Abstract
Hsp90 has a diverse array of cellular roles including protein folding, stress response and signal transduction. Herein we report a novel function for Hsp90 in the ATP-dependent assembly of the 26S proteasome. Functional loss of Hsp90 using a temperature-sensitive mutant in yeast caused dissociation of the 26S proteasome. Conversely, these dissociated constituents reassembled in Hsp90-dependent fashion both in vivo and in vitro; the process required ATP-hydrolysis and was suppressed by the Hsp90 inhibitor geldanamycin. We also found genetic interactions between Hsp90 and several proteasomal Rpn (Regulatory particle non-ATPase subunit) genes, emphasizing the importance of Hsp90 to the integrity of the 26S proteasome. Our results indicate that Hsp90 interacts with the 26S proteasome and plays a principal role in the assembly and maintenance of the 26S proteasome.
Keywords: assembly/Hsp90/maintenance/26S proteasome/yeast
Introduction
The high density of protein molecules in the cytosol increases the likelihood that partially folded or unfolded proteins will undergo off-pathway reactions, such as aggregation, in the protein biosynthetic pathway or by postsynthesis damage. Molecular chaperones recognize proteins with non-native structures, prevent them from irreversible aggregation and assist in their conversion to a functional conformation (Frydman, 2001). On the other hand, the ubiquitin–proteasome pathway plays a pivotal role in selective destruction of misfolded and unassembled proteins (Sherman and Goldberg, 2001). Since chaperones and proteasomes appear to recognize common substrates under non-native states, these two pathways act jointly to prevent aggregation and accumulation of abnormal proteins, thus maintaining protein homeostasis in cells. However, the relationship between molecular chaperones and the ubiquitin–proteasome system is still largely unknown.
Most cellular proteins in eukaryotic cells are targeted for degradation by the 26S proteasome, usually after they have been covalently attached to ubiquitin in the form of a poly-ubiquitin chain functioning as a degradation signal (Pickart, 2001). The 26S proteasome, a eukaryotic ATP-dependent protease, is composed of a catalytic 20S proteasome (alias CP, core particle) and a pair of symmetrically disposed regulatory particles (RP, alias PA700 or the 19S complex) (Baumeister et al., 1998). RP is attached to both ends of the central CP in opposite orientation to form the active 26S proteasome. The 26S proteasome with a molecular mass of ∼2.5 MDa acts as a highly organized apparatus for proteolysis. The 20S proteasome is composed of two copies of 14 different subunits, seven distinct α-type and seven distinct β-type subunits. It is a barrel-like particle formed by the axial stacking of four rings made up of two outer α-rings and two inner β-rings, associated in the order αββα. Three β-type subunits of each inner ring have catalytically active threonine residues at their N-termini, and these active sites reside in a chamber formed by the centers of the abutting β-rings. The regulator RP is a protein complex (>700 kDa) composed of ∼20 subunits, each 25–110 kDa in size (Baumeister et al., 1998). RP consists of two subcomplexes, known as ‘base’ and ‘lid’, which, in the 26S proteasome, correspond to the portions of RP proximal and distal, respectively, to the 20S proteasome (Glickman et al., 1998). The base is mainly composed of up to six ATPases (Rpt, Regulatory particle triple ATPase), while the lid contains multiple non-ATPase subunits (Rpn). The metabolic energy liberated by these Rpt functions is probably utilized for unfolding target proteins so that they can penetrate the channel of the α- and β-rings of the 20S proteasome. However, the roles of many other RP subunits remain undefined.
Of many molecular chaperones, Hsp90 is one of the major species which also requires ATP for its in vivo functions (Panaretou et al.,1998; Young et al., 2001). It is among the most abundant proteins in cells, occupying ∼1–2% of total cellular proteins (Frydman, 2001). The major role of Hsp90 is to manage protein folding, but it also plays a critical role in signal transduction pathways that include mainly steroid receptors and protein kinases (Richter and Buchner, 2001). In Saccharomyces cerevisiae, two Hsp90 species with redundant functions, named Hsp82 and Hsc82, are present, which are equivalent to mammalian Hsp90α and Hsp90β, respectively.
In the present study of Hsp90 in the budding yeast, we unexpectedly noticed that in vivo inactivation of Hsp90 using the temperature-sensitive (ts–) hsp82–4Δhsc82 mutant cells (Kimura et al., 1994) caused almost complete dissociation of the 26S proteasome into its constituents. Furthermore, we found that Hsp90 contributes not only to maintain the functional integrity of the 26S proteasome but also to assist its assembly in vivo and in vitro in an ATP-dependent manner. In addition, we also provide the genetic evidence of in vivo linkage between Hsp90 and the 26S proteasome. Thus the participation of Hsp90 in the 26S proteasome assembly may provide new mechanistic insight into the cooperative interactions between molecular chaperones and proteolysis systems.
Results
Severe thermal stress causes disassembly of the 26S proteasome
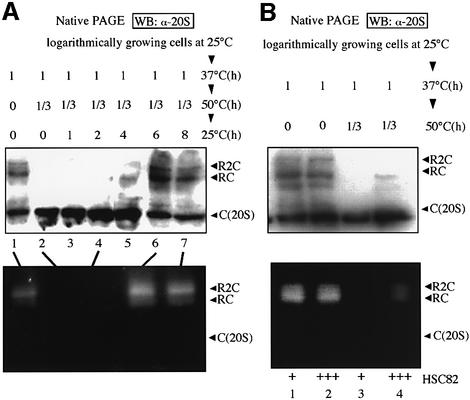
To focus on the relationship between stress response and the cellular proteolysis machinery, we examined the effect of severe heat stress on the functional state of the proteasome, which is subclassified into three species in the budding yeast; i.e. the free 20S proteasome (alias CP and here designated simply as C) and RP associated with both sides of CP (R2C) or one side of CP (RC), as described by Glickman et al. (1998). Wild-type (WT) cells grown at 25°C were first incubated at 37°C for 1 h. This step was essential to allow the cells to survive a subsequent severe thermal insult. The same cells were incubated at 50°C for 20 min and then shifted to normal culture conditions at 25°C. Upon these stresses, after preconditioning at 37°C, more than 80% of the cells were viable, forming colonies when plated at 25°C (Imai and Yahara, 2000).
Native (non-denaturing) polyacrylamide gel electrophoresis (PAGE) analysis and subsequent western blotting using antibodies against the yeast 20S proteasome revealed a marked decrease of both the R2C and RC forms of the 26S proteasome after severe heat shock at 50°C, and a considerable increase in the amount of free 20S proteasome (Figure 1A, lanes 2–4). When these heat-shocked cells were reversed to culture at 25°C, it took ∼6 h for the full recovery of the 26S proteasome (Figure 1A, lane 6). We also examined the peptidase activity of the 26S proteasome by the in-gel overlay assay. Samples of cell extracts were subjected to native PAGE, and peptide-degrading activity was detected by soaking the gel in a solution containing the fluorogenic peptide succinyl- Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin (Suc-LLVY-AMC). As shown in Figure 1A (bottom), the dissociated 20S proteasome due to severe thermal insults was the latent form and the reassembled 26S proteasome (after incubation at 25°C) was functionally active.
Fig. 1. Dissociation and reassembly of the 26S proteasome after severe heat shock. (A) Crude extracts (5 µg) of cells thermally treated as indicated were subjected to native PAGE followed by western blotting (WB) with anti-20S proteasome (top). Cell extracts (20 µg, top, lanes 1, 2, 4, 6 and 7) were loaded onto native PAGE, and thereafter Suc-LLVY-AMC degrading activities were assayed by the in-gel overlay method with 2 mM ATP (bottom). See text for explanation of symbols R2C, RC and C (20S). (B) Effect of overexpression (designated as +++) of Hsc82 on the disassembly of the 26S proteasome by severe heat shock. The analyses were similar to those described in (A), except that cells were grown in SC-U medium and cell extracts were prepared from control cells (YPH500/pYO326, lanes 1 and 3) and Hsc82 overexpressing cells (YPH500/pYO326-HSC82, lanes 2 and 4). Note that the magnitude of the increased level by a plasmid overexpressing Hsp82 was more than 5-fold (Imai and Yahara, 2000).
In addition, we found that overexpression of Hsp90 (Hsc82) partially suppressed the destruction of the 26S proteasome caused by severe thermal insult, as detected by western blot and in-gel overlay analyses (Figure 1B). On the other hand, overexpression of Hsp70 (Ssa1) had no appreciable protective effect on the proteasome disassembly (data not shown).
Disassembly of the 26S proteasome caused by inactivation of Hsp90
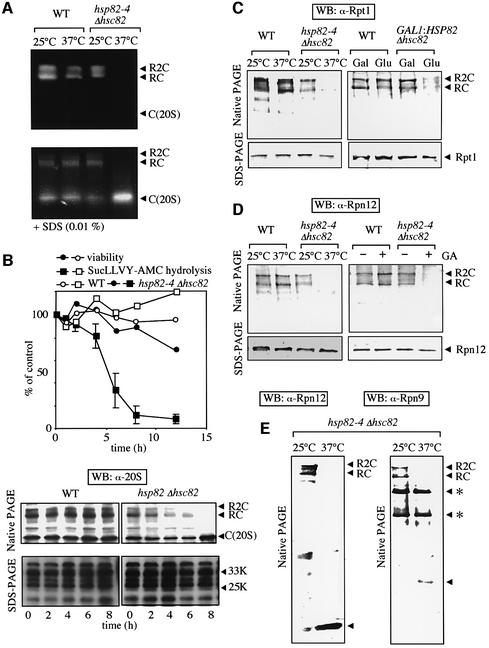
In the next step, we examined the mechanism through which Hsp90 protects against disassembly of the 26S proteasome by severe thermal stress. For this purpose, we analyzed the structure of the 26S proteasome under defective conditions of Hsp90 using temperature-sensitive (ts–) hsp82–4Δhsc82 cells (YOK5) (hereafter, the mutant yeast is simply described as hsp82-4 cells). First, we examined the 26S proteasome by the in-gel peptidase assay. As shown in Figure 2A (top), two slowly migrating active proteasomes, corresponding to R2C and RC, were evident in extracts of WT cells irrespective of the culture temperature (25 or 37°C). However, the signals corresponding to positions of R2C and RC markedly decreased when only samples prepared from hsp82-4 cells that had been cultured for 8 h at non-permissive temperature of 37°C were used (Figure 2A). Activities similar to those of WT cell extracts were observed when extracts of hsp82-4 cells cultured at permissive temperature were used. Moreover, the addition of SDS, which activates the latent 20S proteasome in vitro, caused marked activation of the 20S proteasome, and the magnitude of activation was augmented when we used the crude extracts of hsp82-4 cells that had been cultured for 8 h at 37°C (Figure 2A, bottom). These results indicate that inactivation of Hsp90 causes dissociation of the active 26S proteasome into its constituents containing the 20S proteasome.
Fig. 2. Electrophoretic analyses of the 26S proteasome in hsp82-4 cells. (A) In-gel overlay assay of peptidase activity of the proteasome separated by native PAGE. WT cells (YPH500) and hsp82–4Δhsc82 cells (YOK5H) grown at 25°C were shifted at 37°C and maintained for another 8 h or continued culturing at 25°C. These cell extracts (20 µg) were analyzed as in Figure 1 in the presence of 2 mM ATP (top) or 0.01% SDS (bottom). (B) WT and hsp82-4 cells grown at 25°C were shifted to 37°C and maintained for various times up to 12 h. Cells were sampled at each time point and then cell viability and Suc-LLVY-AMC degrading activity of the 26S proteasome affinity-purified were measured (top). The results are expressed relative to the result at time zero in WT cells. Open and closed squares represent activities of the 26S proteasome from WT and hsp82-4 cells, respectively. Open and filled circles represent viability of WT and hsp82-4 cells, respectively. Identical amounts of cell extracts were loaded onto native PAGE (top, 5 µg) and SDS–PAGE (bottom, 1 µg), followed by immunoblotting with anti-20S proteasome (bottom). (C) Western blotting with anti-Rpt1. The analyses were the same as for (B), except that anti-Rpt1 and 1 µg protein were used for native PAGE (left). The WT and GAL1:HSP82 Δhsc82 cells (5CG2) grown at 30°C in YPGal were transferred to YPD and maintained for another 12 h or continued culturing in YPGal (right). The analyses were the same as for the left panel. (D) Western blotting with anti-Rpn12. The analyses were the same as for (B), except that anti-Rpn12 was used (left) and WT and hsp82-4 cells grown at 25°C were treated (+) or mock treated (–) with GA (18 µM) followed by further culture at 25°C for 3 h (right). (E) Excess loading analyses. The same cell extracts in (D) for hsp82-4 cells were analyzed by native PAGE and western blotting with anti-Rpn12 (left), except that 10 µg protein (lanes 1 and 2) was used. The same analysis was conducted using anti-Rpn9 and 10 µg protein (lanes 3 and 4). The band, indicated by arrowheads in both panels, was specific for antibodies against Rpn9 and Rpn12, respectively. Asterisks in the right panel indicate non-specific bands for anti-Rpn9.
To confirm these observations, we loaded the samples prepared from WT and hsp82-4 cells, which had been incubated for various times under a non-permissive temperature, onto native PAGE and SDS–PAGE, and then conducted western blotting with anti-20S proteasome. In the native PAGE, the three species of the proteasome, i.e. R2C, RC and C, were evident in WT cells even after 8 h incubation (Figure 2B, bottom). In contrast, destruction of both bands with lower electrophoretic mobility, corresponding to the R2C and RC forms of the 26S proteasome, began within only 4 h of inactivation of Hsp90, though it took 8 h for their complete loss in hsp82-4 cells. Consequently, loss of Hsp90 was associated with an increase in free 20S proteasome. However, the total amounts of the 20S proteasome analyzed by western blotting after SDS–PAGE, detected as several bands ranging from 20 to 35 kDa, remained unchanged. Thus it is clear that loss of function of Hsp90 causes dissociation of the 26S proteasome into its constituents, including the 20S proteasome. We then compared peptidase activities of the proteasome and cell viability under the same conditions. Both values were unchanged in WT cells, but the peptidase activities gradually decreased after around 4 h incubation at 37°C and almost completely disappeared upon incubation for 8 h in hsp82-4 cells (Figure 2B, top). It was noteworthy that in these cells, the R2C form disappeared before the RC form and the activities of the proteasome were decreased (4 h after the shift), followed by the loss of the RC form (Figure 2B, top and bottom). Importantly, the loss of proteasome activities in hsp82-4 cells occurred faster than cell death, indicating that the structural abnormality of the 26S proteasome is not due to cell death.
When the same electrophoretic and immunoblotting analyses were conducted using anti-Rpt1 (Figure 2C, left) and anti-Rpn12 (Figure 2D, left), the former is an ATPase base subunit and the latter is a non-ATPase lid subunit of RP (Glickman et al., 1998), two slowly migrating R2C and RC bands were evident in native PAGE in all cases, except hsp82-4 cells, under non-permissive temperature. Again, comparable amounts of Rpt1 and Rpn12 subunits of the RP complex were detected even under culture at 37°C by SDS–PAGE (see bottom panels). Intriguingly, excess loading of samples revealed the presence of Rpn12 and Rpn9, another lid subunit, at rapidly migrating positions, but their electrophoretic mobilities differed from each other (Figure 2E), indicating dissociation of the lid complex under defective Hsp90 conditions of the cells.
We also confirmed the involvement of Hsp90 in the maintenance of the 26S proteasome by shutting off Hsp82 expression using GAL1 promoter. Repression of Hsp82 expression by replacement of galactose with glucose in the media resulted in the disappearance of the 26S proteasome (Figure 2C, right). Moreover, we observed that geldanamycin (GA), an Hsp90 inhibitor, caused loss of the 26S proteasome in hsp82-4 cells even under permissive temperature, although it had no appreciable effects on the proteasomal states in WT cells (Figure 2D, right). These results strongly suggest that Hsp90 is essential for the 26S proteasome. Curiously, we repeatedly observed the sensitivity of hsp82-4, but not WT cells, to GA in both in vivo (Figures 2 and 3) and in vitro (Figure 4) analyses. The exact reason is unclear, but GA may be easily accessible to the active ATPase site of the hsp82-4 protein, perhaps because of its abnormal conformation.
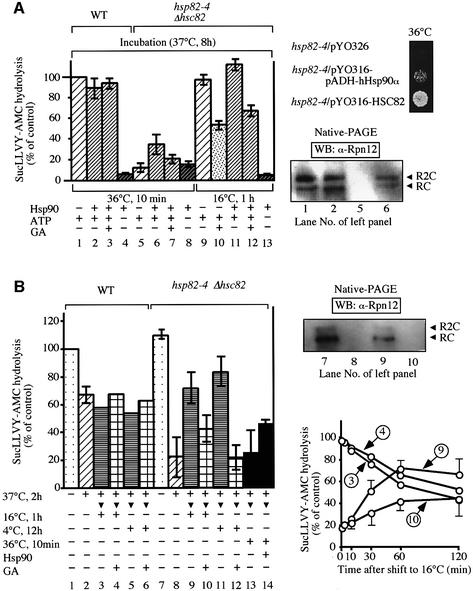
Fig. 3. In vivo analyses of the 26S proteasome under conditions with or without Hsp90 inactivation. (A) Western blotting was carried out with anti-Hsp82 and various proteasomal antibodies for the 26S proteasome (100 ng, left) affinity-purified from the extracts of WT (J106) and hsp82–4Δhsc82 cells (YOK5RH) grown at 25°C by a Ni+-resin column (–, YPH500, WT cells without 6×His-RPT1). Western blotting after unconventional (low) SDS–PAGE (Imai and Yahara, 2000) was conducted to differentiate between Hsc82 and Hsp82 in the crude extracts (1 µg, left). Note that anti-Hsp82 reacted in a fashion similar to Hsc82 and Hsp82. Western blotting with anti-Hsp82 and anti-20S proteasome was carried out for Hsp82-depleted cells (5CG2) whose Hsp90 levels were varied by culturing with different combinations of Gal and Glu (right). The same amounts of cell extracts were loaded onto native PAGE (top, 5 µg) and SDS–PAGE (bottom, 1 µg), followed by immunoblotting with anti-Hsp82 and anti-20S proteasome. (B) WT (left) and hsp82-4 cells (right) grown at 25°C were shifted to 37°C and maintained for an additional 6 h. At time zero, cells were shifted to the indicated temperatures followed by the addition of cycloheximide (CHX, 100 µg/ml). Cells were sampled at each time point and the Suc-LLVY-AMC degrading activity of the 26S proteasome affinity-purified as in (A) was measured. The activities are expressed relative to the activity at time zero in WT cells grown at 25°C. Blue lines, cells grown at 25°C throughout the experimental period; black lines, cells grown at 25°C were shifted to 37°C for 6 h and then to 25°C at time zero; red lines, similar to black lines except that the cells were treated with GA (18 µM) at time zero; green lines, cells grown at 25°C were shifted to 37°C for 6 h and further incubated at 37°C after time zero. Crude extracts (1 µg) of hsp82-4 cells from positions denoted 1, 2 and 3 (right bottom) were separated by native PAGE and analyzed by western blotting with anti-Rpt1 (right top). Data are means ± SEM. (C) ATP requirement for Hsp90-dependent 26S proteasome assembly. WT and hsp82-4 cells grown at 25°C were metabolically poisoned with 10 mM deoxyglucose and 10 mM sodium azide for 1 h at 37°C (designated + for ATP depletion). Cells cultured with YPD media without ATP-depletion are marked –. Thereafter, cells were shifted to YPD media for 30 min at 37°C (left panels) or YPD media containing GA (18 µM) for 30 min at 25°C (right panels). Activities of the 26S proteasome in gel-overlay assay were visualized as in Figure 1 with 2 mM ATP. Note that a considerable amount of Hsp90 is associated with the affinity-purified 26S proteasome fraction.
Fig. 4. In vitro analyses of the 26S proteasome under conditions with or without Hsp90 inactivation. (A) The Suc-LLVY-AMC degrading activities of the affinity-purified 26S proteasome from WT and hsp82-4 cells were measured and expressed as percentages of the control (representing the activity of WT cell extracts without incubation). The extracts (10 mg/ml of protein) from both cells grown at 37°C for 8 h were incubated at 36°C for 15 min or at 16°C for 1 h with or without purified Hsp90 (0.1 mg/ml) and in the presence of an ATP-regeneration system (10 mM creatine phosphate, 5 mM MgCl2 and 10 µg/ml of creatine kinase), an ATP-depletion system (10 mM glucose and 1 µg/ml of hexokinase) or GA (18 µM, left). The cell extracts corresponding to lanes 1, 2, 5 and 6 (left) were subjected to native-PAGE, followed by western blotting with anti-Rpn12 (right bottom). Each strain harboring the human-Hsp90α-expressing plasmid (YOK5H/pRS316-hHsp90α), the Hsc82-expressing plasmid (YOK5/pRS316-HSC82) or the vector alone (corresponding empty vector) was spotted on the same plates and incubated at 36°C for 2 days (right top). (B) In vitro inactivation and reactivation of the 26S proteasome from hsp82-4 cells. The affinity-purified 26S proteasome from the same amounts of cell extracts of WT cells (J106) and hsp82–4Δhsc82 cells (YOK5RH) were maintained at 37°C with an ATP-regenerating system for 2 h and shifted to 16°C for 1 h, 4°C for 12 h or 36°C for 10 min in the presence of an ATP-regenerating system with or without GA (18 µM) or purified Hsp90 (0.1 mg/ml)(left panel). Suc-LLVY-AMC hydrolysis is expressed relative to the activity at time zero of WT cells grown at 25°C. The affinity-purified 26S proteasomes corresponding to lanes 3, 4, 9 and 10 that had been treated at 37°C for 2 h were shifted to 16°C for various times as indicated, and thereafter Suc-LLVY-AMC hydrolysis was assayed as described above (right bottom panel). The affinity-purified 26S proteasomes corresponding to lanes 7–10 were subjected to native PAGE, followed by western blotting with anti-Rpn12 (right top panel). Data in A and B are means ± SEM.
Hsp90-dependent in vivo assembly of the 26S proteasome
We first tested the physical interaction between Hsp90 and the 26S proteasome in vivo. For this purpose, we purified the 26S proteasome in a single step, using a Ni+-resin column, from extracts of WT (J106) and hsp82-4 cells (YOK5RH), whose RPT1 was replaced by 6×His-RPT1. The subunit composition of the enzyme from WT cells resembled that from hsp82-4 cells grown at 25°C (data not shown). In both preparations, the addition of MG132 almost completely inhibited the hydrolysis activity of Suc-LLVY-AMC, revealing no contamination of other protease(s), and the specific activity of the purified 26S proteasome resulted in >10-fold increase in Suc-LLVY-AMC hydrolysis (data not shown). Intriguingly, compared with that WT cells, considerable amounts of Hsp82 were associated with affinity-purified 26S proteasome and larger amounts of Hsp90 were associated with the 26S proteasome from hsp82-4 cells, as judged by the similar contents of various proteasomal subunits (Figure 3A, left), though the total Hsp90 contents in hsp82-4 cell extracts were much less than those of the WT extracts because of the lack of Hsc82 in hsp82-4 cells. Native PAGE and immunoblotting using anti-Hsp82 against Hsp82-depleted cells revealed that anti-Hsp82 reacted strongly with the R2C form than with RC form, but not appreciably with CP, indicating that Hsp82 associates with the 26S proteasome through the RP complex (Figure 3A, right). Intriguingly, even when the amounts of Hsp82 decreased to ∼one-tenth of WT, Hsp82 only bound to R2C and RC (Figure 3A, right), indicating a high affinity of Hsp90 with the 26S proteasome.
We next examined the role of Hsp90 in the in vivo assembly of the 26S proteasome. When both WT and hsp82-4 cells were grown at 25°C, the Suc-LLVY-AMC degrading activity of the 26S proteasome affinity-purified from the same amounts of crude cell extracts was almost similar, even when the peptidase assay was carried out at 37°C in the presence of ATP (time zero in Figure 3B, blue line). However, when hsp82-4 cells were cultured for 8 h after a shift to 37°C, no appreciable amounts of proteasomal proteins were recovered by purifying operation using the same Ni+-resin column, and consequently very little Suc-LLVY-AMC degrading activity was observed in the peptidase assay (data not shown). These results again indicated loss of the 26S proteasome under Hsp90-defective conditions of the cells.
In the next experiments, we examined whether, once disassembled, the proteasome in hsp82-4 cells cultured under a non-permissive temperature could reassemble when the same Hsp90-inactivated cells were shifted to permissive temperature of 25°C. After heat shock at 37°C for 6 h, we added cycloheximide (CHX) to inhibit de novo protein synthesis (time zero in Figure 3B) and then these cells were shifted to 25°C for further culture. The 26S proteasome was affinity-purified from the same amount of cell extracts after reculture for the indicated time intervals. As shown in Figure 3B (right), 2 h after shifting to 25°C a considerable Suc-LLVY-AMC degrading activity was restored (black line). Of note, this restoration was independent of the newly synthesized proteasome because of the presence of CHX, suggesting the reassembly of the 26S proteasome subsequent to the functional recovery of Hsp90. Interestingly, this reactivation was diminished in GA-treated hsp82-4 cells at the time of shift to 25°C and then was maintained at 25°C (red line), indicating that the ATPase function of Hsp90 is necessary for the restoration of the 26S proteasome. In contrast, continued culture in non-permissive temperature at 37°C was not associated with such increment in proteasomal peptidase activity (green line). In parallel analyses using WT cells (Figure 3B, left), irrespective of heat shock, and hsp82-4 cells under permissive temperature (Figure 3B, right, blue line), the Suc-LLVY-AMC degrading activities gradually diminished after addition of CHX. These results suggested that the restoration of peptidase activity is linked to the proper assembly of the 26S proteasome, assuming that the Hsp90 inactivation-induced disassembly of 26S proteasome is reversed by recovery of functional Hsp90. To confirm this attractive conclusion, we analyzed the 26S proteasome 2 h after incubation shown in Figure 3B (right, see arrows) by native PAGE and western blotting using anti-Rpt1. As shown in Figure 3B (right top), changes in activity apparently coincided with reassembly of both the R2C and RC forms of the 26S proteasome.
We next examined the ATP requirement for the Hsp90-dependent reassembly of the 26S proteasome. Depletion of ATP by treatment of cells with metabolic poisons (deoxyglucose and sodium azide) resulted in almost complete dissociation of the 26S proteasome, irrespective of WT and hsp82-4 cells. When these cells were incubated for 30 min at 37°C in YPD media, reassembly of the 26S proteasome appeared in WT cells following culture in glucose-containing (i.e. ATP-generating) YPD media, but not in hsp82-4 cells, indicating the indispensable need for ATP in the Hsp90-dependent reassembly of the 26S proteasome (Figure 3C, left). Moreover, GA suppressed the assembly of the 26S proteasome in hsp82-4 cells, but not WT extracts, at 25°C (Figure 3C, right). Taken together, the above results clearly showed that Hsp90 is required for the in vivo reassembly of the 26S proteasome even when this was partial, and, most importantly, the ATPase function of Hsp90 seems to play a pivotal role in this assembly process.
Hsp90-dependent in vitro reassembly of the 26S proteasome
In the next series of experiments, we examined whether Hsp90-dependent dissociation–reassociation of the 26S proteasome occurs in the in vitro system. Crude cell extracts were prepared from WT and hsp82-4 cells grown at 37°C for 8 h, and subsequently incubated at 36°C for 10 min or 16°C for 1 h in the presence of an ATP-regenerating system, ATP-depleting system or GA, and, in some experiments, purified porcine Hsp90 was supplemented. Thereafter, Suc-LLVY-AMC degrading activity was assayed for the 26S proteasome affinity-purified from these cell extracts as shown in Figure 4A (left). The peptidase activity of the purified 26S proteasome decreased to nearly 10% from hsp82-4 cells upon incubation for 8 h at 37°C compared with 26S proteasome from WT cell extracts, even when ATP was regenerated (see lane 5), but completely disappeared in the absence of ATP irrespective of temperature. Intriguingly, addition of purified Hsp90 caused partial suppression of the reduction in the presence of ATP (lane 6), indicating that Hsp90 is required for the in vitro assembly of functional 26S proteasome. Moreover, this Hsp90 effect was ATP dependent, because no obvious protective effect was detected under ATP-depletion conditions (lane 8). Addition of GA partially suppressed this reactivation (lanes 7, 10 and 12). It is noteworthy that in vitro reincubation of hsp82-4 cell extracts for 1 h at 16°C caused almost complete recovery of the peptidase activity irrespective of Hsp90 supplementation (lanes 9 and 11). These findings suggest that Hsp90 promotes ATP-dependent reassembly of the 26S proteasome that had been dissociated by ts–-dependent Hsp90 inactivation.
We also confirmed that the activity change was proportional to the amounts of the purified 26S proteasome detected by western blotting with anti-Rpn12 (Figure 4A, bottom right ). No appreciable amounts of the 26S proteasome were adsorbed by the Ni+-resin column due to their disassembly in extracts of hsp82-4 cells incubated in vitro at a restricted temperature. However, incubation of the same extracts supplemented with purified Hsp90 caused association of considerable amounts of the 26S proteasome with the affinity column, although the level of the recovered proteasome was somewhat low compared with similarly treated extracts of WT cells. These results indicate that Hsp90 is required for the in vitro reassembly of the 26S proteasome. Consistent with this notion, in vivo forced expression of Hsp90 rescued the growth defect of hsp82-4 cells under non-permissive conditions, although yeast Hsc82 was effective compared with human Hsp90α (Figure 4A, right top).
To assess the above conclusion further, the 26S proteasome was inactivated and reactivated in vitro using the Ni+-resin 26S proteasome purified from WT and hsp82-4 cells grown at 25°C. Note that a considerable amount of Hsp90 is associated with the affinity-purified 26S proteasome fraction (see Figure 3A). Under incubation for 2 h at non-permissive temperature of 37°C, the purified 26S proteasome from hsp82-4 cells showed rapid reduction of the peptidase activity, unlike WT enzymes, even after addition of an ATP-regeneration system (Figure 4B, left panel, lanes 2 and 8). This reduced activity of the 26S proteasome after 2 h of incubation at 37°C was markedly increased after incubation at 16°C for 1 h (lane 9) or at 4°C for 12 h (lane 11) in the presence of an ATP-regeneration system. However, no appreciable restoration of the peptidase activity was observed when an ATP-depletion system was added during incubation at 16°C or 4°C (data not shown), indicating again that metabolic energy is necessary for reactivation of the 26S proteasome. Addition of GA partially inhibited this reactivation (lanes 10 and 12), confirming the importance of ATPase function of Hsp90 for the reactivation, and presumably reassembly, of the 26S proteasome in vitro. In addition, such restoration was observed under in vitro incubation up to 60 min, which was clearly abrogated by addition of GA, although the activities of WT enzymes were gradually decreased, irrespective of GA (right bottom panel). In accordance with these results, when the 26S proteasome from hsp82-4 cells that had been treated at 37°C for 2 h was incubated for 10 min at 36°C with purified Hsp90, the peptidase activity was partly recovered (Figure 4B, left panel, lanes 13 and 14). We also confirmed that the change in activity was proportional to the amount of the 26S proteasome detected by western blotting with anti-Rpn12 (Figure 4B, right top panel).
Genetic interactions between HSC82/HSP82 and genes encoding subunits of regulatory particle
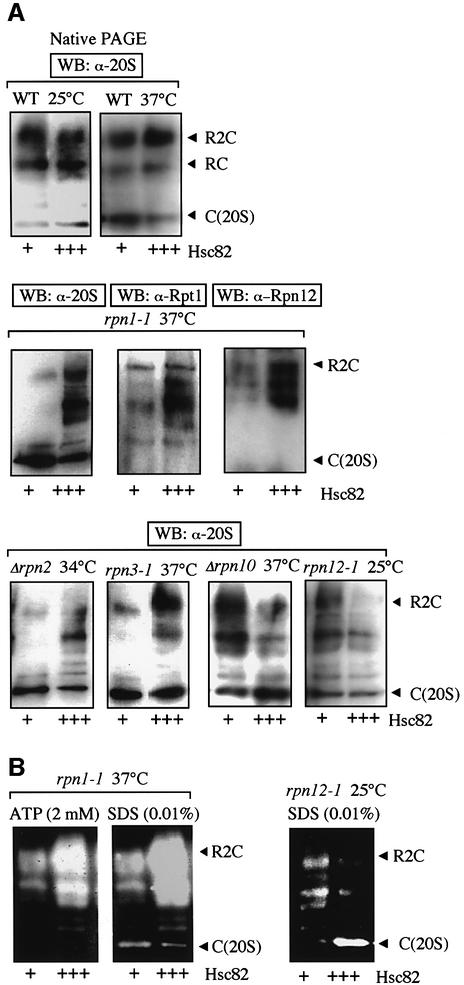
To investigate the above findings in vivo, we examined genetic interactions using various proteasome mutants (Figure 5A). The hsp82–4Δhsc82 double mutant showed severe growth defects when combined with mutants of rpn1-1, Δrpn2 and rpn3-1. Although each parental strain showed no or only a weak growth defect at 30°C, the resultant triple mutants showed no growth at 30°C. When the combination of rpn12-1 was examined, the effect was more severe because growth arrest occurred even when the cells were cultured at 25°C. Combination with Δrpn10 was most surprising; the resultant triple mutant did not grow at all even at 25°C, although Δrpn10 single mutation showed no growth defect even at 37°C (Figure 5B). On the other hand, overexpression of HSC82 suppressed the ts– growth defects of several mutants of the RP subunits of the 26S proteasome, such as rpn1-1 cells, Δrpn2 cells and rpn3-1 cells (Figure 5C, top). Surprisingly, overexpression of Hsc82 inhibited the growth of Δrpn10 cells and rpn12-1 cells when they were cultured at 36°C and 25°C, respectively. At both temperatures, cells lacking the ectopically expressed Hsc82 showed no appreciable growth defect (Figure 5C, bottom). Thus genetic interactions between HSC82/HSP82 and the genes encoding the RP subunits are strong, implying substantial interactions of the Hsp90 and the 26S proteasome.

Fig. 5. Genetic interactions between HSC82/HSP82 and various RPN genes. (A) Growth defects of triple mutants with hsp82–4Δhsc82 together with rpn1-1 (J821), Δrpn2 (J822), rpn3-1 (J823) or rpn12-1 (J8212). Each strain was spotted on YPD and incubated at the indicated temperatures for 2 days. (B) Synthetic lethality in Δrpn10 and hsp82–4Δhsc82 mutations. JD8210 cells were sporulated and dissected. No germination of a Ura+ His+ Leu+ colony corresponding to triple mutants was noted. Germination of small colonies was noted, though rarely (e.g. lanes 3 and 8). (C) Effects of overexpression of Hsc82. Each strain harboring HSC82 (+++) overexpressing plasmid (YPH500/pYO326-HSC82, JR1/pYO323-HSC82, JR2/pYO323-HSC82, JR3/pYO325-HSC82, JR10/pYO326-HSC82, JR12/pYO326-HSC82) and the vector alone (corresponding empty vector) (+) was spotted on SD plates containing appropriate amino acids and incubated at the indicated temperatures for 2 days. Note that the expressed levels of Hsc82 were not affected by these proteasomal mutations (data not shown). (D) Sensitivity to GA in various rpn mutants. Each strain used in A was spotted with or without GA (18 µM) and incubated for 2 days at indicated temperatures.
In addition, even under permissive temperature of 25°C, GA caused severe growth defects in Δrpn10 and rpn12-1 cells (Figure 5D, bottom), though its influence was less in rpn1-1 and Δrpn2 cells (top). On the other hand, Rpn3-1 cells showed obvious growth defect upon addition of GA at 30°C, but not at 25°C (Figure 5D, bottom). These findings support the genetic interactions between HSC82/HSP82 and several proteasome genes.
Effects of Hsc82 overexpression on 26S proteasome assembly in various proteasome mutants
Finally, we examined why overexpression of Hsp90 had opposing effects on cell proliferation, which was dependent on the type of mutation against the RP subunits of the 26S proteasome. Since our results showed that Hsp90 influenced the assembly of the 26S proteasome, we examined the role of Hsp90 on the 26S proteasome assembly. Since HSC82 served as a multicopy suppressor of the temperature-sensitive growth of rpn1-1 cells, Δrpn2 cells and rpn3-1 cells (Figure 5C, top), we examined the molecular species of the proteasome in these cells using native PAGE followed by western blotting using anti-20S proteasome. Overexpression of Hsc82 suppressed the disassembly of the 26S proteasome in rpn1-1 cells, Δrpn2 cells and rpn3-1cells under non-permissive temperatures of 37°C (rpn1-1 cells and rpn3-1 cells) and 34°C (Δrpn2 cells) (Figure 6A, middle and bottom panels), while absence of such overexpression markedly affected the disassembly of the 26S proteasome. In contrast, the inhibitory effects of overexpression of Hsc82 on Δrpn10 cells and rpn12-1cells (Figure 5C) appeared to be the results of disassembly of the 26S proteasome in these cells (Figure 6A, bottom). Note the appearance of several bands in these experiments, which were different from the main three bands observed initially which represented the 20S proteasome and the symmetric and asymmetric forms of the 26S proteasome. These extra bands represented incompletely assembled forms of the 26S proteasome, perhaps out by several components, because reactive bands by western blotting with anti-Rpt1 and anti-Rpn12 differed from each other and those with anti-20S proteasome (Figure 6A, middle). Furthermore, we confirmed peptidase activities of these extra bands by the in-gel peptidase assay (Figure 6B). Since the effects of overexpression of Hsc82 in various proteasome mutants correlate with the presence of the functional 26S proteasome, the above results also suggest that Hsp90 plays an important role in the regulation of assembly and disassembly of the 26S proteasome in vivo.
Fig. 6. Effects of overexpression of Hsc82 on the 26S proteasome assembly in various rpn mutants. (A) WT cells and various proteasome mutant cells harboring control plasmid (+), multicopy plasmid carrying HSC82 (+++), as in Figure 5C, grown at 25°C were shifted to the indicated temperatures and incubated for an additional 8 h. Samples of cell lysates (5 µg) were loaded onto native PAGE and analyzed by western blotting with anti-20S proteasome, anti-Rpt1 or anti-Rpn12. (B) Corresponding cell extracts as in (A) were analyzed by gel-overlay assay as in Figure 1 with 2 mM ATP or 0.01% SDS.
Discussion
The 26S proteasome is an unusually large complex, consisting of two subcomplexes, CP and RP. A major challenge to our understanding of the quaternary structure is how the large complex, composed of many subunits of various sizes, is accurately assembled in the cells. Recent studies of 20S proteasome assembly have shown that a key molecule termed ‘Ump1’ functions as a core factor to gather multiple proteasomal β-subunits (Ramos et al., 1998). At present, however, little is known about the mechanism(s) involved in the assembly of the 26S proteasome, especially the RP complex.
In the present study, we showed that inactivation of Hsp90, using yeast mutants, caused almost complete disassembly of the 26S proteasome, indicating that Hsp90 plays a role in keeping the structural integrity of this large complex (Figure 2). Inactivation of Hsp90 resulted in dissociation of the 26S proteasome into the core 20S particle, which settled into a latent state, as evidenced by the marked activation upon the addition of SDS (Figure 2A). In contrast, Rpn9 and Rpn12 subunits migrated to different positions in native PAGE (Figure 2E), indicating loss of integrity of the lid complex. However, it is not known whether these lid subunits dissociated into their monomeric forms. It is also not known how Hsp90 influences the organization of the base complex, but it is clear at least that the functional loss of Hsp90 triggers disruption of the RP complex. These findings indicate that the RP complex is structurally fragile, requiring continuous supply of a functional Hsp90 to assemble and maintain these complexes, unlike the 20S proteasome, which is apparently stable without functional Hsp90. Thus the assemblies of the CP/20S proteasome and RP complexes are mechanistically different.
The genetic evidence provided in the present study also strongly suggests in vivo linkage between Hsp90 and the 26S proteasome. In fact, we demonstrated that overexpression of Hsc82 suppressed ts– growth defects of rpn1-1 cells, Δrpn2 cells and rpn3-1 cells (Figure 5C) and prevented dissociation of the 26S proteasome, which occurred under conditions otherwise non-permissive for these mutants (Figure 6). Consistent with this genetic interaction, hsp82–4Δhsc82 showed weak synthetic lethality with those proteasomal mutations (Figure 5A). Furthermore, strong synthetic lethality was observed between hsp82–4Δhsc82 and Δrpn10 or rpn12-1. Interestingly, even though the Δrpn10 mutation itself showed no growth defect at all, it exhibited the most severe growth defect when combined with hsp82–4Δhsc82 mutant (Figure 5B and D). However, overexpression of Hsc82 in Δrpn10 cells and rpn12-1 cells enhanced their growth defects and was associated with decreased 26S proteasome levels in these two strains (Figure 6). One possible explanation for these antagonizing effects is that the tight interaction between Hsp90 and some subunits of regulatory particle might hinder them from proper formation of the 26S proteasome, leading to inhibition of cellular proliferation.
We obtained evidence that a larger amount of Hsp90 is associated with the 26S proteasome from hsp82-4 cells grown at permissive temperature, compared with the WT proteasome, irrespective of the lower content of Hsp90 in mutant cell extracts (Figure 3A, left), suggesting that the larger amount of mutant Hsp90 might be required to maintain the 26S proteasome because of its functional impairment. We also found that considerable amounts of Hsp90 are associated with the 26S proteasome through the RP complex, and this association is so tight that it occurs even under low Hsp90 conditions in which many other client proteins were dissociated from Hsp90 (Figure 3A, right). In addition, hsp82-4 cells showed a faster decay of the peptidase activities of the 26S proteasome after addition of CHX even under permissive temperature compared with WT cells (Figure 3B). Taken together, these results suggest that the 26S proteasome is the most important substrate for Hsp90, and Hsp90 is continually required for maintenance of the 26S proteasome.
The energy requirement for 26S proteasome assembly has been recognized since the discovery of the 26S proteasome (Armon et al., 1990; Driscoll and Goldberg, 1990; Chu-Ping et al., 1994), but the molecular mechanisms of ATP consumption have remained elusive. In the present study, we have provided evidence that ATPase of Hsp90 plays an active role in supplying energy required for the 26S proteasome assembly. Thus, we have shown that the dissociated constituents of the 26S proteasome from hsp82-4 cells under non-permissive conditions, reassembled in vivo and in vitro by reactivation of ts– Hsp82 or addition of purified Hsp90 (Figures 3 and 4). Although the reason for not being able to get the full restoration by purified Hsp90 in vitro is not clear at present, it is plausible that Hsp90 acts in concert with various cochaperones, such as p23 (Young et al., 2001), to exert its full activity, some of which might be lost in the purification of Hsp90 by the present technique. More importantly, we demonstrated that ATP is required for Hsp90-dependent reassembly and that inhibition of Hsp90-ATPase function by GA also blocked in part this restoration of the 26S proteasome. Thus we propose that the energy required for the assembly of the 26S proteasome is at least utilized by Hsp90.
An important question is whether the dissociation–association cycle of the 26S proteasome has any physiological significance. An intriguing scenario is that a dissociation–association cycle might be envisaged for the 26S proteasome or it might be regulated to respond to changes in certain environmental circumstances in cells. For example, marked increases have been found in the amounts of the 26S proteasome during the stationary phase compared with those in logarithmically growing yeast cells (Fujimuro et al., 1998). In this regard, it is interesting to note that yeast Hsp90 also increases during the stationary phase (Iida and Yahara, 1984), implying that Hsp90 may contribute to these dynamic alterations, repeating assembly and disassembly in logarithmic/stationary phase shift. In addition, the involvement of an Hsp90 chaperone in the assembly of the 26S proteasome indicates that changes in the physiological state of Hsp90 may alter the amounts of the 26S proteasome. In fact, we initially found that the amounts of the 26S proteasome decreased upon exposure to the thermal insult of 50°C but showed full recovery within 6–8 h after the temperature was reversed to 25°C (Figure 1). In this regard, previous studies reported that the cellular ATP level remained largely unchanged under such severe heat shock conditions (Jamsa et al., 1995), indicating that disassembly of the 26S proteasome is not due to reduced availability of ATP. However, such temporary reductions in the 26S proteasome are conceivable because while Hsp90 is required for the 26S proteasome assembly, it is also responsible for refolding stress-damaged proteins and thereby might be sequestered to those damaged proteins after severe thermal insults. Thus it is conceivable to view the disassembly of the 26S proteasome as a stress response regulated by Hsp90. In other words, our results highlight the importance of Hsp90 in the disassembly–reassembly cycle of the 26S proteasome, although further studies are necessary to determine its precise molecular action.
Materials and methods
Microbiological techniques
Experimental methods for yeast were performed as described (Guthrie and Fink, 1991). Yeast cells were cultured in YPD on logarithmically growing phase, unless otherwise indicated.
Plasmids
Plasmids pYO323-HSC82 and pYO325-HSC82 carry the 4.2 kbp SpeI–SpeI fragments of HSC82 in the XbaI site of pYO323 and pYO325 (Ohya et al., 1991), respectively. Plasmids pYO326-HSC82 (Imai and Yahara, 2000) and pRS316-pADH-hHsp90α were our laboratory stock.
Antibodies and reagents
Rabbit polyclonal anti-Rpn9 and anti-Rpn12 antibodies were a kind gift from Dr A.Toh-e (Tokyo University). Rabbit polyclonal anti-Rpt1 (Takeuchi et al., 1999), anti-20S proteasome (Tanaka et al., 1988) and anti-Hsp82 (Imai and Yahara, 2000) antibodies were used. Purified porcine Hsp90 was our laboratory stock. Suc-LLVY-AMC was obtained from Peptide Institute Inc. CHX, deoxyglucose and azide were purchased from Sigma Chemical Co. (St Louis, MO). GA was obtained from Gibco BRL (Gaithersburg, MD). Protein concentration was determined using BCA protein assay reagent (Pierce Chemical Co.) with bovine serum albumin (Sigma) as the standard.
Strains
The yeast strains used are listed in Table I. Strains referred to in this study were constructed by conventional genetic methods.
Table I. Yeast strains used in this study.
| Strain | Source | Genotype |
|---|---|---|
| YPH500 | MATα ura3 lys2 ade2 trp1 his3 leu2 | Sikorski and Hieter, 1989 |
| YOK5 | MATa ura3 lys2 ade2 trp1 his3 leu2 Δhsc82::URA3 hsp82-4::LEU2 | Kimura et al., 1994 |
| YOK5H | MATa ura3 lys2 ade2 trp1 his3 leu2 Δhsc82::HIS3 hsp82-4::LEU2 | Our laboratory stock |
| YOK5RH | MATa ura3 lys2 ade2 trp1 his3 leu2 Δhsc82::HIS3 hsp82-4::LEU2 6×His-RPT1::URA3 | Present study |
| 5CG2 | MATa ura3 lys2 ade2 trp1 his3 leu2 Δhsc82::URA3 hsp82-4::GAL1-HSP82::LEU2 | Kimura et al., 1994 |
| J106 | MATα 6×His-RPT1::URA3 ura3 lys2 ade2 trp1 his3 leu2 | Takeuchi et al., 1999 |
| YK109 | MATa ura3 lys2 ade2 trp1 his3 leu2 rpn12-1 | Kominami et al., 1997 |
| JR1 | MATa ura3 lys2 ade2 trp1 his3 leu2 rpn1-1 | Present study |
| J821 | MATa ura3 lys2 ade2 trp1 his3 leu2 Δhsc82::HIS3 hsp82-4::LEU2 rpn1-1 | Present study |
| JR2 | MATa ura3 lys2 ade2 trp1 his3 leu2 Δrpn2::URA3 | Present study |
| J822 | MATa ura3 lys2 ade2 trp1 his3 leu2 Δhsc82::HIS3 hsp82-4::LEU2 Δrpn2::URA3 | Present study |
| JR3 | MATa ura3 lys2 ade2 trp1 his3 leu2 rpn3-1::HIS3 | Present study |
| J823 | MATa ura3 lys2 ade2 trp1 his3 leu2 Δhsc82::URA3 hsp82-4::LEU2 rpn3-1::HIS3 | Present study |
| JR10 | MATa ura3 lys2 ade2 trp1 his3 leu2 Δrpn10::HIS3 | Present study |
| JR12 | MATa ura3 lys2 ade2 trp1 his3 leu2 rpn12-1 | Present study |
| J8212 | MATa ura3 lys2 ade2 trp1 his3 leu2 Δhsc82::HIS3 hsp82-4::LEU2 rpn12-1 | Present study |
| J38 | MATα leu2 his3 ura3 trp1 Δrpn10::HIS3 | Takeuchi et al., 1999 |
| JD8210 | MATa/α ura3/ura3 lys2/lys2 ade2/ade2 trp1/trp1 his3/his3 leu2/leu2 Δhsc82::URA3/Δhsc82::URA3 hsp82-4::LEU2/hsp82-4::LEU2 RPN10/Δrpn10::HIS3 | Present study |
| rpn1::URA3 | MATα ura3 lys2 ade2 trp1 his3 leu2 rpn1::URA3 (rpn1-1) | Our laboratory stock |
| W1646–1C | MATα leu2 his3 ura3 trp1 ade2 Δrpn2::URA3 | Our laboratory stock |
| YK137 | MATa leu2 his3 ura3 trp1 rpn3-1::HIS3 | Kominami et al., 1997 |
Electrophoresis
We used 10–20% and 7.5–15% (Figure 2B only) gradient gel for SDS–PAGE and 2–15% polyacrylamide gradient gel (Daiichi Pure Chemical Co.) for native PAGE.
Western blotting
The crude cell extracts were subjected to SDS–PAGE or native PAGE, transferred onto a PVDF membrane. Then the blot was developed with the indicated primary antibodies, horseradish peroxidase conjugated secondary antibodies and the chemiluminescent substrate.
Peptidase activity
Peptidase activity was assayed using Suc-LLVY-AMC as a substrate. Suc-LLVY-AMC (0.1 mM) was incubated with an enzyme source for 10 min at 37°C as described previously (Tanaka et al., 1988). The activities are expressed as averages of three independent experiments or as mean ± SEM. The overlay assay of peptidase activities of the proteasome after native PAGE was described previously (Glickman et al., 1998). Peptidase activity was visualized by irradiating the gel with 380 nm UV light.
Preparation of crude extracts
Yeast cells were harvested and washed once with ice-cold lysis buffer (100 mM Tris–HCl pH 7.6, 2 mM ATP, 0.5 mM EDTA, 2 mM MgCl2 and 2% glycerol), and then disrupted with glass beads in 200 µl of lysis buffer. After removal of unbroken cells and glass beads by brief centrifugation at 100 g, the extracts were clarified by centrifugation twice at 20 000g and 4°C for 10 min. The final supernatant was used as the crude extract.
Affinity-purification of 26S proteasome
Purification of 26S proteasome by Ni-nitrilotriacetic acid (NTA) affinity chromatography was performed. Briefly, the yeast extracts were clarified by centrifugation at 100 000 g at 4°C for 30 min. The supernatant was subjected to ultracentrifugation at 235 000 g and 4°C for 5 h to precipitate the proteasome. The resulting precipitates were gently dissolved in 10 ml of buffer A (20 mM Tris–HCl pH 7.8, 1 mM ATP, 0.1 mM EDTA, 2 mM MgCl2, 100 mM NaCl and 10% glycerol) with an ATP regeneration system. After removal of insoluble materials by centrifugation at 9100 g and 4°C for 10 min, the resulting supernatant was loaded onto a Ni+-NTA–agarose column (Qiagen, Hilden, Germany). After washing the column with buffer A containing 50 mM imidazole, the His-tagged column associated with the proteasome was eluted with elution buffer (buffer A with 200 mM imidazole). In successive experiments, the 26S proteasome was purified from the same volume of cell extracts containing the same amount of protein.
References
- Armon T., Ganoth,D. and Hershko,A. (1990) Assembly of the 26S complex that degrades proteins ligated to ubiquitin is accompanied by the formation of ATPase activity. J. Biol. Chem., 265, 20723–20726. [PubMed] [Google Scholar]
- Baumeister W., Walz,J., Zuhl,F. and Seemuller,E. (1998) The proteasome: paradigm of a self-compartmentalizing protease. Cell, 92, 367–380. [DOI] [PubMed] [Google Scholar]
- Chu-Ping M., Vu,J. H, Proske,R.J., Slaughter,C.A. and DeMartino,G.N. (1994) Identification, purification and characterization of a high molecular weight, ATP-dependent activator (PA700) of the 20S proteasome. J. Biol. Chem., 269, 3539–3547. [PubMed] [Google Scholar]
- Driscoll J. and Goldberg,A.L. (1990) The proteasome (multicatalytic protease) is a component of the 1500-kDa proteolytic complex which degrades ubiquitin-conjugated proteins. J. Biol. Chem., 265, 4789–4792. [PubMed] [Google Scholar]
- Frydman J. (2001) Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem., 70, 603–647. [DOI] [PubMed] [Google Scholar]
- Fujimuro M., Takada,H., Saeki,Y., Toh-e,A., Tanaka,K. and Yokosawa,H. (1998) Growth-dependent regulation of the 26S proteasome assembly in the budding yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun., 251, 818–823. [DOI] [PubMed] [Google Scholar]
- Glickman M.H., Rubin,D.M., Coux,O., Wefes,I., Pfeifer,G., Cjeka,Z., Baumeister,W., Fried,V.A. and Finley,D. (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9 signalosome and elF3. Cell, 94, 615–623. [DOI] [PubMed] [Google Scholar]
- Guthrie C. and Fink,G.R. (eds) (1991) Guide to yeast genetics and molecular biology. Academic Press, San Diego, CA. [Google Scholar]
- Iida H. and Yahara,I. (1984) Durable synthesis of high molecular weight heat shock proteins in G0 cells of the yeast and other eucaryotes. J. Cell Biol., 99, 199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai J. and Yahara,I. (2000) Role of HSP90 in salt stress tolerance via stabilization and regulation of calcineurin. Mol. Cell. Biol., 20, 9262–9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamsa E., Vakula,N., Arffman,A., Kilpelainen,I. and Makarow,M. (1995) In vivo reactivation of heat-denatured protein in the endoplasmic reticulum of yeast. EMBO J., 14, 6028–6033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura Y., Matsumoto,S. and Yahara,I. (1994) Temperature-sensitive mutants of hsp82 of the budding yeast Saccharomyces cerevisiae. Mol. Gen. Genet., 242, 517–527. [DOI] [PubMed] [Google Scholar]
- Kominami K. et al. (1997) Yeast counterparts of subunits S5a and p58 (S3) of the human 26S proteasome are encoded by two multicopy suppressors of nin1-1. Mol. Biol. Cell, 8, 171–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohya Y., Umemoto,N., Tanida,I., Ohta,A., Iida,H. and Anraku,Y. (1991) Calcium-sensitive cls mutants of Saccharomyces cerevisiae showing a Pet– phenotype are ascribable to defects of vacuolar membrane H(+)-ATPase activity. J. Biol. Chem., 266, 13971–13977. [PubMed] [Google Scholar]
- Panaretou B., Prodromou C., Roe S.M., O’Brien R., Ladbury J.E., Piper P.W. and Pearl L.H. (1998) ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J., 17, 4829–4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem., 70, 503–533. [DOI] [PubMed] [Google Scholar]
- Ramos P. C, Hockendorff,J., Johnson,E.S., Varshavsky,A. and Dohmen,R.J. (1998) Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell, 92, 489–499. [DOI] [PubMed] [Google Scholar]
- Richter K. and Buchner,J. (2001) Hsp90: chaperoning signal transduction. J. Cell. Physiol., 188, 281–290. [DOI] [PubMed] [Google Scholar]
- Sherman M.Y. and Goldberg,A.L. (2001) Cellular defences against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron, 29, 15–32. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strain designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi J., Fujimuro,M., Yokosawa,H., Tanaka,K. and Toh-e,A. (1999) Rpn9 is required for efficient assembly of the yeast 26S proteasome. Mol. Cell. Biol., 19, 6575–6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Yoshimura,T., Kumatori,A., Ichihara,A., Ikai,A., Nishigai,M., Kameyama,K. and Takagi,T. (1988) Proteasomes (multi-protease complexes) as 20S ring-shaped particles in a variety of eukaryotic cells. J. Biol. Chem., 263, 16209–16217. [PubMed] [Google Scholar]
- Young J.C., Moarefi,I. and Hartl,F.U. (2001) Hsp90: a specialized but essential protein-folding tool. J. Cell Biol., 154, 267–273. [DOI] [PMC free article] [PubMed] [Google Scholar]