplified by chains of 95 residues. At this point, TM2 is still completely inside the ribosome and the construct essentially behaves like a single-spanning protein. TM1 begins to laterally diffuse away from the translocation site, rendering the cytosolic loop of ∼50% of the nascent chain population accessible to protease digestion.
The next stage occurs with chains of ∼116 residues. Approximately half of TM2 has now emerged from the ribosome. Crosslinking to Sec61β indicates that TM2 is close to the channel. In contrast to the single-spanning protein of about the same length, a significant fraction of the double-spanning chains is protected from protease digestion; puromycin experiments suggest that the cytosolic loop between TM1 and TM2 is shielded by the ribosome. Although it is possible that protease protection of the loop is caused by an altered ribosome-channel junction, we consider this unlikely. The ribosome remains bound to the channel at all times during the biosynthesis of a membrane protein (Mothes et al., 1997), and electron microscopy shows that the structure of the ribosome-channel complex remains the same whether or not a nascent chain is present (Ménétret et al., 2000; Beckmann et al., 2001). In addition, proximity of TM1 to the translocation site is supported by the observed crosslinks between TM1 and Sec61α. It therefore appears that a population of TM1 segments must be close to the channel when TM2 enters the channel.
At a chain length of 126 residues, the entire hydrophobic segment of TM2 is out of the ribosome, but the chain is too short to allow its final NcytClum orientation. The TM segment could either adopt an NlumCcyt orientation inside the channel, or it may be exposed to the cytosol (see scheme in Figure 7). The former possibility is supported by the fact that the cytosolic loop between TM1 and TM2 is protected from proteolysis. In addition, it appears unlikely that the hydrophobic TM segment would favor the aqueous environment of the cytosol. Thus, at this stage, both TM2 and at least some portion of the preceding cytosolic loop are probably inside the channel (Figure 7), restricting the lateral diffusion radius of TM1.
When the chain reaches a length of 137 residues, the second TM segment can begin to reorient to its final NcytClum topology. Again, there may be an equilibrium between different states (see scheme in Figure 7), but a major population of chains must have its TM2 segment in the translocation channel since crosslinks to Sec61α and Sec61β can be detected. Because the C-terminus is relatively short, there are likely to be physical restraints that prevent TM2 from exiting laterally into the lipid phase. Reorientation of TM2 allows the preceding hydrophilic loop to return to the cytosolic compartment and explains why this segment is now accessible to proteolysis. Interestingly, Cys89 in the nascent chain gave Sec61α crosslinks with the 137mer, but not the 126mer, even though in both cases the residue is expected to be inside the channel; the reactive cysteine in Sec61α is thus likely at the luminal side of the channel.
At a chain length of 147 residues, the C-terminal portion following TM2 is long enough to allow TM2 to laterally exit the channel and associate with TM1. Assembly occurs close to the channel, as both TM1 and TM2 can be crosslinked to Sec61α and Sec61β. The connecting loop between TM1 and TM2 is shielded by the ribosome from proteolysis, supporting the notion that both TM segments are close to the channel. This is supported by the peak of Sec61α crosslinks, albeit relatively small, seen at this chain length (Figure 6C). In contrast, TM1 was not in contact with Sec61α in a single-spanning construct of similar length.
Finally, with chains of 158 residues or longer, the complex of TM1 and TM2 begins to diffuse away from the channel. As the luminal loop connecting TM2 with the ribosome at the channel becomes longer, the diffusion radius of the TM segments in the plane of the membrane also increases. Accordingly, the cytosolic loop becomes protease accessible and crosslinks to Sec61α and Sec61β gradually disappear. At 180 residues, long before completion of the chain, both TM segments have completely left the channel environment.
Most of the stages described for the integration of wild-type TM2 are also observed with the other TM2 segments tested (in the H1–A, H1–A+, H1–A2+ proteins). The major difference occurs when the chains reach a length of ∼147 residues, at which TM1 and TM2 of the wild-type protein associate with one another. With the non-interacting TM segments there is neither a second peak of protease protection nor of crosslinks between TM1 and Sec61α. We therefore conclude that in these cases TM2 enters the lipid phase independently of TM1. Whether or not TM2 efficiently partitions into lipid depends on its hydrophobicity. TM2 segments containing one or two positive charges (in the H1–A+ and H1–A2+ proteins) remained close to the translocation channel for an extended range of chain lengths. This is similar to our previous results in which one or two positive charges introduced into the TM1 segment of a single-spanning construct also caused prolonged residence at the channel–lipid interphase (Heinrich et al., 2000). Together, these data indicate that two non-interacting TM segments in a double-spanning membrane protein undergo independent partitioning into the lipid phase, the efficiency of which is determined by their hydrophobicity. If there is no interaction between the TM segments, TM2 needs to be hydrophobic to cause efficient translocation of the polypeptide segment following it. For example, with two positive charges in TM2 (in the H1–A2+ construct), little double-spanning topology was achieved. On the other hand, from the results with the H1–H2H construct it appears that overall hydrophobicity may not be the only property required for TM2 to initiate translocation; perhaps, the Pro residue in this TM segment prevents it from efficiently partitioning into the lipid phase unless it associates with TM1.
From our data it appears that the ‘linear insertion’ and ‘bundling’ models represent the extremes in the integration of multi-spanning proteins. For non-interacting TM segments, integration may indeed occur in a linear and independent fashion, as postulated by the linear insertion model. However, in cases such as wild-type leader peptidase, TM segments are not independently integrated. For none of the cases studied here, the bundling model would apply because both TM segments leave the translocation channel long before termination of translation. The observation that a TM segment can stay close or even return to the translocation channel to associate with another TM segment may explain why multi-spanning membrane proteins can often be expressed as two separate fragments. It seems possible that the TM segments of one fragment can pick up the other fragment from its translocation site to form a membrane-compatible structure.
Our previous results indicated that a TM segment with an NlumCcyt orientation adopts its final topology as soon as the entire segment has emerged from the ribosome, indicating that it enters the translocation channel ‘head first’. The present results suggest that the second TM segment, which ultimately resides in the membrane with the opposite orientation, also initially inserts into the channel ‘head first’. This mechanism ensures that the hydrophobic TM segment is not exposed to the hydrophilic cytosolic compartment. How far the N-terminus of the TM2 segment enters the channel is unclear and may depend on charges, length and folding of the preceding segment. However, consistent with the fact that the N-terminus of a polypeptide emerges first from the ribosome, ‘head first’ insertion of a TM segment into the channel may be the default pathway regardless of the final orientation.
Materials and methods
In vitro mutagenesis
The single-spanning protein is encoded by a modified pAlter plasmid (Promega) as described in Heinrich et al. (2000). Restriction sites for NcoI (CCATGG) and XhoI (CTCGAG) were introduced at base pairs 213–218 and 223–228, respectively. This allowed rapid introduction of different TM segments derived from the asialoglycoprotein receptor or the extended second TM segment from leader peptidase. Consequently, amino acid residues in the loop between TM segments at positions 71 and 72 were changed from lysine and proline to asparagine and histidine, respectively.
For photocrosslinking experiments, a stop codon was introduced in TM1 of H1–H2 at position 28 by converting a leucine codon to UAG. For BMB crosslinking, the cysteine at position 35 of H1–H2 was converted into an alanine, and a cysteine was introduced at position 89. For H1–A constructs, the native cysteine at position 90 was left unchanged. For H1–A+ and H1–A2+ constructs, the leucine at positions 84, and the leucines at positions 84 and 85, respectively, were converted into arginines. All mutants were verified by nucleotide sequencing.
Transcription and translation
Full-length mRNA was generated by in vitro transcription with SP6 RNA polymerase (Ribomax, Promega) of pAlter plasmids coding for H1–H2 and H1–H2H after linearization with BamH1. pAlter plasmids coding for H1–A, H1–A+ and H1–A2+ were linearized with PstI. Truncated mRNAs were generated by transcription of PCR-amplified portions of the gene, using an SP6 primer and various 3′-end primers. The latter also introduced two additional methionine residues to facilitate detection of the proteins.
In vitro translation was carried out in the wheat germ system (Mothes et al., 1997). Ten microliters translation mix contained 12.5 nM SRP, 1.2 equivalents of canine pancreatic RMs and 5 µCi [35S]methionine (Amersham). The samples were incubated for 6–13 min at 26°C, depending on the length of the truncated mRNA, followed by 5 min with 5 mM cycloheximide to stop chain elongation. Full-length proteins were generated by translating full-length mRNA for 30 min. After translation, all samples were centrifuged at 75 000 r.p.m. for 15 min in a Beckman 100.3 rotor and the membrane pellet was resuspended in the same volume of membrane buffer (50 mM HEPES pH 7.6, 250 mM sucrose, 120 mM potassium acetate, 2 mM magnesium acetate and 0.2 mM spermidine).
Photoreactive phenylalanine derivates were incorporated into polypeptide chains by translation of truncated mRNAs containing a stop codon in the presence of 1.5 pmol suppressor tRNA carrying trifluoromethyl-diazirinyl phenylalanine, followed by cycloheximide treatment (Martoglio et al., 1995).
Crosslinking
For crosslinking with BMB, the samples were diluted after translation with 150 µl membrane buffer (Heinrich et al., 2000), and the membranes were sedimented by centrifugation for 15 min at 75 000 r.p.m. in a table-top ultracentrifuge (rotor 100.3; Beckman). They were resuspended in 10 µl membrane buffer, and one half was incubated with 1 mM BMB for 30 min at room temperature. The reaction was stopped with 20 mM β-mercaptoethanol.
Crosslinking with photoreactive phenylalanine derivatives was performed as described in Heinrich et al. (2000). It should be noted that different crosslinking methods were used to probe the proximity of TM1 and TM2 to the Sec61p channel; we have been unable to incorporate photoreactive probes into TM2, because the suppression of stop codons that are too far away from the initiation codon proves to be difficult.
Product analysis
Treatment with proteinase K (0.5 mg/ml for full-length proteins and 0.1 mg/ml for nascent chains) was carried out for 8 min on ice and stopped with 2 mM phenylmethylsulfonyl fluoride.
For alkali extraction, translation samples were diluted with 100 µl of ice-cold 100 mM disodium carbonate/NaOH (pH 12.5) and centrifuged at 75 000 r.p.m. for 10 min in a Beckman 100.3 rotor. Precipitation of tRNA-linked products with CTABr was performed as described by Krieg et al. (1986).
For immunoprecipitation with antibodies against Sec61α or Sec61β, the samples were denatured for 15 min at 40°C in 2% SDS sample buffer lacking dithiothreitol and diluted with 10 volumes 1% Triton X-100, 50 mM Tris–HCl (pH 7.5) and 150 mM potassium acetate. For immunoprecipitation of Sec61α, affinity-purified antibodies covalently coupled to protein A–Sepharose beads were used (60 min at 4°C). In the case of Sec61β, 1 µl of antiserum was used for 2 eq of microsomes, and after 15 min at 4°C, the incubation was continued for 30 min in the presence of 20 µl of protein A–Sepharose beads.
SDS–PAGE was carried out with 7.5–17.5% linear polyacrylamide gels. The samples were prepared for electrophoresis by incubation at 95°C for 10 min.
Acknowledgments
Acknowledgements
We are grateful to Drs K.Cannon and B.Tsai for critical reading of the manuscript. T.A.R. is a Howard Hughes Medical Institute Investigator. The work was supported by a grant from the N.I.H. to T.A.R.
References
- Beckmann R., Spahn,C.M.T., Eswar,N., Helmers,J., Penczek,P.A., Sali,A., Frank,J. and Blobel,G. (2001) Architecture of the protein-conducting channel associated with the 80S ribosome. Cell, 107, 361–372. [DOI] [PubMed] [Google Scholar]
- Blobel G. (1980) Intracellular protein topogenesis. Proc. Natl Acad. Sci. USA, 77, 1496–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G. and Sabatini,D.D. (1970) Controlled proteolysis of nascent polypeptides in rat liver cell fractions. I. Location of the polypeptides within ribosomes. J. Cell Biol., 45, 130–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borel A.C. and Simon,S.M. (1996) Biogenesis of polytopic membrane proteins: membrane segments assemble within translocation channels prior to membrane integration. Cell, 85, 379–389. [DOI] [PubMed] [Google Scholar]
- Gafvelin G. and von Heijne,G. (1994) Topological ‘frustration’ in multi-spanning E.coli inner membrane proteins. Cell, 77, 401–412. [DOI] [PubMed] [Google Scholar]
- Gafvelin G., Sakaguchi,M., Andersson,H. and von Heijne,G. (1997) Topological rules for membrane protein assembly in eukaryotic cells. J. Biol. Chem., 272, 6119–6127. [DOI] [PubMed] [Google Scholar]
- Hegde R.S. and Lingappa,V.R. (1997) Membrane protein biogenesis: regulated complexity at the endoplasmic reticulum. Cell, 91, 575–582. [DOI] [PubMed] [Google Scholar]
- Heinrich S.U., Mothes,W., Brunner,J. and Rapoport,T.A. (2000) The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane segment. Cell, 102, 233–244. [DOI] [PubMed] [Google Scholar]
- High S., Andersen,S.S.L., Görlich,D., Hartmann,E., Prehn,S., Rapoport,T.A. and Dobberstein,B. (1993) Sec61p is adjacent to nascent type-I and type-II signal–anchor proteins during their membrane insertion. J. Cell Biol., 121, 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida Y., Sakaguchi,M., Fukuda,M., Mikoshiba,M. and Mihara,K. (2000) Membrane topogenesis of a type I signal–anchor protein, mouse synaptotagmin II, on the endoplasmic reticulum. J. Cell Biol., 150, 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg U.C., Walter,P. and Johnsson,A.E. (1986) Photocrosslinking of the signal sequence of nascent preprolactin to the 54 kilodalton polypeptide of the signal recognition particle. Proc. Natl Acad. Sci. USA, 83, 8604–8608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. and Addison,R. (1995) A novel integration signal that is composed of two transmembrane segments is required to integrate the Neurospora plasma membrane H+-ATPase into microsomes. J. Biol. Chem., 270, 6935–6941. [DOI] [PubMed] [Google Scholar]
- Malkin L.I. and Rich,A. (1967) Partial resistance of nascent polypeptide chains to proteolytic digestion due to ribosomal shielding. J. Mol. Biol., 26, 329–346. [DOI] [PubMed] [Google Scholar]
- Martoglio B., Hofmann,M.W., Brunner,J. and Dobberstein,B. (1995) The protein-conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell, 81, 207–214. [DOI] [PubMed] [Google Scholar]
- Matlack K.E.S., Mothes,W. and Rapoport,T.A. (1998) Protein translocation—tunnel vision. Cell, 92, 381–390. [DOI] [PubMed] [Google Scholar]
- McGovern K., Ehrmann,M. and Beckwith,J. (1991) Decoding signals for membrane protein assembly using alkaline phosphatase fusions. EMBO J., 10, 2773–2782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ménétret J.F., Neuhof,A., Morgan,D.G., Plath,K., Radermacher,M., Rapoport,T.A. and Akey,C.W. (2000) The structure of ribosome-channel complexes engaged in protein translocation. Mol. Cell, 6, 1219–1232. [DOI] [PubMed] [Google Scholar]
- Mothes W., Prehn,S. and Rapoport,T.A. (1994) Systematic probing of the environment of a translocating secretory protein during translocation through the ER membrane. EMBO J., 13, 3973–3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mothes W., Heinrich,S.U., Graf,R., Nilsson,I., von Heijne,G., Brunner,J. and Rapoport,T.A. (1997) Molecular mechanism of membrane protein integration into the endoplasmic reticulum. Cell, 89, 523–533. [DOI] [PubMed] [Google Scholar]
- Rapoport T.A., Jungnickel,B. and Kutay,U. (1996) Protein transport across the eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu. Rev. Biochem., 65, 271–303. [DOI] [PubMed] [Google Scholar]
- Sabatini D.D., Kreibich,G., Morimoto,T. and Adesnik,M. (1982) Mechanisms for the incorporation of proteins in membranes and organelles. J. Cell Biol., 92, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skach W.R. and Lingappa,V.R. (1993) Amino-terminal assembly of human P-glycoprotein at the endoplasmic reticulum is directed by cooperative actions of two internal sequences. J. Biol. Chem., 268, 23552–23561. [PubMed] [Google Scholar]
- von Heijne G. (1989) Control of topology and mode of assembly of a polytopic membrane protein by positively charged residues. Nature, 341, 456–458. [DOI] [PubMed] [Google Scholar]
- Walter P. and Johnson,A.E. (1994) Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell. Biol., 10, 87–119. [DOI] [PubMed] [Google Scholar]
- Whitley P., Nilsson,L. and von Heijne,G. (1993) Three-dimensional model for the membrane segment of Escherichia coli leader peptidase based on disulfide mapping. Biochemistry, 32, 8534–8539. [DOI] [PubMed] [Google Scholar]