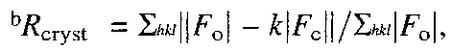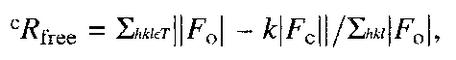Abstract
The Cdc14 family of dual-specificity protein phosphatases (DSPs) is conserved within eukaryotes and functions to down-regulate mitotic Cdk activities, promoting cytokinesis and mitotic exit. We have integrated structural and kinetic analyses to define the molecular mechanism of the dephosphorylation reaction catalysed by Cdc14. The structure of Cdc14 illustrates a novel arrangement of two domains, each with a DSP-like fold, arranged in tandem. The C-terminal domain contains the conserved PTP motif of the catalytic site, whereas the N-terminal domain, which shares no sequence similarity with other DSPs, contributes to substrate specificity, and lacks catalytic activity. The catalytic site is located at the base of a pronounced surface channel formed by the interface of the two domains, and regions of both domains interact with the phosphopeptide substrate. Specificity for a pSer-Pro motif is mediated by a hydrophobic pocket that is capable of accommodating the apolar Pro(P+1) residue of the peptide. Our structural and kinetic data support a role for Cdc14 in the preferential dephosphorylation of proteins modified by proline-directed kinases.
Keywords: Cdc14/dual-specificity phosphatase/proline-directed phosphatase/protein structure/X-ray crystallography
Introduction
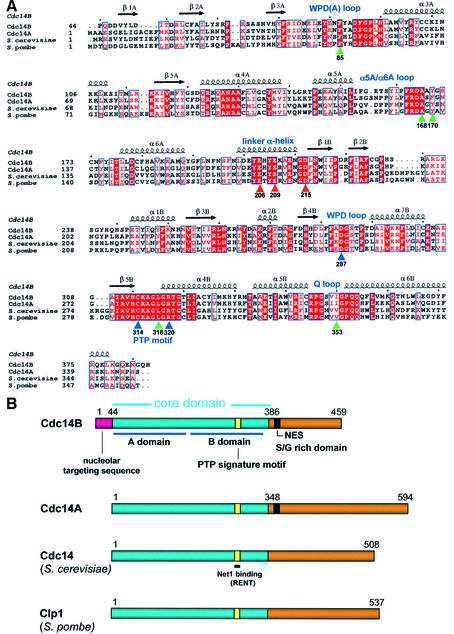
In eukaryotes, normal cell cycle progression and viability rely on the dual-specificity protein phosphatase (DSP) Cdc14 (Wan et al., 1992). Organisms with a mutated Cdc14 gene are unable to complete cytokinesis and/or exit from mitosis (Taylor et al., 1997; Morgan, 1999). The Cdc14 proteins of Saccharomyces cerevisiae, Schizosaccharomyces pombe and recently Caenorhabditis elegans have been extensively studied, and two human isoforms (Cdc14A and Cdc14B) were identified on the basis of sequence similarity to the budding yeast protein. Cdc14A and Cdc14B appear to possess similar biochemical properties to their homologues from other species (Bembenek and Yu, 2001; Kaiser et al., 2002). Cdc14 from diverse species share a conserved core of ∼350 amino acids located towards the N-terminus, and which harbours the conserved protein tyrosine phosphatase (PTP) signature motif HC(X)5R(S/T) (Figure 1). Regions C-terminal to the conserved core are highly divergent and share no structural similarities.
Fig. 1. Structural relationship between eukaryotic Cdc14 proteins. (A) Sequence alignment of budding and fission yeast Cdc14, and human Cdc14A and Cdc14B, within the conserved domain of ∼350 amino acids denoted in blue in (B). Residues that interact with the Pro(P+1) residue of the peptide are indicated by green arrows, residues of the acidic groove by red arrows and critical catalytic site residues by blue arrows. Secondary structural elements in the A- and B-domains are labelled with the suffix A and B, respectively. (B) Schematic of the primary structure of Cdc14 from human and yeast. The conserved domain is shown in blue. Within these regions, human Cdc14B shares 65, 36 and 40% identity with human Cdc14A, S.cerevisiae and S.pombe Cdc14, respectively. The positions of the nucleolar targeting sequence and NES are indicated.
Cdc14 of S.cerevisiae is recognized as the ultimate effector molecule of the mitotic exit network (MEN), a signal cascade that promotes the inactivation of the mitotic cyclin-dependent kinase (Cdk) Cdc28 at the end of anaphase (Traverso et al., 2001). The down-regulation of Cdc28 occurs by Cdc14-mediated dephosphorylation of the Cdk-modified residues of Cdh1, a co-activator of the anaphase promoting complex (APC). Activated (dephosphorylated) Cdh1 binds to the APC forming the APCCdh1 complex, the E3-ubiquitin ligase responsible for the ubiquitylation of Clb2 leading to the destruction of the Clb2/Cdc28 complex (Morgan, 1999). Regulation of Cdc14 activity in S.cerevisiae is achieved by three complex mechanisms controlling subcellular localization. For the majority of the cell cycle, Cdc14 is sequestered in the nucleolus by Net1 of the RENT (regulator of nucleolar silencing and telophase) complex (Visintin and Amon, 2000; Traverso et al., 2001). At anaphase, the FEAR (Cdc fourteen early anaphase release) network (Stegmeier et al., 2002) and later the MEN (Jaspersen et al., 1998; Geymonat et al., 2002) promote the release of Cdc14 into the cytoplasm, initially to further regulate its own translocation from the nucleolus, and then to dephosphorylate, hence activating Cdh1, and promote the destruction of Clb2. Inactivation of Cdk activity is further augmented by Cdc14-mediated dephosphorylation of two other Cdk substrates. Dephosphorylation of Sic1 prevents its degradation, hence promoting inhibitory interactions with Cdc28, whereas dephosphorylation of the transcription factor Swi5 stimulates Sic1 gene expression.
In contrast to budding yeast, the Cdc14 homologue of S.pombe Clp1 (also termed Flp1) is not required for cyclin degradation or the activation of the APC, and thus does not appear to promote mitotic exit (Cueille et al., 2001). However, Clp1 does interact with the fission yeast homologues of the MEN that are termed the SIN (septation initiation network). This network co-ordinates cytokinesis during nuclear division, and Clp1 localizes to both the mitotic spindle and the contractile ring. Clp1 differs from S.cerevisiae Cdc14 by regulating the G2/M transition. Cells deleted for Clp1 enter mitosis prematurely, whereas overexpression of the phosphatase delays mitotic entry by preventing dephosphorylation of Cdc2 on Tyr15 (Trautmann et al., 2001). Interactions with the cytoskeleton to facilitate cytokinesis also apply to the recently characterized Cdc14 of C.elegans, CeCDC14, which is essential for the localization of key components to the central spindle in anaphase and the midbody in telophase. Depletion of CeCDC14 by RNAi in embryos resulted in lethality as a consequence of poor central spindle formation and cytokinesis, rather than a defect in mitosis or the nuclear cycle. It is proposed that successful central spindle formation is dependent on CeCDC14 dephosphorylation of one or several Cdk-modified substrates (Gruneberg et al., 2002).
The roles of the two human isoforms of Cdc14 are poorly understood, but the potential for the regulation of both mitotic exit and cytokinesis is likely. In situ observations of fluorescently labelled Cdc14A in human cells show a localization and recruitment to interphase centrosomes following nuclear export. This release is mediated by a nuclear export signal (NES) in Cdc14A, located just beyond the C-terminal reaches of the phosphatase domain. Mutation of the NES results in the retention of Cdc14A in the nucleolus. Conversely, the N-terminal 44 amino acids are responsible for localizing Cdc14B to the nucleolus throughout interphase (Kaiser et al., 2002) (Figure 1B). Overexpression of Cdc14A results in premature centrosome splitting and an excessive number of mitotic spindles, whereas down-regulating endogenous Cdc14A using siRNA impairs centrosome splitting and prevents successful cytokinesis (Mailand et al., 2002). While the range of substrates for Cdc14A or Cdc14B is not well defined, in vitro studies have shown that Cdc14A has a clear preference for substrates of proline-directed kinases (Cdks and MAP kinases) modified at pSer/pThr-Pro motifs (Kaiser et al., 2002; Trautmann and McCollum, 2002). This is consistent with the observation that, just as the yeast Cdc14 dephosphorylates a Cdk-modified Cdh1, human Cdc14A can activate the human Cdh1 to constitute the APCCdh1 in vitro (Bembenek and Yu, 2001). Cdc14B may play an important role within the nucleolus during interphase and, as the nuclear membrane dissolves at prophase, Cdc14B will be released into the cytoplasm and may act on a wider range of Cdk- or MAPK-modified substrates during mitosis.
The diverse roles of Cdc14 to regulate cytokinesis and mitosis suggest that Cdc14 will act on different targets in each case to effect a subtly different result. However, the underlying element of continuity is the dephosphorylation of Cdk-modified substrates (Visintin et al., 1998; Trautmann and McCollum, 2002). A comprehensive understanding of the mechanisms of catalysis, and specificity for Cdk-modified substrates by Cdc14, requires structural investigation. To address this question, we have determined crystal structures of the core domain of human Cdc14B in both the apo state, and as a complex with a phosphopeptide substrate, at 2.2 Å resolution. These are the first reported X-ray crystallographic data for Cdc14. The overall structure illustrates a novel fold of two DSP domains arranged in tandem that may have evolved from an early gene duplication event of an ancestral DSP gene. The structure of Cdc14B demonstrates the molecular basis of its specificity for substrates with pSer-Pro and pThr-Pro motifs that are common to Cdk- and MAP kinase-modified proteins.
Results
Structure determination
To understand the three-dimensional (3D) structure of human Cdc14B (Mr 53 kDa), we expressed the full-length protein using the insect cell/baculovirus system, and purified the protein to near homogeneity. This form of the protein did not readily crystallize, although the appearance of small Cdc14B crystals were noted in hanging drops from an individual preparation of the protein after a period of 3 months. Analysis of the protein mass in the protein/crystal drop using SDS–PAGE revealed spontaneous and partial degradation of Cdc14B to a size of ∼40 kDa, suggesting that the crystals grew from a truncated form of the protein. Elective limited proteolysis was used to delineate the structurally stable domain that corresponded to the spontaneously truncated protein. Limited proteolysis of full-length Cdc14B using three different proteases yielded a stable product of ∼40 kDa, similar in size to the truncated form of Cdc14B obtained by spontaneous degradation. Edman sequencing revealed the N-terminus as Pro44, whereas an estimation of the C-terminus was based on the C-terminal boundary of the conserved catalytic domains of Cdc14A, Cdc14B and S.cerevisiae Cdc14. The resultant protein (residues Pro44–His386) when purified had a molecular mass, as judged by SDS–PAGE, equivalent to the partially degraded Cdc14B obtained by limited trypsinolysis and, moreover, readily crystallized. Significantly, this region of Cdc14B corresponds to the segment of sequence conservation within Cdc14 sequences from diverse species, and therefore represents the Cdc14 catalytic core (Figure 1).
Determination of the structure of wild-type apo Cdc14B was performed using the single anomalous dispersion method utilizing tungstate, a phosphate mimic and catalytic site inhibitor, as a heavy atom derivative. The concentration of tungstate used to derivatize Cdc14B was estimated from the concentration required to inhibit the Cdc14 catalytic activity towards p-nitrophenol-phosphate (pNPP; data not shown). The structure of wild-type apo Cdc14B was solved to 2.5 Å resolution, the diffraction limit of these crystals. Subsequently, we obtained crystals of a Cdc14B–phosphopeptide complex by substituting serine for the catalytic Cys314 residue. These crystals diffracted to 2.2 Å and were solved by molecular replacement using the apo Cdc14B structure (Table I). In both structures, residues Pro44–Lys379 are well defined in the electron density maps, whereas the C-terminal seven residues are disordered. Apo and complex Cdc14B share virtually identical conformations (see below). Because the higher resolution of the Cdc14–peptide complex resulted in a better model for the protein, we use this form as the basis of the description of molecular structure.
Table I. Crystallographic data and refinement statistics.
| Crystal parameters | Native | Tungstate | Peptide |
|---|---|---|---|
| Space group |
|
|
|
| a (Å) | 114.82 | 114.74 | 114.76 |
| b (Å) | 52.08 | 51.93 | 53.15 |
| c (Å) | 65.19 | 64.96 | 64.17 |
| β (°) | 118.24 | 118.63 | 117.48 |
| Z |
1 |
1 |
1 |
| Data collection and processing statistics |
|
|
|
| Resolution (Å) | 2.5 | 2.6 | 2.2 |
| Measurements (n) | 38 931 | 30 373 | 62 805 |
| Unique (n) | 11 713 | 10 480 | 17 049 |
| % complete | 99.2 (97.1) | 99.5 (99.6) | 97.2 (92.2) |
| Rsyma | 0.106 (0.345) | 0.119 (0.331) | 0.073 (0.24) |
|
I/σI |
6.2 |
6.8 |
7.1 (2.9) |
| Refinement statistics |
|
|
|
| Resolution (Å) | 2.5 | 2.6 | 2.2 |
| No. of reflectionswork (%) | 10 814 (91.6) | 9931 (84.5) | 16 051 (91.1) |
| No. of reflectionsfree (%) | 589 (5.0) | 553 (4.7) | 997 (5.7) |
| Atoms (protein and ligands) | 2744 | 2748 | 2771 |
| Water molecules | 57 | 24 | 143 |
| Rfreeb | 0.282 | 0.280 | 0.254 |
| Rfactorc | 0.204 | 0.210 | 0.208 |
| R.m.s.d. bond lengths (Å) | 0.011 | 0.0079 | 0.013 |
| R.m.s.d. bond angles (°) | 1.55 | 1.30 | 1.53 |
where Ii(h) and I(h) are the ith and mean measurements of the intensity of reflection h.
where Fo and Fc are the observed and calculated structure factor amplitudes of reflection h, and k is a weighting factor.
where Fo and Fc are the observed and calculated structure factor amplitudes of reflection h, and k is a weighting factor.
Z, number of molecules in the asymmetric unit.
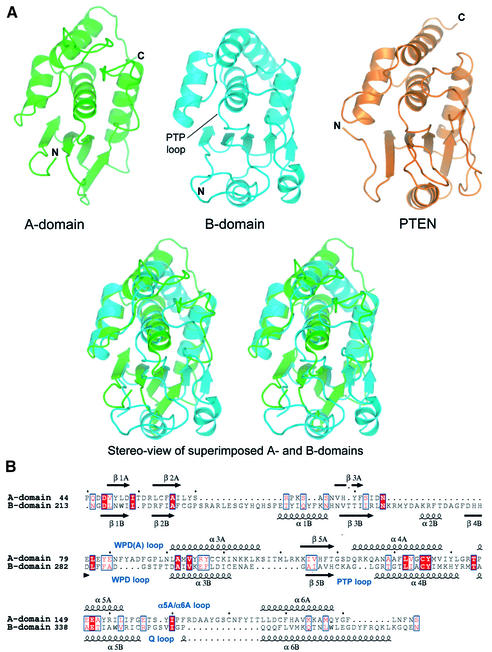
Cdc14 is composed of two structurally equivalent domains
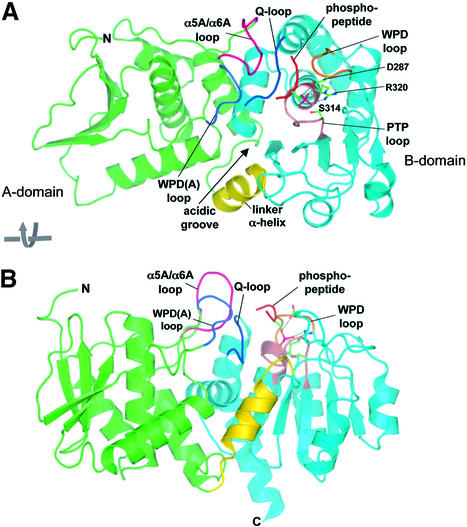
The molecular architecture of Cdc14B is composed of two similar sized domains arranged in tandem, associated via an extensive interface to form a single globular whole (Figure 2). Strikingly, both domains adopt a DSP-like fold. A linker α-helix (residues 199–212) connects the two domains. The conserved PTP signature motif (Cys-[X]5-Arg) that defines the catalytic centre of all PTP-family members is located within the C-terminal domain (B-domain, residues 213–379) and, together with the location of the phosphopeptide substrate in the catalytically inactive C314S mutant, identified the position of the catalytic site of Cdc14. As expected, tungstate bound to this site. Although the centre of the catalytic site is formed from B-domain, two loops from the N-terminal domain (A-domain) also contribute to the catalytic site, facilitating peptide substrate specificity (see below).
Fig. 2. Ribbon diagram of Cdc14B. Two orthogonal views showing the overall structure of the Cdc14–phosphopeptide complex. The A- and B-domains are green and cyan, respectively, and the inter-domain α-helix is yellow. There is a large solvent-accessible surface area of 2108 Å2 buried between the two domains. The phosphopeptide substrate is shown as a red coil, and key catalytic site loops are labelled. Figures were made with PyMOL (http://www.pymol.org).
The conformation of apo wild-type Cdc14B is virtually identical to both the Cdc14B–tungstate complex and the Cdc14B–phosphopeptide complex. Equivalent Cα atoms of apo Cdc14B and the Cdc14–peptide complex superimpose within an r.m.s.d. of 0.46 Å, and there is no indication of relative domain movements on association of peptide. The structure of apo Cdc14B that we describe here is the first example of a DSP crystallized in the absence of an oxy-anion bound to the catalytic site. Significantly, the conformation of the invariant WPD (Trp-Pro-Asp) loop, connecting β4 and α3, which bears the essential and invariant general acid/base Asp287 residue, adopts the closed, catalytically competent conformation in both apo and complex states. This finding demonstrates, that for Cdc14, in contrast to all known tyrosine specific PTPs, the binding of substrate is not required to induce closure of the WPD loop (Jia et al., 1995).
The B-domain contains the catalytic centre and is structurally related to PTEN
The architecture of the B-domain is highly reminiscent of other DSPs (Figures 2 and 3) (Barford et al., 1998). These proteins share the general characteristic of having a central mainly parallel β-sheet of five strands, with two α-helices on one side of the sheet. The fifth and middle β-strand leads into the conserved PTP signature motif that forms the base of the catalytic site, which in turn is connected to one of four α-helices that pack onto the opposite side of the β-sheet. A search of the protein database (PDB; Berman et al., 2000) using the DALI server (Holm and Sander, 1996) revealed that surprisingly the B-domain of Cdc14 is most similar to the phosphoinositol 3,4,5 tris-phosphate (PIP3) phosphatase PTEN (Lee et al., 1999) (Figure 3A), and the phosphatase domain of the mRNA capping enzyme (Changela et al., 2001) (Table II). A structural feature critical for the ability of PTEN to dephosphorylate the D3 position of its negatively charged PIP3 substrate are two conserved lysine residues within the PTP motif: (HCKAGKGR; lysines in bold) and a His residue in the WPD loop (Lee et al., 1999). Interestingly, the PTP motif of Cdc14 (HCKAGLGR) is also reminiscent of PTEN, although the His residue of the WPD loop of PTEN is a glycine (Gly288) in Cdc14, and therefore it is unlikely that Cdc14 functions to dephosphorylate lipid substrates. The most closely related protein phosphatases to Cdc14 are kinase-associated phosphatase (KAP) (Song et al., 2001) and vaccinia H1-related phosphatase (VHR) (Yuvaniyama et al., 1996) (Table II).
Fig. 3. Structural relatedness of the A- and B-domains of Cdc14B. (A) Comparison of structures of the A- and B-domains of Cdc14B and the phosphatase domain of PTEN. In the upper panel, the three domains are shown in the same orientation, and a stereoview of the A-domain (green) and B-domain (blue) superimposed is shown in the lower panel. (B) Structure-based sequence alignment of domains A and B of Cdc14B. Equivalent secondary structural elements are suffixed with ‘A’ and ‘B’ for domains A and B, respectively.
Table II. Highest structural relationships between domains of Cdc14B and protein structures deposited at the PDB as defined by DALI.
| Protein 1 | Protein 2 | No. of alignedresidues | R.m.s.d. Cα (Å) | Z-scorea |
|---|---|---|---|---|
| A-domain | PTEN | 126 | 3.0 | 9.9 |
| VHR | 124 | 3.0 | 9.8 | |
| B-domain | 119 | 2.6 | 9.6 | |
| Cdc14B | ||||
| mRNA capping | 121 | 2.7 | 9.5 | |
| protein | ||||
| KAP | 123 | 3.0 | 9.0 | |
| B-domain | PTEN | 150 | 2.0 | 19.5 |
| mRNA capping | 150 | 2.2 | 18.7 | |
| protein | ||||
| KAP | 153 | 2.7 | 18.2 | |
| VHR | 141 | 2.4 | 16.6 | |
| PTP1B | 151 | 2.5 | 13.7 |
aZ-scores above two are significant (Holm and Sander, 1996).
The A-domain has a DSP-like fold
The 3D architecture of the A-domain (residues 44–198) bears a remarkable resemblance to the B-domain of Cdc14. As shown in Figure 3A, the secondary structural elements of the A-domain superimpose closely onto the conserved core elements of the B-domain, and the two domains share the same secondary structure topology and polypeptide connectivities. Overall, the Cα atoms of 119 equivalent residues superimpose within an r.m.s.d. of 2.6 Å and the Z-score, a measure of the structural similarity in standard deviations above the expected value between two molecules, is 9.6 (Table II). Interestingly, this analysis indicated that the PTP/DSP family is structurally unique, such that a similar topology does not occur in other proteins. These findings suggest that the A-domain of Cdc14 resulted from divergent evolution from an ancestral PTP/DSP family member, possibly from a gene duplication event of the existing catalytically active B-domain. The structural similarity between the A- and B-domains of Cdc14B is not reflected in any sequence similarity. A structure-based alignment of the A- and B-domains indicates only 11% sequence identity (Figure 3B). Importantly, none of the catalytic site residues, including the catalytic site Cys and Arg residues, characteristic of PTP/DSPs, is present in the A-domain. Significantly, the structure of the A-domain suggests that it would be unable to bind phosphate in the equivalent region of the molecule to the phosphate-binding cradle formed by the PTP signature motif of the B-domain. In the A-domain, an insertion of two residues at the N-terminus of α4A, equivalent to the α4B helix which forms the base of the catalytic site in the B-domain (Figure 3B), alters the conformation of the A-domain so that it no longer forms a phosphate-binding cradle. Consistent with the notion that the A-domain is incapable of binding a phosphate moiety, we observed tungstate at 25 mM bound only to the catalytic site of the B-domain.
Other variations between the A- and B-domains include a 13 residue insertion in the α5A/α6A loop, which contributes to the peptide-binding groove, and the counterpart to the WPD loop of the B-domain is four residues longer in the A-domain (Figure 3B). Finally, there are no equivalents of the α1 and α2 helices, and β4 strand, conserved in the B-domain of Cdc14B and other DSPs.
The peptide-binding groove is selective for proline-directed peptides
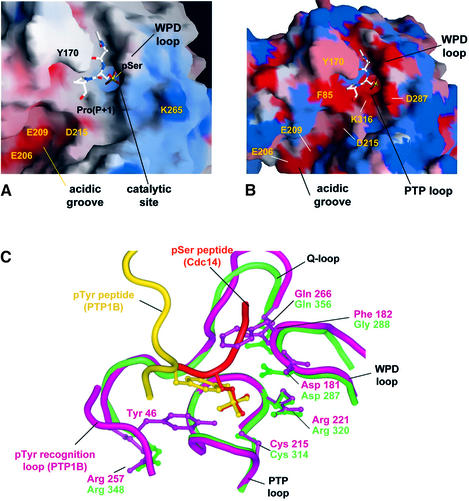
A unique feature of the catalytic site of Cdc14B is its location within a long groove (25 Å long and 10 Å wide), at the interface of the A- and B-domains. Residues of two loops of the A-domain, the extended WPD(A) and α5A/α6A loops, create one side of the groove (Figures 2, 4 and 5A). The WPD and Q-loops of the B-domain form the opposite face of the channel, whereas the inter-domain linker α-helix is positioned at the entrance to one end of the channel. Significantly, this region of the linker α-helix is rich in acidic residues (Glu206, Glu209 and Asp215) that cluster to generate a pronounced acidic groove leading to the catalytic site (Figure 5A). Cdc14 is genetically and biochemically linked to the dephosphorylation of Cdk substrates (Visintin et al., 1998; Kaiser et al., 2002), suggesting that the phosphatase must be capable of dephosphorylating phosphoserine/threonine residues located immediately N-terminal to a proline residue. Moreover, because Arg and Lys residues are usually located at the P+2 and P+3 positions C-terminal to Cdk sites of phosphorylation (Songyang et al., 1994; Holmes and Solomon, 1996; Kreegipuu et al., 1999), it is likely that Cdc14 will display some selection for phosphopeptides with basic residues C-terminal to the phospho-amino acid. It is, therefore, tempting to suggest that the cluster of acidic residues at the catalytic groove of Cdc14 may function to confer this selectivity.
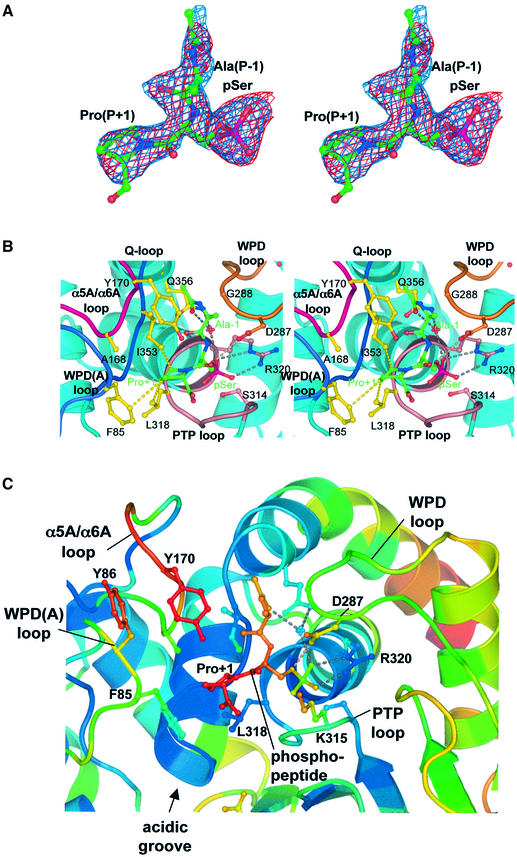
Fig. 4. Catalytic site of the Cdc14B–phosphopeptide complex. (A) Stereoview of 2Fo – Fc (cyan) and Fo – Fc (red) stimulated annealing omit electron density maps of the phosphopeptide, contoured at 1σ and 3σ, respectively. (B) Stereoview showing details of the catalytic site–peptide interactions, indicating the hydrophobic pocket that accommodates the Pro(P+1) residue. Hydrogen bonds are shown as dotted grey lines, and the van der Waals contacts between the peptide Pro(P+1) residue and Cdc14B are shown as yellow dotted lines. (C) Expanded view of the catalytic site with atoms coloured according to atomic B-factors with colours ramped from blue to red to denote B-factors ranging from to 13 to 52 Å2.
Fig. 5. Molecular surface of the Cdc14 catalytic channel with bound phosphopeptide substrate. (A) Electrostatic potential of Cdc14 catalytic channel, negative and positive electrostatic potentials are shown as red and blue, respectively. (B) Surface plot showing conservation of residues at the catalytic site of Cdc14 based on a sequence alignment of budding and fission yeast Cdc14 and human Cdc14A and Cdc14B. The colours are ramped from red to blue to denote invariant to unconserved residues. Figures drawn using SHARP (Nicholls et al., 1991). (C) Comparison of Cdc14–phosphopeptide complex (green) and PTP1B–IRK complex (purple) showing the PTP, WPD and Q-loops and phosphopeptide substrates of both proteins super imposed. The pTyr recognition loop is unique to PTP1B, and Tyr46 of the pTyr loop and Phe182 of the WPD loop of PTP1B create the selectivity for pTyr residues. A shallow catalytic channel in Cdc14 for pSer/Thr peptides is possible because of a Gly in the WPD loop in place of Phe and the absence of the pTyr loop.
To address the basis of Cdc14–substrate recognition, we co-crystallized a catalytically inactive Cys314 to Ser mutant of Cdc14 with a phosphopeptide of sequence A-pS-P-R-R-R, comprising the generic features of a Cdk substrate: a proline at the P+1 position and basic residues at P+2 to P+4. The structure of the Cdc14–phosphopeptide complex is shown in Figures 2, 4 and 5. Only the three residues A-pS-P are clearly delineated in electron density omit maps (Figure 4A). Density corresponding to the C-terminal basic residues is not visible, suggesting that these amino acids adopt multiple conformations when bound to Cdc14B. Atomic temperature factors of the peptide are in the same range as surface residues of the enzyme (Figure 4C). In the Cdc14–phosphopeptide complex, the Pro residue of the peptide is clearly defined as being in the trans isomer. With this conformation, residues C-terminal to the pSer-Pro motif will be directed into the acidic groove at the catalytic site and, importantly, a peptide with a cis proline would be unable to engage with the catalytic site due to a steric clash with the sides of the groove. This finding suggests that the pSer/pThr-Pro specific cis–trans peptidyl prolyl isomerase Pin1 may function to facilitate Cdc14 activity (Lu et al., 2002).
Interactions of the substrate phosphoserine residue with the catalytic site are reminiscent of phospho-amino acids bound to other protein phosphatases (Jia et al., 1995; Salmeen et al., 2000; Song et al., 2001); its phosphate moiety is coordinated by residues of the PTP loop, positioning it adjacent to the nucleophilic thiol group of Cys314 (Figures 4B and 5C). Similarly to PTP1B, the carboxylate group of the general acid Asp287 (Asp181 of PTP1B) is placed to donate a hydrogen bond to the Oγ atom of the pSer substrate. Interestingly, the peptide orientation is opposite to that of peptides bound to the phosphotyrosine-specific PTP1B. In PTP1B, Asp48 of the pTyr recognition loop forms bidendate interactions to the amide nitrogen atoms of the pTyr and P+1 residues, helping to define the substrate peptide orientation (Jia et al., 1995; Salmeen et al., 2000). There is no equivalent to the pTyr recognition loop of pTyr-specific PTPs in Cdc14, and interactions between main-chain groups of the peptide and the catalytic site do not appear to be a significant feature of the Cdc14–phosphopeptide complex. Moreover, residues of PTP1B that define the depth of the catalytic site pocket, and hence the selection for pTyr substrates, i.e. Tyr46 of the pTyr recognition loop and Phe182 of the WPD loop, are absent from Cdc14 (Figure 5C).
Apart from interactions involving the phospho-amino acid residue, the only other significant contacts formed between the phosphatase and peptide are contributed by the P+1 proline residue. The prolyl ring of Pro (P+1) is located in a hydrophobic pocket created by the side chains of Phe85 and Tyr170 of the A-domain WPD(A) and α5A/α6A loops, respectively, and two B-domain residues: Leu318 of the PTP loop and Ile353 of the Q-loop (Figures 4 and 5). Significantly, the aliphatic side chains of Phe85, Leu318 and Ile353 form optimal van der Waals contacts with the Pro(P+1) prolyl ring (Figure 4B). The location of this hydrophobic pocket immediately adjacent to the catalytic site for the phospho-amino acid confers the selectivity of Cdc14 for peptides with proline residues at the P+1 position. The tertiary nitrogen of the proline amide has no hydrogen-bonding potential, allowing it to be accommodated within an apolar environment. In contrast, the requirement to satisfy the hydrogen-bonding potential of the amide nitrogen of all other amino acids would select against peptides with non-proline residues at this position. Interestingly, a similar non-polar pocket at the P+1 position of the substrate peptide-binding site in Cdk2 determines its Pro-directed substrate selectivity (Brown et al., 1999). The conservation of residues that define the P+1 pocket in Cdc14 genes from diverse species suggests a common mechanism by which Cdc14 phosphatases are directed towards proline-containing peptides (Figures 1 and 5B). Finally, in the Cdc14–peptide complex, the side chain of Ala(P-1) is directed into solvent (Figures 4B and 5A), indicating that Cdc14 would be capable of dephosphorylating substrates modified by Cdks bearing any residue at this position. This finding correlates both with observations of Holmes and Solomon (1996), who noted that the activity of Cdk1 and Cdk2 towards model peptide substrates was insensitive to the residue at P-1, and empirical observations of in vivo Cdk substrates demonstrating no discernible amino acid selectivity at this position (Kreegipuu et al., 1999).
Steady-state kinetic parameters for the phosphatase activity of Cdc14B
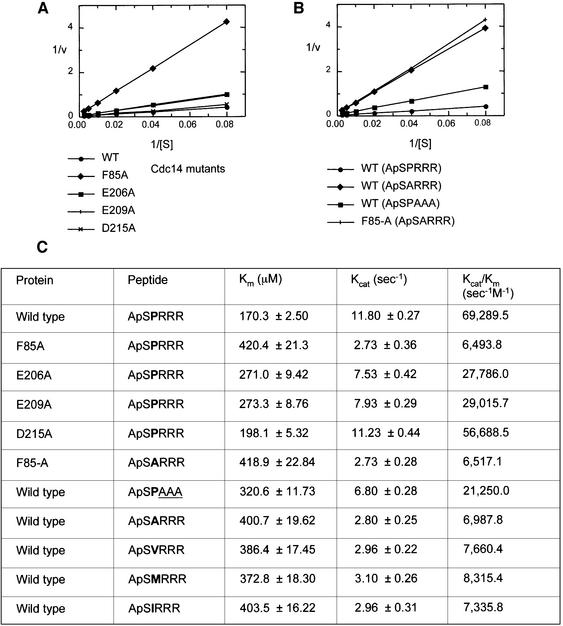
To assess the substrate selectivity of Cdc14, and the role of individual catalytic site residues for substrate catalysis and specificity, steady-state kinetic experiments were performed to compare the activities of wild-type and mutant forms of Cdc14 using various phosphopeptide substrates. Four catalytic site residues of Cdc14, Phe85 of the peptide–proline-binding pocket and Glu206, Glu209 and Asp215 of the acidic groove, were individually mutated to alanine, and the kinetic parameters of each mutant towards the phosphopeptide substrate acetyl-Ala-pSer-Pro-Arg-Arg-Arg-amide were determined. The release of phosphate was measured by an increase in absorbance at 360 nm using a coupled spectrophotometric assay (see Materials and methods). The time-dependent release of phosphate in the full time course reaction with wild-type Cdc14B conformed to typical Michaelis–Menten kinetics (data not shown).
Cdc14 is a less efficient phosphatase than the tyrosine-specific PTPs towards their optimal phosphotyrosine substrates (Figure 6). The kcat/Km value, a measure of both substrate affinity and catalytic turnover, is 69 290 s–1 M–1, some 25- to 400-fold lower than the equivalent value for PTP1B towards phosphopeptides modelled on its substrates, the insulin receptor kinase activation segment (Salmeen et al., 2000) and the kinase domain of the EGFR (Zhang et al., 1993), respectively. The reduced catalytic efficiency is mainly a consequence of the ∼20- to 80-fold reduced affinity of Cdc14 for its substrates relative to PTP1B, because both enzymes dephosphorylate their substrates with similar kcat values. The catalytic activity of Cdc14 is more reminiscent of the DSP VHR, which dephosphorylates pTyr185 of the ERK2 activation segment with similar catalytic parameters as Cdc14 (Denu et al., 1995; Schumacher et al., 2002).
Fig. 6. Kinetic properties of Cdc14B. (A) Lineweaver–Burke plot of the initial velocities of dephosphorylation of phosphopeptide ApSPRRR by wild-type and mutants of Cdc14B. (B) Lineweaver–Burke plot of the initial velocities of dephosphorylation of an optimal phosphopeptide (ApSPRRR) compared with an example of a peptide with a substitution at the P+1 position (ApSARRR) and a peptide with substitutions at the basic residues of P+2 to P+4 (ApSPAAA) by wild-type Cdc14B. The activity of the F85-A mutant of Cdc14B towards ApSARRR is also shown. (C) Table of kinetic constants for the dephosphorylation of phosphopeptide substrates by wild-type and mutant forms of Cdc14B. Amino acid substitutions in the P+1 position are shown in bold, and substitutions in the P+2 to P+4 position are underlined.
To investigate the selectivity of Cdc14 to dephosphorylate phosphopeptides harbouring a proline residue at the (P+1) position, the kinetic parameters of peptides with a substitution of the Pro residue were determined (Figure 6B and C). A peptide with Ala at the P+1 position is dephosphorylated 10-fold less efficiently than the optimal peptide substrate. The reduced activity of Cdc14 towards this peptide probably results from both loss of hydrophobic interactions between the enzyme and substrate, and the introduction of an unsatisfied hydrogen bond to the main-chain amide of an alanine residue at the P+1 position. Significantly, however, three other peptides with larger aliphatic amino acids substituted for Pro(P+1), namely Val, Ile and Met, which would be predicted to retain some hydrophobic contacts to the P+1 pocket, were also dephosphorylated between 8- and 9-fold less efficiently than the proline-containing peptide (Figure 6C). These results demonstrate selectivity for peptides with a Pro residue at P+1, which probably results from a combination of the absence of hydrogen-bonding potential of its apolar main-chain amide, and the ability to form optimized hydrophobic contacts to the apolar P+1 peptide-binding pocket.
The Cdc14 crystal structure indicates that the hydrophobic side chain of Phe85 packs onto the aliphatic prolyl ring of the peptide Pro(P+1) residue (Figure 4B) to create an optimal environment for a Pro residue at this position. Consistent with this notion, substituting an Ala for Phe85 reduces the catalytic efficiency (kcat/Km) some 11-fold towards the optimal peptide substrate, to a value similar to that for the dephosphorylation of a peptide with an Ala at the P+1 position by the wild-type protein (Figure 6C). Although these data imply that crucial binding interactions between the peptide Pro(P+1) residue and Cdc14B are made by Phe85, the crystal structure indicates that Leu318 and Ile353 also make important contributions to the Pro(P+1)-binding site. Acidic residues Glu206, Glu209 and Asp215 cluster at the entrance of the catalytic site, suggesting a potential to interact with basic residues of a peptide. However, kinetic data showing that substituting Ala residues at these positions decreases the rates of peptide dephosphorylation by only 2-fold supports the structural observations that these residues are not critical for peptide binding. These findings correlate with additional data that replacing the C-terminal arginine residues of the peptide with alanines reduces kcat/Km by only 3-fold (Figure 6).
Discussion
The discovery that Cdc14 is composed of a tandem DSP/PTP fold was unexpected, because the N-terminal domain shares no sequence similarity with other DSPs. The absence of all PTP signature motif residues suggests that this domain does not function catalytically. Instead, its association with the catalytic site of the B-domain to create a prominent catalytic site channel indicates a role in defining substrate specificity. Tandem PTP domains are also a feature of the receptor-like PTPs. In these instances, it is the N-terminal D1 domain that is catalytically active, whereas D2 usually lacks catalytic activity, while significantly retaining many of the catalytic PTP signature motif residues. The association of D1 and D2 of leukocyte antigen-related (LAR) is quite different from the association of the A- and B-domains of Cdc14 (Nam et al., 1999). The two enzymes employ different interfaces within each domain for their association, and D2 of LAR is remote from the D1 catalytic site, as is D1 from the putative catalytic site of D2. Therefore, unlike Cdc14, the association of D1 and D2 in LAR does not appear to generate a protein substrate-binding site. Finally, the extensive and rigid hydrophobic inter-domain interface of Cdc14 differs from the smaller and flexible interface of D1 and D2 of LAR (Nam et al., 1999).
The catalytic site of Cdc14, located in a channel formed at the interface of its two domains, provides the specificity for pSer and pThr residues located immediately before a proline residue at the P+1 position. The structure of the Cdc14–phosphopeptide complex suggests that residues other than proline would encounter an energetic cost by binding in a pocket that does not compensate the hydrogen-bonding potential of their amide nitrogen atoms, in addition to loss of optimal hydrophobic contacts to the prolyl side chain. Consistent with this prediction, Cdc14 dephosphorylates peptides with Ala, Ile, Val or Met substitutions of the Pro residue with 8- to 10-fold reduced catalytic efficiency. The pronounced conserved negatively charged channel in the vicinity of the Pro(P+1)-binding site suggests the potential to interact with the basic residues that occur at the P+2 and P+3 sites C-terminal to Cdk sites of phosphorylation (Songyang et al., 1994; Holmes and Solomon, 1996; Kreegipuu et al., 1999). However, interactions of Arg residues of the peptide and the channel were not observed in our crystal structure, and both mutations of these acidic residues of Cdc14, and replacing the peptide-Arg amino acids with Ala, only marginally decreased the catalytic efficiency of the phosphatase towards its substrate. Moreover, we note that the sequence of our peptide represents a generic Cdk phosphopeptide. Cdks are capable of phosphorylating Ser/Thr-Pro motifs in the context of variability in the sequence of C-terminal basic residues. It is, therefore, important for the role of Cdc14 that its catalytic site is capable of accommodating substrates defined by a pSer/Thr-Pro motif N-terminal to basic residues in a variety of sequence contexts. The acidic groove may function to allow structurally diverse basophilic peptide substrates with a phosphoSer/Thr-Pro motif to be accommodated at the catalytic site. Moreover, it is also possible that Cdc14 displays a higher affinity for selective substrates with particular sequences of basic residues C-terminal to the pSer-Pro motif. Further studies are required to establish this. The significantly lower affinity of Cdc14 towards its phosphopeptide substrates compared with PTP1B suggests that the cellular localization of Cdc14 will play a significant part in determining the timing and selectivity of its phosphatase activity.
The high level of similarity amongst Cdc14 protein sequences suggests that the structure of the enzyme, and the catalytic groove will be conserved from yeast to humans (Figures 1A and 5B). In budding yeast Cdc14, the catalytic groove is likely to perform an additional functional role to facilitate interactions with the inhibitory Net1 subunit of the RENT complex that retains Cdc14 in the nucleolus during interphase (Visintin and Amon, 2000; Traverso et al., 2001).
Materials and methods
Protein expression, purification and crystallization
The full-length human Cdc14B cDNA was amplified by PCR and cloned into the BamHI and HindIII site of pFASTBAC Htb (Invitrogen), which provides a His6 tag at the N-terminus of the protein that can be removed by TEV protease cleavage. Expression of Cdc14B was performed in Sf9 cells in SF900II media (Invitrogen) for 72 h at 27°C. The protein was purified using a combination of TALON (Clontech) metal affinity chromatography, ammonium sulfate precipitation, S200 size exclusion chromatography and finally S75 size exclusion chromatography (Amersham Biotech.). The His6 tag was removed from the protein following the ammonium sulfate precipitation by cleavage with TEV protease. The protein was isolated from the final purification step in 10 mM Tris–HCl pH 8.0, 750 mM NaCl, 5% glycerol, 3 mM dithiothreitol (DTT) and 1 mM EDTA. Limited proteolysis experiments were performed using trypsin, subtilisin and thermolysin. Cdc14B (2 mg/ml) in a buffer of 90 mM Tris–HCl pH 8.5, 750 mM NaCl, 4 mM DTT, 2 mM CaCl2 was digested for 30 min with each of 0.1, 0.5, 1, 5, 10 or 20 µg/ml protease. The ∼40 kDa major product of the digests was characterized by N-terminal sequencing, indicating an N-terminus at residue Pro44. The C-terminal boundary was estimated as His386 on the basis of molecular weight and secondary structure predictions. A pFASTBAC vector containing the coding sequence for Pro44–His386 was constructed and termed pFB-Cdc14B. A number of mutations were made to pFB-Cdc14B using standard site-directed mutagenesis techniques. Cdc14B and Cdc14B mutants were expressed in Sf9 cells and purified as described for the full-length protein, except that the ammonium sulfate precipitation step was omitted. Crystals of Cdc14B were obtained by hanging drop vapour diffusion by mixing 1 µl of 5 mg/ml protein with 1 µl of well solution (of 700 µl) comprising 100 mM Tris-acetate pH 8.0, 30% polyethylene glycol (PEG) 4000, 0.2 M MgCl2 and 7 mM DTT incubated at 20°C. These crystals were improved by microseeding into 100 mM Tris-acetate pH 8.0, 23% PEG 4000, 0.2 M MgCl2, 7 mM DTT. Crystals containing a bound substrate were produced by mixing a phosphopeptide (acetyl-Ala-pSer-Pro-Arg-Arg-Arg-amide) with protein at a 3:1 molar ratio and crystallizing in 0.1 M Tris-acetate, 30% PEG 1500 at 20°C. For data collection at 100 K, crystals were transferred to a cryoprotectant buffer of 100 mM Tris–HCl pH 8.0, 20% PEG 4000, 10% MPD, 0.4 M NaCl, 0.21 M MgCl2 and 5% glycerol. At a concentration of 20 mM Na2WO4, the activity of Cdc14B using 5 mM pNPP as a substrate is completely inhibited. We therefore used Na2WO4 to form a Cdc14B–tungstate derivative by supplementing the cryoprotectant with 25 mM Na2WO4.
Structure determination
The 39.7 kDa Cdc14B fragment crystallized in space group C2 with one molecule per asymmetric unit. The structure was determined by the SAD method using WO4-labelled crystals from data collected at the ID14 beamlines, ESRF, Grenoble, France. All crystallographic data were processed using MOSFLM (CCP4, 1994) and SCALA (CCP4, 1994). WO4 heavy atom coordinates were identified using SOLVE (Terwilliger and Berendzen, 1999) and the parameters were refined and protein phase calculations were performed using SHARP (De la Fortelle and Bricogne, 1997). The improved phases from SOLOMON (Abrahams and Leslie, 1996) were used to calculate electron density maps that were readily interpretable. Crystallographic statistics are listed in Table I. Electron density map interpretation and model building were performed using the program O (Jones et al., 1991) and the structure was refined by CNS (Brünger et al., 1998). Electron density maps for the structure of the phosphopeptide substrate bound to Cdc14B Cys314→Ser were generated by refining the model of the apo enzyme against the data from the substrate-bound crystals using CNS.
Cdc14B phosphatase assays
Kinetic parameters for the dephosphorylation of phosphopeptides by Cdc14B and Cdc14B mutants were assessed using the EnzChek Phosphate Assay Kit (Molecular Probes Inc.) with a Shimadzu spectrophotometer. The liberation of phosphate was measured by an increase in absorbance at 360 nm, as described by the manufacturer in a buffer of 50 mM Tris–HCl pH 7.5, 750 mM NaCl, 1 mM MgCl2, 0.6 mM DTT, 0.2 mM EDTA, 1% (v/v) glycerol at 20°C. The activities of the wild-type, Phe85→Ala, Glu206→Ala, Glu209→Ala and Asp215→Ala mutants of Cdc14B were assessed against the phosphopeptide substrate acetyl-A-pS-P-R-R-R-amide, and wild-type enzyme was also assayed towards peptides with sequences acetyl-A-pS-A-R-R-R-amide, acetyl- A-pS-V-R-R-R-amide, acetyl-A-pS-I-R-R-R-amide, acetyl-A-pS-M-R-R-R-amide and acetyl-A-pS-P-A-A-A-amide. Finally, the Cdc14B F85A mutant was assayed using the acetyl-A-pS-A-R-R-R-amide peptide. Each reaction was carried out in a volume of 100 µl in a quartz cuvette containing 500 nM appropriate Cdc14B enzyme and 0–400 µM phosphopeptide substrate. A data set of initial velocities was obtained by varying the concentration of phosphopeptide substrate at a fixed concentration of Cdc14B and the values of kinetic constants (Km and Vmax) were established by plotting these data in a double reciprocal (Lineweaver–Burke) plot. Velocity measurements at each substrate concentration were measured in triplicate and the reciprocal of the average velocity was used for each data point.
Accession codes
Coordinates and structure factors for the three structures of Cdc14B have been deposited with the PDB and assigned ID codes 1ohc, 1ohd and 1ohe for apo-Cdc14b, and the tungstate and peptide complexes, respectively.
Acknowledgments
Acknowledgements
We thank Pawel Dokurno and Vivenne Thompson for contributions to this project, and staff at the ESRF for access to synchrotron facilities. The work was funded in D.B.’s laboratory by a grant from Cancer Research UK.
References
- Abrahams J.P. and Leslie,A.G.W. (1996) Methods used in the structure determination of the bovine mitochondrial F1 ATPase. Acta Crystallogr. D, 52, 30–42. [DOI] [PubMed] [Google Scholar]
- Barford D., Das,A.K. and Egloff,M.-P. (1998) The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu. Rev. Biophys. Biomol. Struct., 27, 133–164. [DOI] [PubMed] [Google Scholar]
- Bembenek J. and Yu,H. (2001) Regulation of the anaphase-promoting complex by the dual specificity phosphatase Cdc14A. J. Biol. Chem., 276, 48237–48242. [DOI] [PubMed] [Google Scholar]
- Berman H.M., Westbrook,J., Feng,Z., Gillialnd,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown N.R., Noble,M.E., Endicott,J.A. and Johnson,L.N. (1999) The structural basis for specificity of substrate and recruitment peptides for cyclin-dependent kinases. Nat. Cell Biol., 1, 438–443. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Changela A., Ho,C.K., Martins,A., Shuman,S. and Mondragon,A. (2001) Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme. EMBO J., 20, 2575–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cueille N., Salimova,E., Esteban,V.,Blanco,M., Moreno,S.,Bueno,A. and Simanis,V. (2001) Flp1, a fission yeast orthologue of the S.cerevisiae Cdc14 gene, is not required for cyclin degradation or rum1p stabilization at the end of mitosis. J. Cell Sci., 114, 2649–2664. [DOI] [PubMed] [Google Scholar]
- De la Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy atom refinement in the MIR and MAD methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Denu J.M., Zhou,G., Wu,L., Zhao,R., Yuvaniyama,J., Saper,M.A. and Dixon,J.E. (1995) The purification and characterization of a human dual-specific protein tyrosine phosphatase. J. Biol. Chem., 270, 3796–3803. [DOI] [PubMed] [Google Scholar]
- Geymonat M., Jensen,S. and Johnston,L. (2002) Mitotic exit: the Cdc14 double cross. Curr. Biol., 12, R482. [DOI] [PubMed] [Google Scholar]
- Gruneberg U., Glotzer,M., Gartner,A. and Nigg,E.A. (2002) The CdCDC-14 phosphatase is required for cytokinesis in the Caenorhabditis elegans embryo. J. Cell Biol., 158, 901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1996) Alignment of three-dimensional protein structures: network server for database searching. Methods Enzymol., 266, 653–662. [DOI] [PubMed] [Google Scholar]
- Holmes J.K and Solomon,M.J. (1996) A predictive scale for evaluating cyclin-dependent kinase substrates. J. Biol. Chem., 271, 25240–25246. [DOI] [PubMed] [Google Scholar]
- Jaspersen S.L., Charles,J.F., Tinker-Kulberg,R.L. and Morgan,D.O. (1998) A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell, 9, 2803–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z., Barford,D., Flint,A.J. and Tonks,N.K. (1995) Structural basis for phosphotyrosine peptide recognition by protein tyrosine phosphatase 1B. Science, 268, 1754–1758. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 50, 157–160. [DOI] [PubMed] [Google Scholar]
- Kaiser B.K., Zimmerman,Z.A., Charbonneau,H. and Jackson,P.K. (2002) Disruption of centrosome structure, chromosome segregation and cytokinesis by misexpression of human Cdc14A phosphatase. Mol. Biol. Cell, 13, 2289–2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreegipuu A., Blom. N. and Brunak,S. (1999) PhosphoBase, a database of phosphorylation sites: release 2.0. Nucleic Acids Res., 27, 237–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.O., Yang,H., Georgescu,M.M., Di Cristofano,A., Maehama,T., Shi,Y., Dixon,J.E. Pandolifi,P. and Pavletich,N.P. (1999) Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell, 99, 323–334. [DOI] [PubMed] [Google Scholar]
- Lu K.P., Liou,Y.C. and Zhou,X.Y. (2002) Pinning down proline-directed phosphorylation signaling. Trends Cell Biol., 12, 164–172. [DOI] [PubMed] [Google Scholar]
- Mailand N., Lukas,C., Kaiser,B.K., Jackson,P.K., Bartek,J. and Lukas,J. (2002) Deregulated human Cdc14A phosphatase disrupts centrosome separation and chromosome segregation. Nat. Cell Biol., 4, 317–322. [DOI] [PubMed] [Google Scholar]
- Morgan D.O. (1999) Regulation of the APC and the exit from mitosis. Nat. Cell Biol., 1, E47–E53. [DOI] [PubMed] [Google Scholar]
- Nam H.J., Poy,F., Krueger,N.X., Saito,H. and Frederick,C.A. (1999) Crystal structure of the tandem phosphatase domains of RPTP LAR. Cell, 97, 449–457. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Salmeen A., Andersen,J.N., Myers,M.P., Tonks,N.K and Barford,D. (2000) Molecular basis for the dephosphorylation of the activation segment of the insulin receptor by protein tyrosine phosphatase 1B. Mol. Cell, 6, 1401–1412. [DOI] [PubMed] [Google Scholar]
- Schumacher M.A., Todd,J.L., Rice,A.E., Tanner,K.G. and Denu,J.M., (2002) Structural basis for the recognition of a bisphosphorylated MAP kinase peptide by human VHR protein phosphatase. Biochemistry, 41, 3009–3017. [DOI] [PubMed] [Google Scholar]
- Song H., Hanlon,N., Brown,N.R., Noble,M.E., Johnson,L.N. and Barford,D. (2001) Phosphoprotein–protein interactions revealed by the crystal structure of kinase-associated phosphatase in complex with phosphoCDK2. Mol. Cell, 7, 615–626. [DOI] [PubMed] [Google Scholar]
- Songyang Z., Blechner,S., Hoagland,N., Hoekstra,M.F., Piwnica-Worms,H. and Cantley,L.C. (1994) Use of an oriented peptide library to determine the optimal substrates of protein kinases. Curr. Biol., 4, 973–982. [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Visintin,R. and Amon,A. (2002) Separase, polo kinase, the kinetochore protein Slk19 and Spo12 function in a network that controls Cdc14 localization during early anaphase. Cell, 108, 207–220. [DOI] [PubMed] [Google Scholar]
- Taylor G.S., Lui,Y., Baskerville,C. and Charbonneau,H. (1997) The activity of Cdc14p, an phospho-meric dual specificity protein phosphatase from Saccharomyces cerevisiae, is required for cell cycle progression. J. Biol. Chem., 272, 24054–24063. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverso E.E., Baskerville,C., Liu,Y., Shou,W., James,P., Deshaies,R.J. and Charbonneau,H. (2001) Characterization of the Net1 cell cycle-dependent regulator of the Cdc14 phosphatase from budding yeast. J. Biol. Chem., 276, 21924–21931. [DOI] [PubMed] [Google Scholar]
- Trautmann S. and McCollum,D. (2002) Cell cycle: new functions for Cdc14 family phosphatases. Curr. Biol., 12, R733–R735. [DOI] [PubMed] [Google Scholar]
- Trautmann S., Wolfe,B.A., Jorgensen,P., Tyers,M., Gould,K.L. and McCollum,D. (2001) Fission yeast Clp1p phosphatase regulates G2/M transition and coordination of cytokinesis with cell cycle progression. Curr. Biol., 11, 931–940. [DOI] [PubMed] [Google Scholar]
- Visintin R. and Amon,A. (2000) The nucleolus: the magician’s hat for cell cycle tricks. Curr. Opin. Cell Biol., 12, 372–377. [DOI] [PubMed] [Google Scholar]
- Visintin R., Craig,K., Hwang,E.S., Prinz,S., Tyers,M. and Amon,A. (1998) The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell, 2, 709–718. [DOI] [PubMed] [Google Scholar]
- Wan J., Xu,H. and Grunstein,M. (1992) CDC14 of Saccharomyces cerevisiae. Cloning, sequence analysis and transcription during the cell cycle. J. Biol. Chem., 267, 11274–11280. [PubMed] [Google Scholar]
- Yuvaniyama J., Denu,J.M., Dixon,D.E. and Saper,M.A. (1996) Crystal structure of the dual specificity protein phosphatase VHR. Science, 272, 1328–1331. [DOI] [PubMed] [Google Scholar]
- Zhang Z.H., Thieme-Sefler,A.M., Maclean,D., McNamara,D.J., Dobrusin,E.M., Sayer,T.K. and Dixon,J.E. (1993) Substrate specificity of the protein tyrosine phosphatases. Proc. Natl Acad. Sci. USA, 90, 4446–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]