Abstract
To study the relationship between DNA replication and transcription in vivo, we investigated Hox gene activation in two vertebrate systems: the embryogenesis of Xenopus and the retinoic acid-induced differentiation of pluripotent mouse P19 cells. We show that the first cell cycles following the mid- blastula transition in Xenopus are necessary and sufficient for HoxB activation, whereas later cell cycles are necessary for the correct expression pattern. In P19 cells, HoxB expression requires proliferation, and the entire locus is activated within one cell cycle. Using synchronous cultures, we found that activation of HoxB genes is colinear within a single cell cycle, occurs during S phase and requires S phase. The HoxB locus replicates early, whereas replication is still required for maximal expression later in S phase. Thus, induction of HoxB genes occurs in a DNA replication-dependent manner and requires only one cell cycle. We propose that S-phase remodelling licenses the locus for transcriptional regulation.
Keywords: chromatin domains/DNA replication/Hox/mouse/Xenopus
Introduction
It has long been thought that transcription and DNA replication might be mechanistically linked in eukaryotic cells to coordinate formation of chromatin with gene activity on replicating DNA and to avoid initiating DNA replication within transcribing genes (Wolffe, 1991). DNA replication is coupled to chromatin reformation and thus provides an opportunity for transcriptional reprogramming. Early evidence for a coupling of replication and transcription came from observations that transcriptionally active genes replicate early and vice versa (Goldman et al., 1984), and studies using genome-wide microarray analysis of transcription and timing of DNA replication in Drosophila and yeast have addressed the generality of this notion. Whereas, in yeast, no correlation was observed between replication timing and transcriptional activity (Raghuraman et al., 2001), the opposite result was obtained in Drosophila (Schübeler et al., 2002). DNA microinjected into human cells is also more likely to be transcribed in early rather than late S phase (Zhang et al., 2002). Thus, coregulation of DNA replication and transcription may have evolved in higher eukaryotes to coordinate cell proliferation and patterning during development.
In both invertebrates and vertebrates, cell differentiation can in some cases be uncoupled from proliferation (Harris and Hartenstein, 1991; de Nooij and Hariharan, 1995), whereas in other cases cell division is necessary to specify a change in cell fate (Ambros, 1999; Mennerich and Braun, 2001). DNA replication may provide a mechanism to link proliferation to such patterning decisions by activating or repressing transcription. Some in vitro experiments have suggested that this may be the case (Crowe et al., 2000). We have chosen to investigate this question in vivo in a developmentally regulated locus, the HoxB locus.
The Hox loci are contiguous sets of genes encoding homologous transcription factors involved in patterning of various tissues in all animals (Krumlauf, 1994). Hox gene transcriptional activation occurs in vertebrate embryos according to temporal and spatial colinearity, which dictate that activation timing and anterior boundaries of expression follow the order of the genes along the complex (Duboule and Morata, 1994; Krumlauf, 1994; Gaunt and Strachan, 1996). Many cis elements that regulate transcription of different Hox genes have been identified (for reviews, see Duboule, 1998; Gellon and McGinnis, 1998), but there is as yet no mechanistic explanation for colinearity. Although vertebrate Hox activation has been proposed to be controlled by proliferation, which would mediate progressive opening of the Hox locus (Duboule, 1994), this has not yet been tested. Nevertheless, cell division may be associated with Hox patterning (Ohsugi et al., 1997), and disrupting the somitic cell cycles of chick embryos produces homeotic defects (Primmett et al., 1989). Temporal colinearity may be an intrinsic feature of embryonic expression of Hox genes, since it also occurs, over the space of a week, on HoxB induction by retinoic acid (RA) treatment of pluripotent human NT2 cells (Simeone et al., 1990), although the relationship of this activation with cell proliferation is not known.
In this paper, we show that Hox gene activation is dependent on DNA replication in two different systems. We found that, in Xenopus, all HoxB genes studied are activated in a short time window at the mid-blastula transition (MBT). Inhibition of DNA replication before the MBT prevents HoxB activation, whereas inhibition one or two cell cycles after the MBT alters the Hox expression pattern. Using synchronous P19 cells induced by RA, we demonstrate that expression of the entire HoxB locus can occur within one cell-cycle period after stimulation and is sensitive to inhibition of DNA replication and that colinear activation of Hox expression occurs during S phase. We found that the HoxB locus replicates early, whereas colinear activation occurs later, implying that DNA replication licenses the locus for transcriptional control. We propose that the domain structure and chromatin of the HoxB locus has evolved for coregulation of DNA replication and transcription, providing support that such coregulation, as implied from microarray analysis in Drosophila, also applies to vertebrates.
Results
HoxB genes are expressed between the MBT and gastrulation in a replication-dependent manner
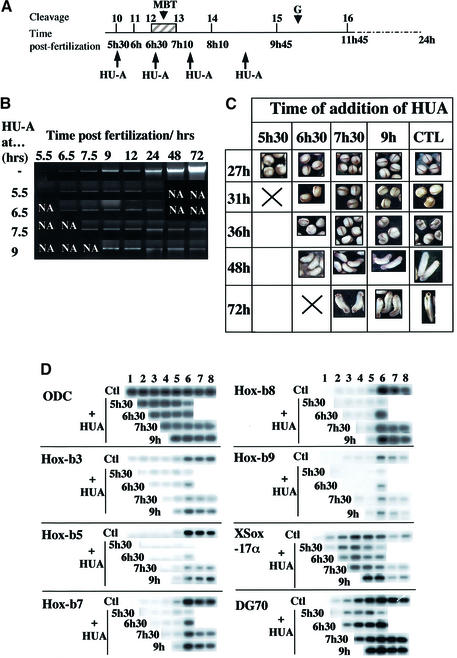
In Xenopus, it has been observed that embryos exposed to aphidicolin at late blastula, mid-gastrula or neurula stages continue to differentiate and that the expression of various embryo-specific genes was unaffected by the arrest of DNA synthesis (Rollins and Andrews, 1991; Harris and Hartenstein, 1991). Although replication-inhibited embryos do not develop correctly, neural determination can still take place. We wished to precisely define the moment at which differentiation becomes independent of DNA replication, so we targeted the first cell cycles after the MBT (Figure 1A). Xenopus embryos undergo the first cleavage after 90 min and the following 11 synchronous divisions every 30 min (Newport and Kirschner, 1982a; Boterenbrood et al., 1983). Cleavage 12–13, 7 h after fertilization, is marked by an abrupt transition, the MBT, with an increase of the cell-cycle length and the onset of zygotic transcription (Newport and Kirschner, 1982a,b). Cell cycles 12, 13 and 14 last 40, 60 and 75–100 min, respectively, with increasing asynchrony (Satoh, 1977; Newport and Kirschner, 1982a; Boterenbrood et al., 1983; Masui and Wang, 1998). We used a combination of hydroxyurea and aphidicolin (HU-A) that rapidly and completely inhibits DNA replication in embryos (Harris and Hartenstein, 1991), which we added at short intervals around the MBT (Figure 1A). Figure 1B shows the increase in genomic DNA content due to DNA replication in embryos and shows that HU-A treatment inhibits DNA replication within 60–90 min. If HU-A is added 90 min before the MBT, embryos do not develop further and disaggregate a day later. However, morphological aspects of neurulation occurred normally if HU-A was added only 30 min before the MBT (Figure 1C), allowing the MBT cell cycle to take place (Figure 1B). Adding HU-A 30 min after the MBT allows development until the early tailbud stage, but the embryos are truncated and have less definition in the head, although they do have cement glands. Adding HU-A 2 h after the MBT permits tailbud progression, but head development is affected and embryos are curved (Figure 1C). Neural precursors can differentiate but not divide in such embryos, allowing generation of all types of neurones, although fewer in number (Harris and Hartenstein, 1991).
Fig. 1. Inhibition of DNA replication in Xenopus embryos and Hox expression. (A) Summary of this experiment: schema showing cell-cycle stages and timing during early Xenopus development and timing of hydroxyurea and aphidicolin (HU-A) addition. MBT, mid-blastula transition; G, onset of gastrulation. (B) HU-A was added to developing Xenopus embryos before or just after the MBT at the indicated times (left) and maintained throughout development. Genomic DNA was quantitatively prepared from embryos frozen during the time course at the indicated times (top), and DNAs from 0.5 embryo equivalents were loaded on a gel and analysed by gel electrophoresis and ethidium bromide staining. NA, not applicable, since these embryos (i) at these time points are from the control group and shown in the control lanes, thus no DNA was loaded in these lanes (left) or (ii) started to disaggregate by 24 h (right). (C) Phenotype of embryos photographed at the indicated times after fertilization. (D) Total HoxB expression in replication-inhibited embryos. At the indicated times from the five different time courses, five embryos were lysed for total RNA preparation, and semiquantitative RT–PCR and Southern blotting was performed for the indicated Hox genes, the ornithine decarboxylase gene (ODC) and the genes XSox17α and DG70. Time points were (1) 5 h 30, (2) 6 h 30, (3) 7 h 30, (4) 9 h, (5) 12 h, (6) 24 h, (7) 48 h and (8) 72 h.
These data confirm and extend previous studies (Harris and Hartenstein, 1991; Rollins and Andrews, 1991) showing that extensive morphogenesis can occur in the absence of DNA replication in post-blastula embryos. We also show that HU-A added during early cleavages blocks the cell cycle and further development after the MBT and that very few cell cycles after the MBT, possibly only one, are sufficient to permit neural determination. Allowing DNA replication for a slightly longer time allows a dramatic increase in morphological development. Intervals of no more than one cell cycle are probably responsible for differences in patterning between groups of progressively later-inhibited embryos.
From these data, we expected that HoxB expression might occur if DNA replication is permitted for only one or two cell cycles after the MBT. Expression of the Hoxb3–Hoxb9 domain in HU-A-treated and control embryos was measured by RT–PCR. Figure 1D shows that constitutively expressed ornithine decarboxylase mRNA did not significantly vary whether or not HU-A was present. In contrast, HoxB mRNA expression was induced from 12 h after fertilization (Figure 1D; data not shown), within the fourth cell cycle after the MBT. HoxB genes are partly (Hoxb3 and Hoxb5) or normally (Hoxb7–Hoxb9) induced if DNA replication is inhibited after the MBT (HU-A added at 6 h 30 or 7 h 30), whereas all are repressed when HU-A is added during the early cleavage stage (5 h 30). Similar results were obtained from three experiments. We conclude that HoxB transcription occurs if the cell cycle corresponding to the MBT, or one cell cycle after, is permitted and that expression of HoxB genes occurs within a short window and does not require further rounds of replication.
We wondered whether this cell-cycle linkage and subsequent uncoupling is unique to Hox genes or whether other developmental genes are similarly regulated. We therefore looked at the expression of other genes activated around or shortly after the MBT. Figure 1D shows data for two of these: DG70, expressed at gastrulation with similar timing to HoxB genes; and XSox17α, involved in endoderm formation. The cell-cycle dependence of their expression represents the two situations we found (coupling or not). For XSox17α, addition of HU-A 90 min before the MBT does not prevent normal induction of gene expression, showing that some zygotic gene expression has no need for cell-cycle progression for its programming. However, DG70 expression is greatly reduced if passage through the cell cycle at the MBT is blocked. Thus, in general, determination of gene expression affected by the cell cycle occurs very early in Xenopus: one or two cell cycles permits high level expression significantly later in development.
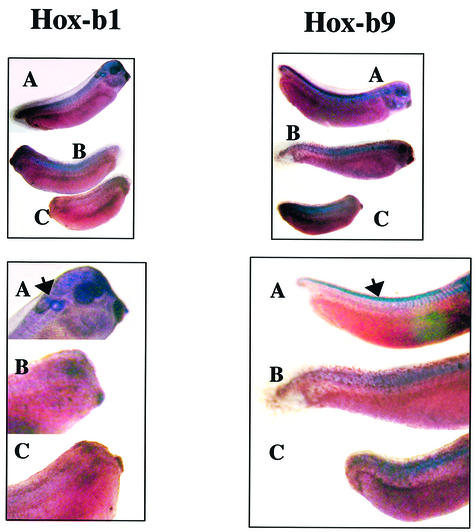
The experiment described above did not address whether HoxB expression was correctly localized. In normal tailbud embryos, Hoxb1 is expressed in the hindbrain around the otic vesicle and in somites, and Hoxb9 is expressed in the neural tube and in somites (Godsave et al., 1994). Spatial localization of HoxB expression in replication-inhibited embryos and controls is shown in Figure 2. Embryos in which DNA replication was inhibited by HU-A at 30 min (Figure 2B) or 90 min after the MBT (Figure 2C) express both Hox genes in the area where somites condense, whereas neither was detectable in the central nervous system (CNS), even though neural precursors are present (Harris and Hartenstein, 1991).
Fig. 2. HoxB expression in the CNS is disrupted in replication-inhibited Xenopus embryos. In situ hybridization (see Materials and methods) for Hoxb1 (left) and Hoxb9 mRNA (right) in normal (A) or replication- inhibited embryos to which hydroxyurea and aphidicolin was added at 9 h (B) or 7.5 h (C) after fertilization and maintained in the medium. Embryos were fixed at 72 h after fertilization. HoxB expression was fixed for all embryos after the same length of time of the staining reaction. The lower images are close-ups of the same embryos (left, A and C, and right B) or embryos treated in exactly the same way (left, B, and right, A and C) as those in the upper images. The arrows point to normal expression of the two Hox genes in the CNS (see text for details).
Induction of HoxB genes in embryonal carcinoma cells occurs in a period of time corresponding to one cell cycle and requires S phase
The induction of HoxB genes during early Xenopus development suggested a cell-cycle dependence of their expression. However, in embryos, due to asynchronous cell divisions among blastomeres after the MBT (Satoh, 1977), a strict relationship between cell-cycle progression and Hox expression cannot be established. We therefore used pluripotent embryonal carcinoma (EC) cells to further study this relationship.
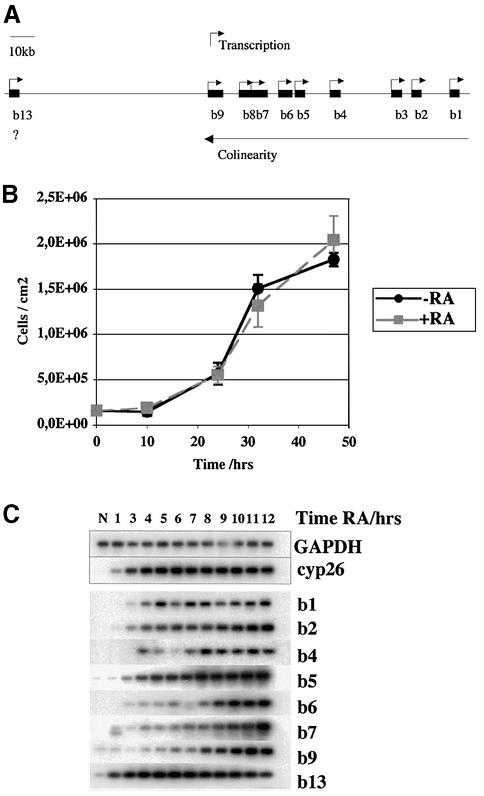
Figure 3A shows the main characteristics of the mouse HoxB locus. RA can induce Hox expression in EC cells, and this activation occurs with temporal colinearity over the space of a week for HoxB genes in human NT2 EC cells (Simeone et al., 1990), suggesting that such cells may retain a similar Hox regulation to that of mouse embryos. We chose the mouse EC cell line P19, in which RA can stimulate differentiation from any point in the cell cycle (Berg and McBurney, 1990) and induce HoxB expression. In P19 cells, HoxB genes were maximally induced at 0.5 µM RA (data not shown). The main questions we addressed were whether HoxB expression occurs in dividing cells and with what timing, whether it depends on cell division, whether P19 cells show temporal colinearity and, if so, whether this is related to the cell cycle.
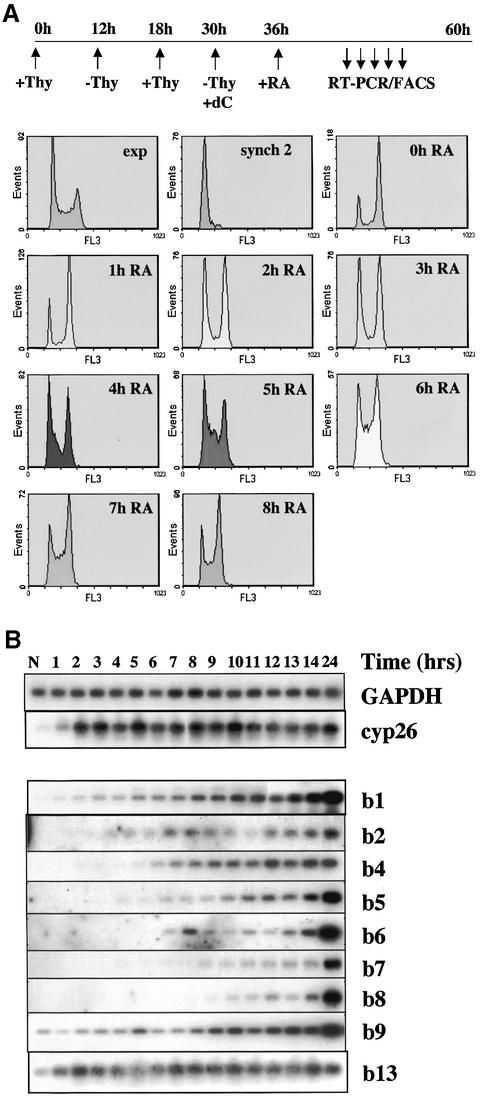
Fig. 3. Expression of all HoxB genes in P19 cells is activated during the first cell cycle after induction. (A) Schema showing the position, size, orientation and colinearity of gene transcription in the mouse HoxB locus. (B) Cell proliferation continues after retinoic acid (RA) stimulation. P19 cells were seeded at 10% confluence, incubated overnight and then either stimulated (+RA) or not (-RA) with 0.5 µM RA, and triplicate samples were trypsinized and counted. (C) HoxB activation occurs within one cell cycle. P19 cells were grown to early exponential phase, aggregated in suspension overnight, aggregates stimulated by RA, and extracts prepared during a 12 h time course for subsequent analysis by semiquantitative RT–PCR and Southern hybridization.
Figure 3B shows that RA-treated P19 cells continue to proliferate for at least three cell divisions, at a similar rate to control cells (∼10 h per cell cycle). Flow cytometry analysis of DNA content showed a similar cell-cycle distribution to controls (data not shown). Addition of RA rapidly induces the control Cyp26 gene, encoding a cytochrome-p450 RA hydroxylase (Sonneveld et al., 1999), as expected (Figure 3C), whereas constitutively expressed GAPDH did not vary. Initiation of expression of the entire HoxB locus occurs within 4 h, and genes are well expressed within 9–10 h, thus in the time corresponding to less than one cell cycle (Figure 3C). Highest levels of expression were obtained after 24 h (data not shown; see also Figure 4A). We could not detect temporal colinearity under these conditions.
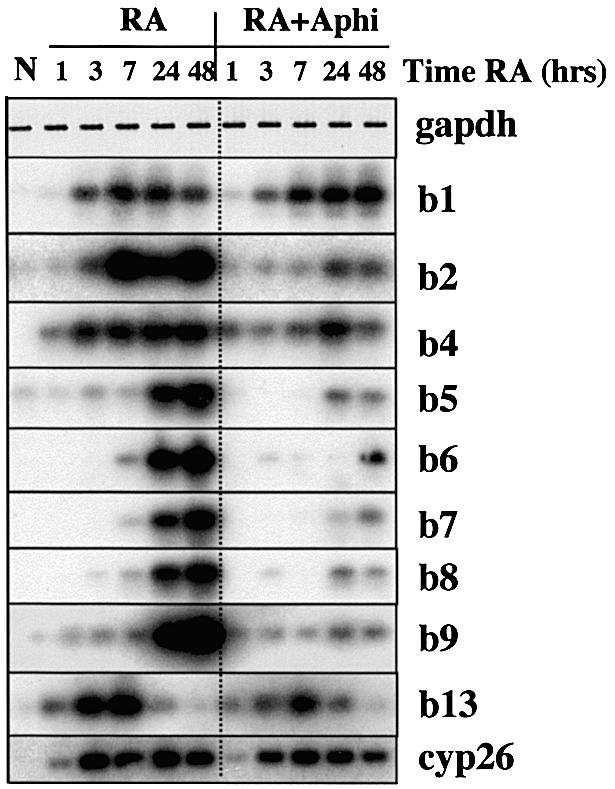
Fig. 4. Inhibition of DNA replication disrupts HoxB expression in P19 cells. P19 cell aggregates were treated with retinoic acid (RA) with or without aphidicolin (Aphi) and samples were lysed for semiquantitative RT–PCR at the indicated times and detected by Southern blotting using labelled probes for the relevant HoxB genes (abbreviated to b1, b2, etc.) and Cyp26 (for GAPDH, staining of the gel is shown). No expression was detected without cDNA input in the PCR (data not shown). N, non-treated P19 cell aggregates.
Since RA-activated HoxB expression requires less than one cell cycle, we asked whether it requires passage through the cell cycle by treating cells with 15 µM aphidicolin at the same time as RA. We verified by flow cytometry that aphidicolin blocks DNA replication in these cells (data not shown). A time course of activation of HoxB genes in the absence or presence of aphidicolin is shown in Figure 4. In untreated cells, HoxB mRNAs are not detectable, and after RA addition they become detectable after 1–3 h, increasing throughout the experiment to 48 h, apart from Hoxb13, which remains stable or decreases by 10 h (as in Figure 3B). In the presence of aphidicolin, Hoxb1 and Hoxb13 at the 3′ and 5′ ends of the domain (Figure 3A) are activated with normal timing, as is Cyp26. However, expression of the Hoxb2–Hoxb9 genes was significantly reduced by aphidicolin treatment (Figure 4; see also Figures 6C, 9C and 10B). Quantifica tion of these results is provided in Supplementary data (available at The EMBO Journal Online). We also reproducibly observed a gradient of sensitivity to aphidicolin, with expression of the more internal domain Hoxb6–Hoxb8 the most sensitive to inhibition of DNA replication, followed by Hoxb4 and Hoxb5, then Hoxb2 and Hoxb9, and finally Hoxb1 and Hoxb13 at opposite ends of the domain either weakly or not at all inhibited by aphidicolin (see also Figure 10B). The overall profile of this inhibition was similar in six separate experiments.
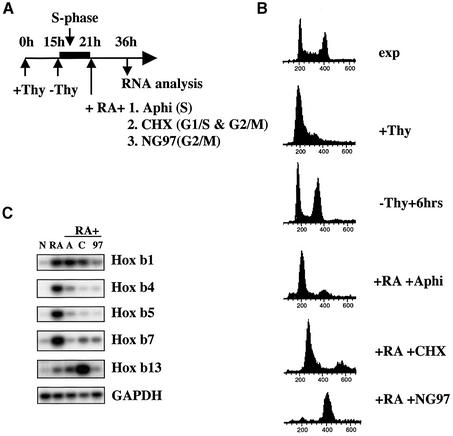
Fig. 6. Passage through DNA replication, rather than mitosis, is required for correct HoxB expression. (A) Exponentially growing (exp) P19 cell aggregates were pre-synchronized with 2 mM thymidine (+Thy) for 15 h and then released for 6 h (-Thy) before adding retinoic acid (RA) for a further 15 h, either alone or in combination with aphidicolin (Aphi), cycloheximide (CHX) or NG97. (B) Progression through the cell cycle was checked by flow cytometry. (C) Southern blots of semiquantitative RT–PCR products from the same experiment. N, non-treated aggregates; RA alone, or with aphidicolin (A), cycloheximide (C) or NG97 (97).
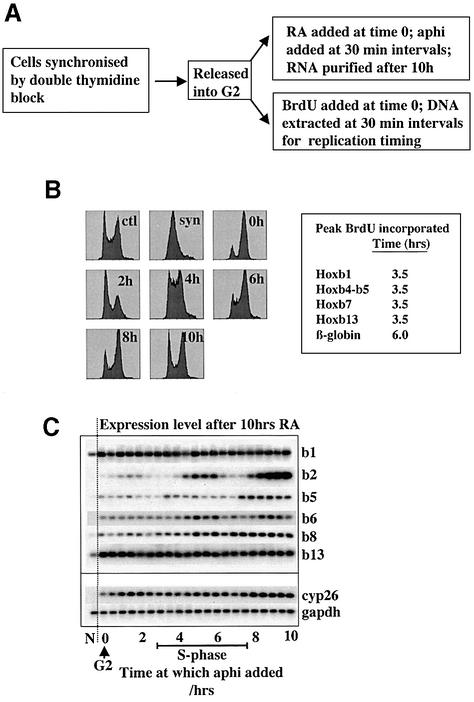
Fig. 9. S-phase transitions in dependence of HoxB expression on DNA replication. Parallel cultures of double thymidine synchronized P19 cell aggregates (syn) were released into G2 for 6 h and retinoic acid (RA) was added, corresponding to time zero (0 h), to one population and bromodeoxyuridine (BrdU) to the other. The time course was then followed for 10 h. (A) Summary of experiment (aphi, aphidicolin). (B) Left: samples were also withdrawn and fixed for flow cytometry every 2 h to monitor cell-cycle progression (ctl, exponentially growing control cells). Right: summary of replication timing analysis for the loci shown, as determined by quantitative anti-BrdU chromatin immunoprecipitation and PCR, from parallel time course with samples every 30 min. (C) RA was added at 0 h to all samples; aphidicolin was then added to samples at half-hour intervals within the 10 h time course, and RNA was extracted from all samples at the end of the 10 h, for analysis of gene expression by semiquantitative RT–PCR and Southern blotting. N, non-RA-treated cells.
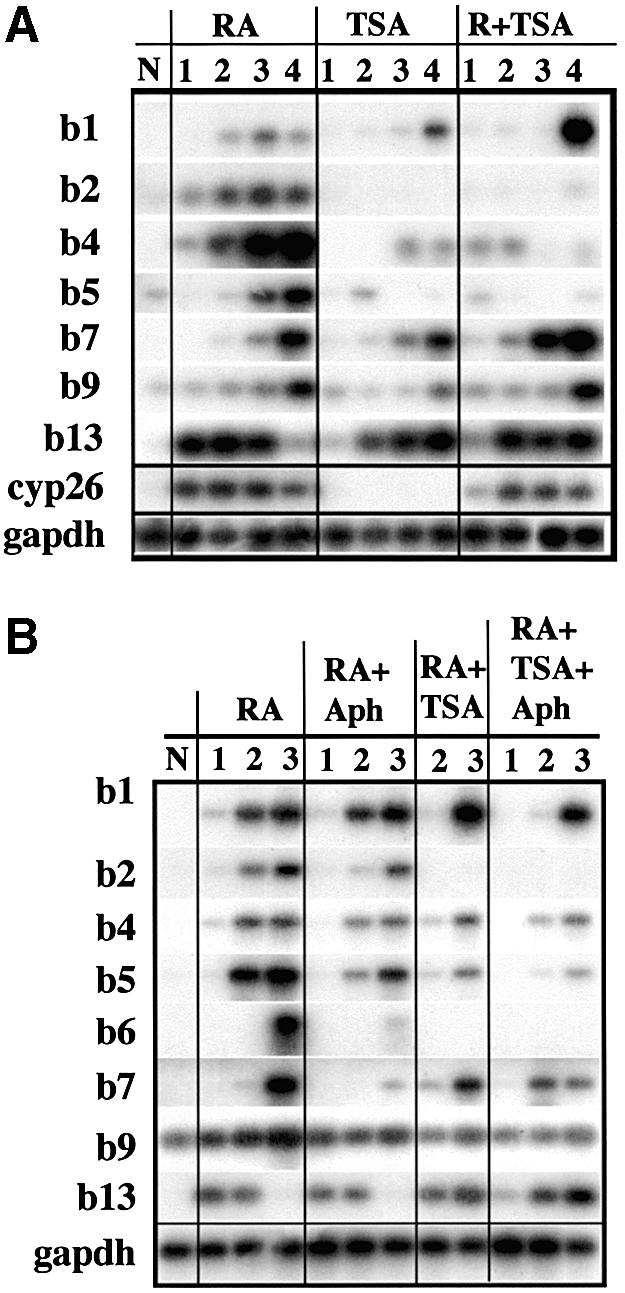
Fig. 10. Inhibition of histone deacetylation and methylation affects HoxB expression but does not mimic retinoic acid (RA) induction. (A) Southern blots of semiquantitative RT–PCR products from an experiment in which P19 aggregates were treated with RA alone, trichostatin (TSA) alone or both (R+TSA). N, non-treated cells. Time points were (1) 1 h, (2) 3 h, (3) 9 h and (4) 24 h. No products were detectable in the PCR blank (data not shown). (B) An experiment similar to that in (A) to test whether TSA could restore normal HoxB expression to P19 cells blocked by aphidicolin. Aggregates were treated with RA alone or in combination with aphidicolin (+Aph), TSA or both. Time points were (1) 2 h, (2) 8 h and (3) 24 h. N, non-treated cells.
This inhibition of HoxB expression could be due to either mRNA destabilization in the cytoplasm or reduced nuclear transcription. To address this issue we performed two experiments. In the first, HoxB mRNA was induced by 18 h RA alone, then transcription was blocked by actinomycin D and the effect of aphidicolin on the stability of HoxB mRNA followed for 8 h. Figure 5A shows that aphidicolin had little or no effect on the stability of HoxB mRNA. We then analysed the effect of aphidicolin on accumulation of nuclear transcripts (Figure 5B; see also Materials and methods). Nuclear HoxB transcripts were absent in untreated P19 cells and detectable after RA treatment, as expected, whereas aphidicolin inhibits RA induction of the Hoxb6–Hoxb7 domain (Figure 5B), but, interestingly, transcription of Hoxb1 and Hoxb13 are again not inhibited, but rather increased, by aphidicolin treatment in P19 cells.
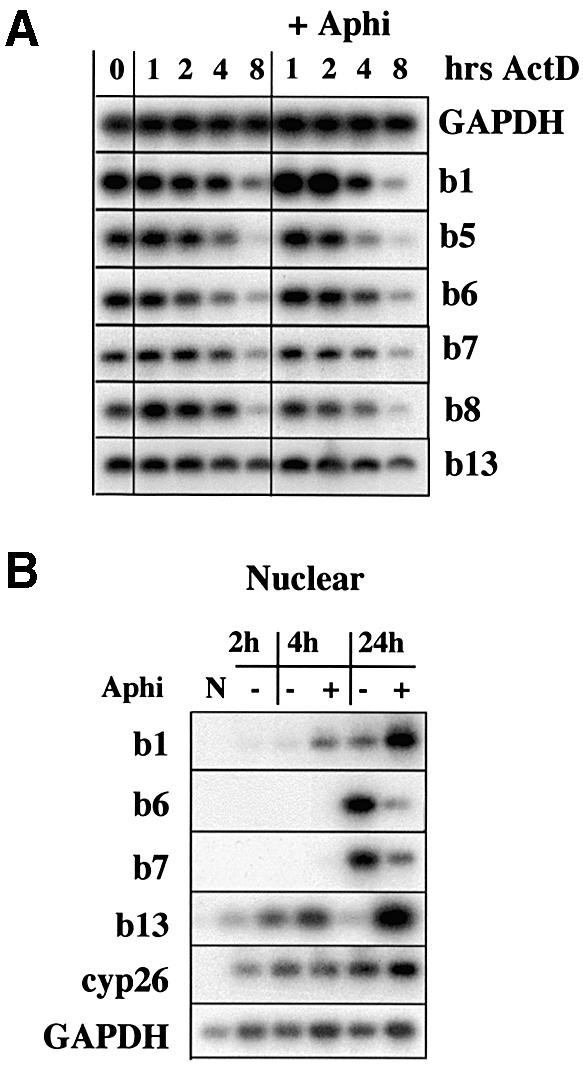
Fig. 5. Inhibition of DNA replication disrupts HoxB transcription in P19 cells. (A) Aphidicolin does not destabilize HoxB mRNA. P19 aggregates were stimulated by retinoic acid (RA) for 18 h and duplicate samples were incubated either with actinomycin D (ActD) alone (left) or actinomycin D and aphidicolin (+Aphi, right) for an 8 h time course. Samples were prepared for semiquantitative RT–PCR and Southern hybridization at the indicated times. (B) Experiment performed as in Figure 4, but cells were lysed for nuclear transcript purification, followed by semiquantitative RT–PCR and Southern blotting. N, non-treated P19 aggregates.
The expression of central HoxB genes is induced in one cell cycle and depends on DNA replication
Aphidicolin blocks the DNA polymerase machinery (Sheaff et al., 1991) and thus blocks the cell cycle in S phase, consequently also preventing progression through the rest of the cell cycle in most of the cells of asynchronous populations. To see whether initiation of DNA replication is required for HoxB expression, a double block protocol was necessary. P19 cells were first pre-synchronized with 2 mM thymidine, released and then placed in the presence of RA with either aphidicolin, NG97, a cdk inhibitor (see Materials and methods) which we found blocks P19 cells at G2–M (D.Fisher, unpublished data), or cycloheximide (CHX), which blocks protein synthesis and thus blocks the cell cycle in both G1 and G2 (Figure 6A). The three treatments block DNA synthesis by arresting the cells, respectively, at the G1–S transition (+Aphi), or in G1 and G2 (+CHX) or at the G2–M transition (+NG97) (Figure 6B), whereas aphidicolin does not block the G2–M transition.
All three treatments significantly inhibited RA-induced expression of Hoxb4, Hoxb5 and Hoxb7 (Figure 6C). Thus, induction of HoxB requires initiation of S phase rather than mitosis, since cells can undergo mitosis in the presence of aphidicolin after release into G2, yet Hoxb4, Hoxb5 and Hoxb7 expression is still inhibited. Hoxb1 and Hoxb13 are again less sensitive to inhibition of DNA synthesis, although they are sensitive to a block of the G2–M transition (Figure 6C). We also reproducibly observed that Hoxb13 was overexpressed in the presence of CHX. We conclude that RA induction of the more central HoxB cluster depends on DNA replication, whereas passage through mitosis is sufficient for induction of the two genes bordering this cluster (Hoxb1 and Hoxb13).
Detection of temporal colinearity depends on synchronous DNA replication
In human NT2 cells, colinear HoxB activation occurred over the space of 1 week (Simeone et al., 1990). However, we failed to detect temporal colinearity in P19 cells, which activate the entire locus in a few hours, less than one cell cycle (Figure 3C). We found a dependence of HoxB expression on DNA replication. As such, timing of normal activation of HoxB genes could also be linked to DNA replication. If so, colinearity of expression might be masked in asynchronous populations.
To test this possibility, RA-induced HoxB expression was analysed in P19 cells synchronized by a double thymidine block (Figure 7A). Cells were released into G2 and induced with RA, and progression through the cell cycle was followed by flow cytometry (Figure 7A). In these conditions, a striking colinearity of HoxB expression from Hoxb1–Hoxb8 was unveiled (Figure 7B). Hoxb1 is detectable 1 h after induction, and Hoxb2–Hoxb8 is successively detectable 4–8 h after induction, corresponding to the time at which cells undergo S phase. Hoxb9 may also follow temporal colinearity, but its constitutive basal level of expression partly masks it. Hoxb13 did not follow colinearity. The colinearity of Hoxb13, although previously reported (Zeltser et al., 1996), has not been extensively studied. In mice, this gene is ∼100 kb upstream of the main HoxB locus, which itself spans ∼100 kb, and as such may belong to a different chromatin domain. This agrees with the fact that deletion of the entire locus from Hoxb1 to Hoxb9 does not affect transcription of Hoxb13 (Medina-Martinez et al., 2000). We conclude that temporal colinearity of expression of the Hoxb1–Hoxb9 locus does occur after RA induction. Furthermore, this temporal colinearity occurs within one single cell cycle. Finally, colinearity appears to be lost for Hoxb13.
Fig. 7. Temporal colinearity of HoxB expression occurs within one cell cycle, and most HoxB genes are activated during S phase. Aggregates of P19 cells were synchronized by a double thymidine (Thy) block, as schematized in (A) and described in Materials and methods. Progression through the cell cycle was checked by flow cytometry. RA, retinoic acid. (B) Samples were taken at 1 h intervals for semiquantitative RT–PCR and Southern blotting.
Replication timing of the HoxB locus and timing of requirement for DNA replication
The observation that transcriptionally active genes replicate early in S phase and that non-expressed genes replicate later suggests a link between replication timing and transcriptional regulation in metazoans (Gilbert, 2002; Schübeler et al., 2002). We asked whether such a link could contribute to HoxB colinearity. The above results suggested that colinearity might be related to replication timing, in which case a progression of replication timing through the locus might be observed. Alternatively, colinearity may be revealed in synchronously dividing cells due to early DNA replication licensing the domain for transcriptional regulatory mechanisms, which determine colinearity independently.
This experiment was performed in exponentially growing monolayer P19 cells to analyse the undifferentiated state. Cells were labelled for 45 min with bromodeoxyuridine (BrdU), and after fixation they were sorted by FACS analysis into four different S-phase groups: early, early–mid, mid–late and late (Figure 8A, pools 1–4). Genomic DNA was purified from sorted cells and control unlabelled cells, sonicated to 1 kb and used for immunoprecipitation with anti-BrdU antibodies to recover replicating DNA. As control loci, we used adenosine deaminase (ADA), expressed in all cells, and β-globin, expressed only in haematopoietic cells. Figure 8B shows that the entire HoxB locus replicates early: the strongest signal is obtained in either the early or early–mid compartments. There may be more than one replication origin, or perhaps replication forks move very slowly in this region, leading to differences in timing. The controls ADA and β-globin replicate early and late, respectively, as expected. We conclude that the HoxB locus is already organized as an early replicating chromatin domain in pluripotent exponentially growing P19 cells.
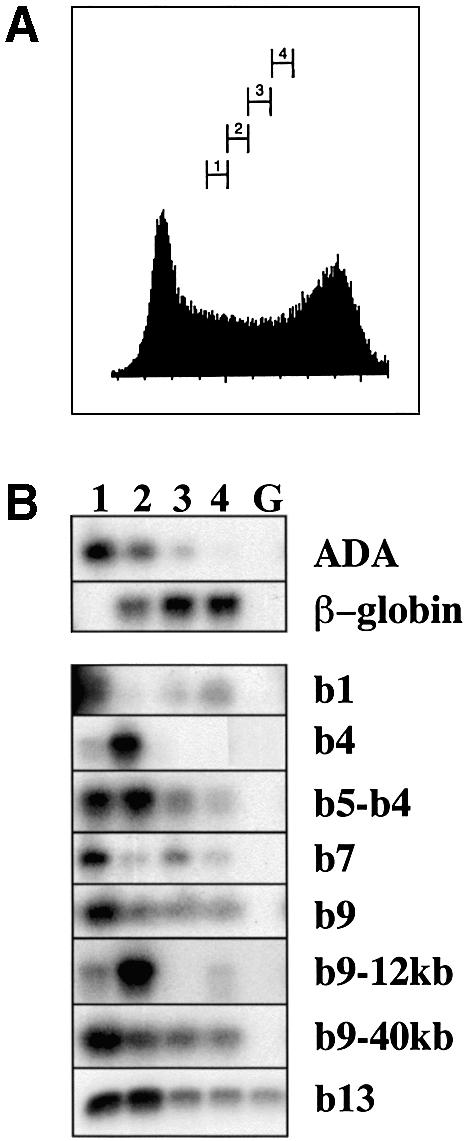
Fig. 8. The HoxB locus replicates early in untreated P19 cells. Exponentially growing monolayer P19.6 cells were treated with a 45 min bromodeoxyuridine (BrdU) pulse and fixed and sorted by flow cytometry (A) into four S-phase compartments (1–4). (B) Replicated DNA was recovered by anti-BrdU immunoprecipitation from these four samples (1–4) and the equivalent quantity of unlabelled P19 genomic DNA (G). Semiquantitative PCR was performed using primers corresponding to the Hox genes shown on the right (b1–b9), or a region of the Hox locus at a set distance from Hoxb9 (–40 kb and –12 kb) or regions of the adenosine deaminase (ADA) and β-globin loci.
We wished to find out whether DNA replication of the HoxB genes themselves is necessary for their transcriptional upregulation, or whether the latter is tied to some other aspect of S phase. Because it is not possible to specifically abolish DNA replication of one locus, we wanted to see whether there is a correlation between replication timing and the moment at which DNA replication is dispensable for high levels of HoxB expression. If replication of the gene itself is required to stimulate gene expression, then, after it is replicated, inhibition of replication by aphidicolin should no longer affect the subsequent expression level. If, alternatively, some other aspect of S-phase biochemistry is required, there might be no such abrupt transition in aphidicolin sensitivity of gene expression. This could only be performed in synchronized cells, with replication timing analysed in parallel by BrdU labelling of parallel cultures. This experiment is presented in Figure 9A. RA was added to cells synchronized in G2, and replication timing and the point at which aphidicolin is no longer effective at blocking subsequent HoxB expression was followed.
Figure 9B shows that S phase occurs from shortly after 2 h to between 6 and 8 h; this was confirmed by hybridizing genomic DNA from the parallel culture with anti-BrdU antibodies (data not shown). Replication timing analysis, by chromatin immunoprecipitation and quantitative PCR of BrdU-labelled DNA at half-hour intervals, confirmed that, as shown in Figure 8, the HoxB locus replicates early: peak values for all regions tested were from 3 to 3.5 h (Figure 9B, right; data not shown). In parallel samples, aphidicolin was added at half-hour intervals, and all samples were left until the end of the experiment, at 10 h after adding RA, before determining the final Hoxb expression level. For Hoxb1, Hoxb13 and Cyp26, as expected, aphidicolin had no effect on final expression level, whereas for Hoxb2, Hoxb5, Hoxb6 and Hoxb8 there was a clear and abrupt transition, if not two transitions, in the level of Hoxb expression subsequently achieved (Figure 9C). This may correspond to sequential replication of the two alleles or, alternatively, to activation of transcriptional derepression at two specific points in S phase, one early and one late. Also, at either one or two points in S phase, if DNA replication is inhibited at this point the resulting HoxB expression is less than if replication is inhibited either earlier or later, which could be explained by an S-phase position-specific abolition of transcription coupled with a constant rate of mRNA degradation. Interestingly, this occurs just before the point at which DNA replication is required for the transition to a higher level of HoxB expression and is particularly intriguing in the light of the observed wave-like expression behaviour of Hox genes in embryos, which is dynamic, covering one somite segmentation round of ∼2 h (Zakany et al., 2001).
These experiments confirm that DNA replication plays an important part in HoxB expression, where polar repressor–enhancer interactions may be revealed by chromatin remodelling during DNA replication.
Inhibition of histone deacetylation and methylation affects HoxB expression but does not mimic RA induction
Histone acetylation is a general mechanism increasing chromatin accessibility that is regulated by the competing activity of histone acetylases and histone deacetylases. We analysed whether this regulation may contribute to HoxB expression. We observed that trichostatin (TSA), a histone deacetylase inhibitor, can partly induce the HoxB cluster, without RA addition (Figure 10A). Of this cluster, Hoxb1, Hoxb7, Hoxb9 and Hoxb13 are the most responsive to TSA, whereas Hoxb2, Hoxb4 and Hoxb5 are less sensitive to TSA induction. The control genes GADPH, a constitutively expressed gene, and the RA-responsive Cyp26 are not affected by TSA (Figure 10A). We conclude that favouring histone acetylation with TSA is sufficient to induce the expression of some of the HoxB genes but does not reproduce RA induction. However, TSA probably does not mimic endogenous regulation of histone acetylation.
When coupled to RA induction (Figure 10), histone deacetylase inhibition increases the expression of the border genes of the cluster (Hoxb1 and Hoxb13) but reproducibly inhibits the induction of the internal genes of the cluster (Hoxb2 and Hoxb4–Hoxb6). This general effect is more pronounced if DNA synthesis is blocked by aphidicolin (Figure 10B). We conclude that inhibition of histone deacetylases, although it can act independently from RA induction, does not reverse the inhibition of HoxB expression by DNA synthesis inhibition. Nevertheless, this does not necessarily mean that RA-induced HoxB activation is dependent on DNA replication independently of changes in histone acetylation.
Another epigenetic modification that could influence HoxB expression is DNA methylation. We observed that 5′-azacytidine, a methylation inhibitor, alone does not activate any expression of HoxB genes, although it strongly enhances expression of the border genes of the cluster (Hoxb1 and Hoxb9) in the presence of RA (data not shown). We analysed the global methylation status of five regions located between Hoxb1 and Hoxb13 using digestion by methylation sensitive (HpaII) or insensitive (MspI) enzymes. Although differences of methylation status of these regions were observed, this did not change on RA treatment (see Supplementary data). We conclude that RA-induced HoxB expression in P19 cells does not rely on global DNA demethylation of the HoxB locus.
Discussion
HoxB gene induction requires at least one cell cycle in both Xenopus development and mouse P19 cells
We report that inhibition of the cell cycle disrupts HoxB activation in two different embryonal model systems (Xenopus embryos and P19 EC cells) and provides evidence that Hox expression is linked to DNA replication.
In Xenopus embryos, we find significant differences in subsequent embryonic patterning using only small differences in timing of inhibition of DNA replication, which shows that patterning decisions in Xenopus are tied to the early cell cycles following the MBT. Arresting DNA replication at or before the MBT prevents activation of Hox genes, probably by blocking activation of the zygotic genome, whereas permission for DNA replication during MBT or the next cell cycle allows HoxB activation. However, HoxB expression in replication-inhibited embryos occurs only in somites and is not detectable in the CNS. This suggests that DNA replication may be required at a later stage for HoxB patterning of the CNS, even though neural precursors are present and can differentiate (Harris and Hartenstein, 1991).
Mouse P19 EC cells can act analogously as neural precursors (MacPherson et al., 1997), and we find that proliferation of these cells is also required for normal levels of HoxB activation. In aggregated P19 EC cells, RA stimulates neural differentiation, and cells continue to proliferate. All HoxB genes are activated in the first cell cycle after RA stimulation, and DNA replication is required for normal HoxB expression.
Hox gene expression and DNA replication: a license to express?
Temporal colinearity of HoxB expression in P19 cells is clear only when cells are synchronized within the cell cycle, suggesting that HoxB activation is linked to progression through the cell cycle. If HoxB genes are activated in a defined order during the cell cycle, each successive gene will be activated at the same time in all cells in synchronous cultures, which would explain why differences in timing of activation (i.e. temporal colinearity) are clearly visible.
The colinearity of expression of the HoxB domain within a single cell cycle has a striking relationship with its sensitivity to inhibition of DNA replication. Thus, the later the genes are expressed, the more they are sensitive to inhibition of DNA synthesis. The peak of sensitivity is for paralogues 6–8, whereas the border paralogues Hoxb9 and Hoxb13 at the 5′ end and Hoxb1 at the 3′ end are less sensitive. All HoxB genes, other than Hoxb13 (which does not obey temporal colinearity in these cells) and Hoxb1, are activated in S phase when stimulated in G2. Hoxb1 and Hoxb13 are activated before S phase and thus not surprisingly do not require S phase for correct activation.
G1–S progression is not sufficient for Hoxb2–Hoxb9 expression, since aphidicolin, which specifically blocks DNA polymerase function, prevents HoxB expression in G2-synchronized cells. This is not due to the requirement for RA itself to act at a particular cell-cycle stage, since a transient pulse of RA can induce neural differentiation from any phase of the cell cycle (Berg and McBurney, 1990), and in our experiments the control gene Cyp26, as well as Hoxb1 and Hoxb13, are activated normally in replication-inhibited cells.
These results are compatible with current models for temporal colinearity. Colinearity of HoxD genes in the mouse embryo appears to be due in part to a polar release from transcriptional repression by a distal enhancer (Kondo and Duboule, 1999; Kmita et al., 2002). Furthermore, HoxB activation along the anterior–posterior axis in chick embryos suggests that progressive opening of the HoxB locus occurs in all regions of the neural tube but genes are only expressed if cis-acting factors are present (Bel-Vialar et al., 2002). Thus, colinearity might be achieved by two component steps: regulated derepression of the locus by DNA replication, making it permissive to regulated expression of specific transcription factors.
Two recent papers demonstrate a differential regulation of anterior and posterior HoxB genes in the response to RA and FGF signalling (Bel-Vialar et al., 2002; Osterveen et al., 2003). FGF can stimulate expression of Hoxb6–Hoxb9 partly through effects on cdx expression and partly by rendering this part of the locus more accessible to Cdx activation. Furthermore, in general, anterior HoxB transgenes integrated at ectopic locations recapitulate endogenous expression patterns, whereas posterior HoxB transgenes do not, suggesting that higher-order organization of the HoxB locus is required for posterior HoxB expression. Our experiments suggest a similar difference in accessibility of the chromatin to transcriptional activation, since posterior HoxB genes are more dependent on DNA replication than anterior genes. It would be interesting to know whether the synergy between FGF and cdx expression requires proliferation, and whether expression from ectopically integrated HoxB transgenes requires DNA replication.
We propose that the gradient of sensitivity to aphidicolin observed according to the position of HoxB paralogues on the chromatin domain is related to polar competition between transcriptional repressor and activator elements, in which DNA replication strongly favours expression of repressed chromatin by creating nascent DNA upon which new chromatin is formed or by restructuring the domain during S phase. In P19 cells the HoxB locus may be in a predetermined state, since it replicates early and is mainly unmethylated and activation of the entire locus is rapid, yet DNA replication is still required to allow normal Hox gene expression. Possibly, the HoxB locus in P19 cells is organized into chromatin in such a way that passage through a single cell cycle allows relief of repression over the entire locus in much the same way that a single DNA replication derepresses chromatin to allow long-range enhancer function in mouse embryos (Forlani et al., 1998).
In some vertebrates, cell-cycle lengths show an anterior–posterior gradient (long to short) (Sanders et al., 1993), and there is a caudal–rostral wave of initiation of HoxB expression in the neural tube in these embryos (Gaunt and Strachan, 1996). Furthermore, cell-cycle synchrony occurs in developing somites (Primmett et al., 1989), which also show waves of HoxB expression (Zakany et al., 2001). As such, the spatial and temporal organization of proliferation could provide a template for developmental gene expression in which staggered DNA replication along a developing axis relieves gene expression from transcriptional silencing.
Materials and methods
Xenopus embryos
Xenopus eggs were fertilized in vitro, dejellied in 2% cysteine and maintained in 0.2× MBSH at 23°C. A mixture of aphidicolin (150 µM, from 20 mg/ml stock in DMSO) and hydroxyurea (20 mM) was applied to sibling embryos at the times indicated. Whole-mount in situ hybridizations were performed according to Harland (1991), with modifications (Islam and Moss, 1996). The Hoxb1 and Hoxb9 clones used for preparing the probes were gifts from Jonathon Slack (Bath, UK). Genomic DNA was prepared by homogenizing four embryos frozen in liquid nitrogen at the time points indicated and, after thawing, in 0.3 ml of SETS (150 mM NaCl, 10 mM EDTA, 50 mM Tris pH 7.5, 0.5% SDS) and subsequent proteinase K/RNase/phenol treatment as described previously (Sambrook et al., 1989), carefully ensuring at each step that the maximum DNA was retained. DNAs from each sample were resuspended in 50 µl, and 6 µl were loaded on an agarose gel.
Cell culture and manipulation of P19 cells
P19.6 cells were a gift from Pierre Chambon (Strasbourg, France). Manipulation and induction of differentation were performed according to Rudnicki and McBurney (1987). RA was added to the culture medium at 0.5 µM. We verified RA dose dependence for HoxB expression and found no difference between monolayers and aggregates of P19 cells. Monolayers were used for the experiment in Figure 3B, in which quantitative recovery and counting of cells was essential, and in Figure 8, in which we wished to study cells as undifferentiated as possible, but aggregates were used in experiments involving HoxB expression analysis (all other experiments), since this is the standard procedure for neural differentiation of P19 cells. Aphidicolin was used at 15 µM, and cells were either pretreated for 1 h before RA or treated simultaneously with RA, as indicated. NG97 was a gift from Laurent Meijer (Roscoff, France) and was used at 10 µM. CHX was used at 1 µg/ml. Synchronous culture was performed by treating with 2 mM thymidine for 15 h, washing twice for 5 min with PBS and twice for 10 min with culture medium and releasing for 6 h into G2. For the double thymidine block, after release into G2, 2 mM thymidine was re-applied and the cells incubated for 15 h, before washing as above and release into thymidine-free medium containing 25 µM deoxycytidine. Cells were fixed for flow cytometry by trypsinizing ∼106 cells, washing once with culture medium and twice with PBS, resuspending in 90% ethanol/PBS and storing at –20°C. Flow cytometry was performed by rehydrating the cells in PBS, washing three times in PBS/1%BSA, resuspending in 1 ml PBS/BSA with 100 µg/ml RNase A and 50 µg/ml propidium iodide and analysing using an EPICS or FACscan cytometer.
DNA replication timing
By cell sorting: 2 × 108 exponentially growing cells to which 50 µM BrdU was added for 45 min were fixed for flow cytometry; 60 000 cells were sorted from each of four S-phase compartments. By synchronous culture: 50 µM BrdU was added to synchronized P19 cell aggregates, and DNA was prepared from 106 cells at 30 min intervals using DNAzol (Invitrogen). DNA replication timing was analysed by semiquantitative PCR from substrates prepared by anti-BrdU (Becton Dickinson) immunoprecipitation exactly as described for the β-globin locus (Cimbora et al., 2000). A control sample contained an equal quantity of genomic DNA from P19 cells grown without BrdU.
RT–PCR analysis of gene expression
RNA was prepared from ∼106 cells or five embryos using the SV RNA isolation kit (Promega). RT was performed using 2 µg of total RNA with Superscript II according to the manufacturer’s instructions (Life Technologies). First-strand cDNAs were diluted to 100 µl, and 2 µl were used as substrates for PCR. Semiquantitative PCR was performed as follows. For each primer set, a mixture of all first-strand cDNAs for each experiment were subjected to 15–40 cycles of PCR to determine the optimum number of cycles for mid-exponential amplification (1 ng of product per 20 µl of reaction). This number of cycles was then performed on individual samples. Sequences of PCR primers are available on request. Products were separated by gel electrophoresis, Southern blotted with radioactive probes and quantified by PhosphorImager, using standard procedures.
Analysis of nuclear transcript levels
Aggregates of 106 P19 cells after different treatments were rinsed twice in cold PBS, and nuclei were isolated by osmotic lysis on ice in 2 ml of 10 mM Tris pH 7.5, 3 mM MgCl2, 10 mM NaCl and 0.3% NP-40. After 5 min, cells were lysed by eight strokes in a Dounce homogenizer, centrifuged at 4°C for 10 min, washed twice in 20 mM Tris, pH 8.0, 140 mM KCl, 10 mM MgCl2, 1 mM MnCl2, 1 mM DTT and 20% glycerol and snap frozen in liquid nitrogen in 100 µl of 50 mM Tris pH 8.3, 5 mM MgCl2, 0.1 mM EDTA and 40% glycerol. In vitro transcript extension was performed on thawed samples by the addition of 125 µl of 2× reaction buffer (300 mM KCl, 5 mM MgCl2, 10 mM Tris pH 7.9) and 25 µl of rNTPs (2.5 mM), with aphidicolin (15 µM) or the equivalent dilution of DMSO. After incubation at 30°C for 20 min, 10 µl of DNase was added and incubation continued at 37°C for 10 min. RNAs were prepared by stopping the reactions with 1.5 ml of Tri-reagent (Molecular Research Centre) and following the manufacturer’s protocol. One-tenth of each RNA was used for RT, as above, and PCRs were normalized with respect to GAPDH.
Supplementary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
Thanks to those researchers who sent us reagents (see Materials and methods) and to Philippe Jay and Anne-Marie Martinez for comments on the manuscript. This work was supported by grants from the CNRS, Association pour la Recherche sur le Cancer, and the Human Frontiers Science Program.
References
- Ambros V. (1999) Cell cycle-dependent sequencing of cell fate decisions in Caenorhabditis elegans vulva precursor cells. Development, 126, 1947–1956. [DOI] [PubMed] [Google Scholar]
- Bel-Vialar S., Itasaki,N. and Krumlauf,R. (2002) Initiating Hox gene expression: in the early chick neural tube differential sensitivity to FGF and RA signaling subdivides the HoxB genes into two distinct groups. Development, 129, 5103–5115. [DOI] [PubMed] [Google Scholar]
- Berg R.W. and McBurney,M.W. (1990) Cell density and cell cycle effects on retinoic acid-induced embryonal carcinoma cell differentiation. Dev. Biol., 138, 123–135. [DOI] [PubMed] [Google Scholar]
- Boterenbrood E.C., Narraway,J. and Hara,K. (1983) Duration of cleavage cycles and asymmetry in the direction of cleavage waves prior to gastrulation in Xenopus laevis. Roux Arch. Dev. Biol., 192, 216–221. [DOI] [PubMed] [Google Scholar]
- Cimbora D.M., Schübeler,D., Reik,A., Hamilton,J., Francastel,C., Epner,E.M. and Groudine,M. (2000) Long-distance control of origin choice and replication timing in the human β-globin locus are independent of the locus control region. Mol. Cell. Biol., 20, 5581–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe A.J., Piechan,J.L., Sang,L. and Barton,M.C. (2000) S-phase progression mediates activation of a silenced gene in synthetic nuclei. Mol. Cell. Biol., 20, 4169–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Nooij J.C. and Hariharan,I.K. (1995) Uncoupling cell fate determination from patterned cell division in the Drosophila eye. Science, 270, 983–985. [DOI] [PubMed] [Google Scholar]
- Duboule D. (1994) Temporal colinearity and the phylotypic progression: a basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Development Suppl., 135–142. [PubMed] [Google Scholar]
- Duboule D. (1998) Vertebrate Hox gene regulation: clustering and/or colinearity? Curr. Opin. Genet. Dev., 8, 514–518. [DOI] [PubMed] [Google Scholar]
- Duboule D. and Morata,G. (1994) Colinearity and functional hierarchy among genes of the homeotic complexes. Trends Genet., 10, 358–364. [DOI] [PubMed] [Google Scholar]
- Forlani S., Bonnerot,C., Capgras,S. and Nicolas,J.F. (1998) Relief of a repressed gene expression state in the mouse 1-cell embryo requires DNA replication. Development, 125, 3153–3166. [DOI] [PubMed] [Google Scholar]
- Gaunt S.J. and Strachan,L. (1996) Temporal colinearity in expression of anterior Hox genes in developing chick embryos. Dev. Dyn., 207, 270–280. [DOI] [PubMed] [Google Scholar]
- Gellon G. and McGinnis,W. (1998) Shaping animal body plans in development and evolution by modulation of Hox expression patterns. BioEssays, 20, 116–125. [DOI] [PubMed] [Google Scholar]
- Gilbert D.M. (2002) Replication timing and transcriptional control: beyond cause and effect. Curr. Opin. Cell Biol., 14, 377–383. [DOI] [PubMed] [Google Scholar]
- Godsave S., Dekker,E.J., Holling,T., Pannese,M., Boncinelli,E. and Durston,A. (1994) Expression patterns of HoxB genes in the Xenopus embryo suggest roles in anteroposterior specification of the hindbrain and in dorsoventral patterning of the mesoderm. Dev. Biol., 166, 465–476. [DOI] [PubMed] [Google Scholar]
- Goldman M.A., Holmquist,G.P., Gray,M.C., Caston,L.A. and Nag,A. (1984) Replication timing of genes and middle repetitive sequences. Science, 224, 686–692. [DOI] [PubMed] [Google Scholar]
- Harland R.M. (1991) In situ hybridization: an improved whole-mount method for Xenopus embryos. Methods Cell Biol., 36, 685–695. [DOI] [PubMed] [Google Scholar]
- Harris W.A. and Hartenstein,V. (1991) Neuronal determination without cell division in Xenopus embryos. Neuron, 6, 499–515. [DOI] [PubMed] [Google Scholar]
- Islam N. and Moss,T. (1996) Enzymatic removal of vitelline membrane and other protocol modifications for whole mount in situ hybridization of Xenopus embryos. Trends Genet., 12, 459. [DOI] [PubMed] [Google Scholar]
- Kmita M., Fraudeau,N., Herault,Y. and Duboule,D. (2002) Serial deletions and duplications suggest a mechanism for the collinearity of Hoxd genes in limbs. Nature, 420, 145–150. [DOI] [PubMed] [Google Scholar]
- Kondo T. and Duboule,D. (1999) Breaking colinearity in the mouse HoxD complex. Cell, 97, 407–417. [DOI] [PubMed] [Google Scholar]
- Krumlauf R. (1994) Hox genes in vertebrate development. Cell, 78, 191–201. [DOI] [PubMed] [Google Scholar]
- MacPherson P.A, Jones,S., Pawson,P.A., Marshall,K.C. and McBurney,M.W. (1997) P19 cells differentiate into glutamatergic and glutamate-responsive neurons in vitro. Neuroscience, 80, 487–499. [DOI] [PubMed] [Google Scholar]
- Masui Y. and Wang,P. (1998) Cell cycle transition in early embryonic development of Xenopus laevis. Biol. Cell, 90, 537–548. [PubMed] [Google Scholar]
- Medina-Martinez O., Bradley,A. and Ramirez-Solis,R. (2000) A large targeted deletion of Hoxb1–Hoxb9 produces a series of single-segment anterior homeotic transformations. Dev. Biol., 222, 71–83. [DOI] [PubMed] [Google Scholar]
- Mennerich D. and Braun,T. (2001) Activation of myogenesis by the homeobox gene Lbx1 requires cell proliferation. EMBO J., 20, 7174–7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newport J. and Kirschner,M. (1982a) A major developmental transition in early Xenopus embryos: I. Characterization and timing of cellular changes at the midblastula stage. Cell, 30, 675–686. [DOI] [PubMed] [Google Scholar]
- Newport J. and Kirschner,M. (1982b) A major developmental transition in early Xenopus embryos: II. Control of the onset of transcription. Cell, 30, 687–696. [DOI] [PubMed] [Google Scholar]
- Ohsugi K., Gardiner,D.M. and Bryant,S.V. (1997) Cell cycle length affects gene expression and pattern formation in limbs. Dev. Biol., 189, 13–21. [DOI] [PubMed] [Google Scholar]
- Osterveen T., Niederreither,K., Dollé,P., Chambon,P., Meijlink,F. and Deschamps,J. (2003) Retinoids regulate the anterior expression boundaries of 5′ Hoxb genes in posterior hindbrain. EMBO J., 22, 262–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primmett D.R., Norris,W.E., Carlson,G.J., Keynes,R.J. and Stern,C.D. (1989) Periodic segmental anomalies induced by heat shock in the chick embryo are associated with the cell cycle. Development, 105, 119–130. [DOI] [PubMed] [Google Scholar]
- Raghuraman M.K. et al. (2001) Replication dynamics of the yeast genome. Science, 294, 115–121. [DOI] [PubMed] [Google Scholar]
- Rollins M.B. and Andrews,M.T. (1991) Morphogenesis and regulated gene activity are independent of DNA replication in Xenopus embryos. Development, 112, 559–569. [DOI] [PubMed] [Google Scholar]
- Rudnicki M.A. and McBurney,M.W. (1987) Cell culture methods and induction of differentiation of embryonal carcinoma cell lines. In Robertson,E.J.(ed.), Teratocarcinomas and Embryonic Stem Cells: A Practical Approach, IRL Press, Oxford, UK, pp. 19–50. [Google Scholar]
- Satoh N. (1977) ‘Metachronous’ cleavage and initiation of gastrulation in amphibian embryos. Dev. Growth Differ., 19, 111–117. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [Google Scholar]
- Sanders E.J., Varedi,M. and French,A.S. (1993) Cell proliferation in the gastrulating chick embryo: a study using BrdU incorporation and PCNA localization. Development, 118, 389–399. [DOI] [PubMed] [Google Scholar]
- Schübeler D., Scalzo,D., Kooperberg,C., Van Steensel,B., Delrow,J. and Groudine,M. (2002) Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat. Genet., 32, 438–442. [DOI] [PubMed] [Google Scholar]
- Sheaff R., Ilsley,D. and Kuchta,R. (1991) Mechanism of DNA polymerase α inhibition by aphidicolin. Biochemistry, 30, 8590–8597. [DOI] [PubMed] [Google Scholar]
- Simeone A., Acampora,D., Arcioni,L., Andrews,P.W., Boncinelli,E. and Mavilio,F. (1990) Sequential activation of HOX2 homeobox genes by retinoic acid in human embryonal carcinoma cells. Nature, 346, 763–776. [DOI] [PubMed] [Google Scholar]
- Sonneveld E., van den Brink,C.E., Tertoolen,L.G., van der Burg,B. and van der Saag,P.T. (1999) Retinoic acid hydroxylase (CYP26) is a key enzyme in neuronal differentiation of embryonal carcinoma cells. Dev. Biol., 213, 390–404. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P. (1991) Implications of DNA replication for eukaryotic gene expression. J. Cell Sci., 99, 201–206. [DOI] [PubMed] [Google Scholar]
- Zakany J., Kmita,M., Alarcon,P., de la Pompa,J.L. and Duboule,D. (2001) Localized and transient transcription of Hox genes suggests a link between patterning and the segmentation clock. Cell, 106, 207–217. [DOI] [PubMed] [Google Scholar]
- Zeltser L., Desplan,C. and Heintz,N. (1996) Hoxb-13: a new Hox gene in a distant region of the HOXB cluster maintains colinearity. Development, 122, 2475–2484. [DOI] [PubMed] [Google Scholar]
- Zhang J., Xu,F., Hashimshony,T., Keshet,I. and Cedar,H. (2002) Establishment of transcriptional competence in early and late S phase. Nature, 420, 198–202. [DOI] [PubMed] [Google Scholar]