Abstract
Bcl-2 family proteins are key regulators of apoptosis. Both pro-apoptotic and anti-apoptotic members of this family are found in mammalian cells, but only the pro-apoptotic protein Debcl has been characterized in Drosophila. Here we report that Buffy, the second Drosophila Bcl-2-like protein, is a pro-survival protein. Ablation of Buffy by RNA interference leads to ectopic apoptosis, whereas overexpression of buffy results in the inhibition of developmental programmed cell death and γ irradiation-induced apoptosis. Buffy interacts genetically and physically with Debcl to suppress Debcl-induced cell death. Genetic interactions suggest that Buffy acts downstream of Rpr, Grim and Hid, and upstream of the apical caspase Dronc. Furthermore, overexpression of buffy inhibits ectopic cell death in diap1 (th5) mutants. Taken together these data suggest that Buffy can act downstream of Rpr, Grim and Hid to block caspase-dependent cell death. Overexpression of Buffy in the embryo results in inhibition of the cell cycle, consistent with a G1/early-S phase arrest. Our data suggest that Buffy is functionally similar to the mammalian pro-survival Bcl-2 family of proteins.
Keywords: apoptosis/cell survival/Drosophila/programmed cell death/RNA interference/TUNEL
Introduction
A balance between cell proliferation and apoptosis is essential for development of the multicellular organism. Superfluous or damaged cells must be removed by apoptosis, whilst those cells required for subsequent stages of development are protected by cell survival factors (reviewed in Baehrecke, 2002). In the nematode Caenorhabditis elegans, four genes, egl-1, ced-3, ced-4 and ced-9, are essential for regulating apoptosis (reviewed in Horvitz, 1999). Of these, EGL-1, CED-3 and CED-4 are required for cell death to occur, whereas CED-9 is essential for cell survival. In C.elegans, EGL-1 functions upstream of CED-9, while CED-9 interacts with and regulates CED-4-mediated CED-3 activation (Hengartner, 2001).
Not surprisingly, the pathways of cell death are considerably more complex in mammals, where EGL-1, CED-3 and CED-9 are represented by multiple family members (reviewed in Baehrecke, 2002). There are several mammalian homologs of CED-3, some of which have essential functions in apoptosis (reviewed in Cryns and Yuan, 1998). The mammalian homolog of the CED-4 adaptor protein, Apaf-1, is essential for the activation of caspase-9. The mammalian homologs of CED-9, including Bcl-2, Bcl-xL and Bcl-w, act as inhibitors of caspase activation and function upstream of Apaf-1 (reviewed in Cory and Adams, 2002). The mammalian EGL-1 homologs share a small region of homology (BH3 domain) with CED-9/Bcl-2 proteins and act as pro-apoptotic proteins upstream of Bcl-2 (Conradt and Horvitz, 1998).
Drosophila appears to have cell death machinery of intermediary complexity compared with that of C.elegans and mammals. Thus far, Drosophila has yielded seven caspases and a single CED-4/Apaf-1 homolog, termed Dark/Dapaf-1/HAC-1, which interacts with the Drosophila caspase Dronc and is required for its activation (reviewed in Richardson and Kumar, 2002). EGL-1-related, pro-apoptotic, BH3-only proteins have not yet been identified in Drosophila. Like mammals, Drosophila contains IAP (inhibitor of apoptosis) pro-survival proteins, which bind to and inhibit caspases (reviewed in Deveraux and Reed, 1999). Two IAP homologs have been reported in Drosophila—Diap1/Thread and Diap2 (reviewed in Hay, 2000)—which are antagonized by the IAP inhibitors Reaper (Rpr), Hid (Head involution defective/Wrinkled), Grim (reviewed in McCall and Steller, 1997) and Sickle (Christich et al., 2002; Srinivasula et al., 2002; Wing et al., 2002). The recent analysis of the human genome sequence shows that there are no strongly related homologs of rpr, hid or grim (Aravind et al., 2001); however, the mammalian apoptosis inducer Smac/Diablo appears to act as a functional homolog of Rpr, Hid or Grim, as it functions to neutralize caspase inhibitory function of the IAP protein family (Du et al., 2000; Verhagen et al., 2000).
This paper focuses on the role of the Bcl-2 family of proteins in Drosophila programmed cell death. Accumu lated evidence suggests that in mammalian cells, mitochondrial Bcl-2 prevents the release of cytochrome c, required for the formation of the Apaf-1 apoptosome and therefore caspase activation (Zou et al., 1997). Conversely, the pro-apoptotic Bcl-2 proteins promote mitochondrial permeability and cytochrome c release. Life or death of the cell is determined by whether the balance is tipped towards the pro-survival or the pro-apoptotic Bcl-2 members (reviewed in Cory and Adams, 2002). In Drosophila, there are two homologs of the Bcl-2/Ced-9 family of programmed cell death (PCD) proteins, Debcl/dBorg-1/dRob-1 and Buffy/dBorg-2 (Brachmann et al., 2000; Colussi et al., 2000; Igaki et al., 2000). Although both Debcl and Buffy share the BH1, BH2, BH3 and C-terminal transmembrane domains of the Bcl-2 family of proteins, they appear to lack the N terminal BH4 domain. In mammals, the BH4 domain distinguishes the pro-apoptotic Bcl-2 family members, e.g. Bax and Bok, from the anti-apoptotic members, e.g. Bcl-2, Bcl-xL and Bcl-w (reviewed in Cory and Adams, 2002). Based upon this, both Debcl and Buffy were expected to be pro-apoptotic. Although both Debcl and Buffy are most closely related to the mammalian pro-apoptotic Bok, only Debcl has been shown to have a pro-apoptotic function in Drosophila, since ectopic overexpression of debcl in transgenic flies results in ectopic PCD and functional knockout of Debcl by RNA interference (RNAi) leads to an inhibition of cell death (Brachmann et al., 2000; Colussi et al., 2000; Igaki et al., 2000).
Here we provide the first evidence that Buffy is a pro-survival relative of Bcl-2/Ced-9. Buffy is required for cell survival and can prevent developmental and irradiation-induced cell death. We also show that Buffy overexpression prevents cell cycle progression and results in the accumulation of cells in G1, like its mammalian pro-survival counterpart Bcl-2 (O’Reilly et al., 1996). Thus, both pro-survival and cell cycle functions of Bcl-2 have been evolutionarily conserved in Buffy, suggesting that Buffy is the Drosophila homolog of the pro-survival Bcl-2 proteins.
Results
The buffy expression pattern correlates with debcl expression and apoptotic domains
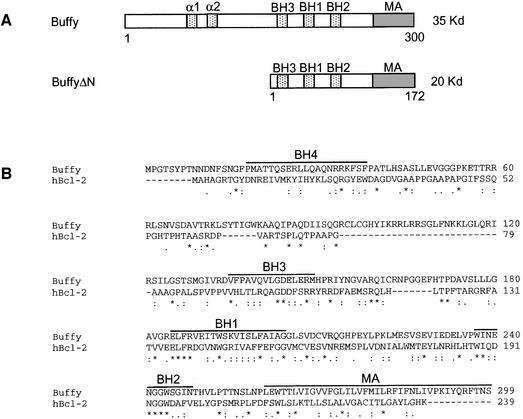
Buffy encodes a protein that is 19% identical and 56% similar to human Bcl-2 over a 239 amino acid region and shares several conserved motifs with mammalian Bcl-2, including the BH1, BH2 and BH3 domains, and a C-terminal hydrophobic membrane anchor (Figure 1A and B). Although an N-terminal BH4 domain present in pro-survival Bcl-2 proteins is not obvious in the Buffy sequence, there are two α-helical domains in the N-terminal region that might be functionally similar to the BH4 domain.
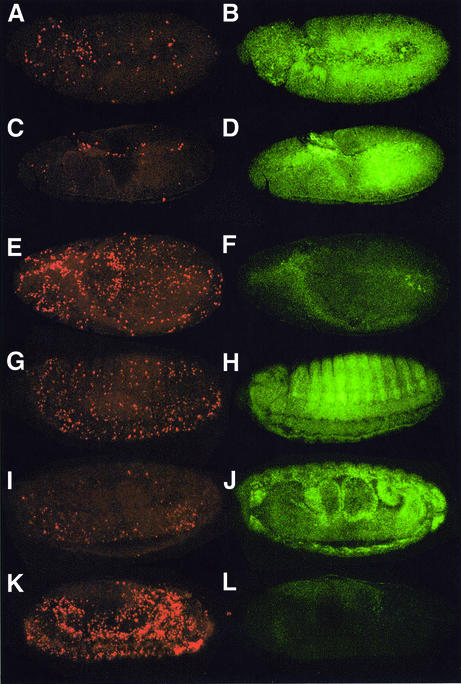
Fig. 1. (A) Protein structure of Buffy. The positions of the BH1, BH2 and BH3 domains and a C-terminal hydrophobic membrane anchor are shown. Two upstream α-helical domains are also indicated. The deletion mutant BuffyΔN, used to generate UAS-buffyΔN transgenic flies, lacks the first 128 amino acids. (B) Alignment of the predicted protein sequences of Buffy and human Bcl-2 [DDBJ/EMBL/GenBank accession no. AAA35591; performed using the program Clustal_W (http://www.ebi.ac.uk/clustalw/)]. Identical residues are indicated by an asterisk (*), conserved residues by a colon (:), and similar residues by a full point (.). Buffy and Bcl-2 share 19% identity and 56% similarity over a 239 amino acid region. The positions of the BH1, BH2, BH3 and BH4 domains of Bcl-2, based on BH domain consensus sequences (http://www.expasy.org/prosite/), are indicated by lines above the sequence. Buffy contains conserved BH1, BH2 and BH3 domains, but lacks a conserved BH4 domain. The putative membrane anchor (MA) of Buffy is shown. (C) RT–PCR analysis of buffy expression. After reverse transcription, PCR was performed using buffy specific primers spanning an intron to amplify a 465-bp fragment. Cytochrome c (cyt c) was used as a control from the same cDNA samples. In situ hybridization analysis using DIG-labeled probes (D–S). Drosophila embryos with an antisense buffy probe: (D) stage 5; (E) germ band extended/stage 10; (F) germ band retracted stage 13 (red arrows show segmental pattern; mg = midgut, hg = hindgut); (G) stage 16 (red arrows show epidermis of the gut; p = pharynx, c = clypeolabrum); (H) stage 16 with an antisense debcl (I) stage 16 embryo hybridized with sense buffy (J) stage 10a ovaries with antisense buffy (nc = nurse cells, ec = egg chamber); (K) stage 10a ovaries with sense probe; (L) third instar larval midgut with an antisense buffy probe; (M) third instar larval midgut with the sense probe; (N) third instar salivary glands with antisense buffy; (O) third instar salivary glands with the sense control probe; (P) third instar larval brain lobes with an antisense buffy probe; (Q) third instar larval brain lobes with a sense buffy probe; (R) third instar larval eye discs with an antisense buffy probe; and (S) third instar larval eye discs with a sense buffy probe.
To examine the expression of buffy mRNA during development, we initially carried out northern blot and RT–PCR analysis (Figure 1C; data not shown). Due to the low level of buffy mRNA expression, the 1.2-kb buffy transcript was scarcely detectable upon northern analysis (data not shown). By RT–PCR, however, buffy mRNA was detected at all developmental stages, with the strongest expression detected from the late larval/early pupal stages (Figure 1C).
The spatial distribution of buffy mRNA was determined using in situ hybridization. The buffy transcript was expressed at very low levels and, as with debcl mRNA, detection required indirect tyramide-amplification (TSA™), as described previously (Colussi et al., 2000). In situ hybridization analysis of Drosophila embryos revealed buffy transcript in non-cellularized, stage 5 embryos. Since zygotic transcription does not occur prior to stage 5, this represents maternally deposited mRNA (Figure 1D). General ubiquitous expression was observed in germ band extended, stage 10 embryos (Figure 1E). Later in embryogenesis the pattern of buffy mRNA becomes more restricted, with staining in the midgut, the hindgut and a segmental pattern throughout the epidermal tissue (Figure 1F). buffy message becomes more restricted at stage 16 of embryogenesis and is prominent in the epidermis of the gut and regions of the head, including the pharynx and clypeolabrum [compare Figure 1G with I (sense control)]. buffy mRNA was detected in the same pattern as the pro-apoptotic Drosophila Bcl-2-related gene debcl (Colussi et al., 2000). The similarity between the expression patterns of debcl and buffy was particularly striking in stage 16 embryos (compare Figure 1G with H). Such similar expression patterns suggest that coordinated expression may be important for regulating cell death. The patterns of buffy and debcl expression correlate with regions of cell death in the developing embryo (Colussi et al., 2000).
During oogenesis, the nurse cells dump their cytoplasm into the oocyte, a process coordinated with nurse cell apoptosis, and are regulated by apoptotic stimuli (reviewed in Buszczak and Cooley, 2000). buffy mRNA was abundant in the nurse cell chambers from stage 10a ovaries [(compare Figure 1J with K (sense control)], which undergo apoptosis at stage 10b. During early pupal stages, most larval tissues are histolysed, an extensive apoptotic process regulated by pulses of the steroid hormone ecdysone (reviewed in Baehrecke, 2000). During third instar, buffy mRNA was strongest in larval midgut (Figure 1L) and salivary glands (Figure 1N), tissues destined for histolysis in pupariation (Jiang et al., 1997). buffy mRNA was also detected (albeit at lower levels) in larval tissues that are remodeled into the adult tissue during pupal development—a process requiring a balance between apoptosis and cell survival—including the brain lobes (Truman et al., 1994) (Figure 1P) and eye imaginal discs (Miller and Cagan, 1998) (Figure 1R). buffy, therefore, is expressed throughout development, in the same pattern as the pro-apoptotic gene debcl and in tissues susceptible to apoptosis.
Buffy interacts with the pro-apoptotic Drosophila Bcl-2 homolog, Debcl
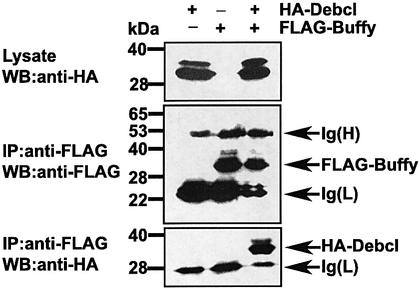
The mammalian pro-apoptotic Bcl-2 proteins function by binding and sequestering pro-survival Bcl-2 members (reviewed in Cory and Adams, 2002). Debcl binds most mammalian pro-survival Bcl-2 proteins, including Bcl-2 and Bcl-XL, but not their pro-apoptotic counterparts (Colussi et al., 2000). In order to determine whether Debcl heterodimerizes with Buffy, we carried out co-immunoprecipitation experiments (Figure 2). FLAG-tagged Buffy was coexpressed with HA-tagged Debcl in 293T cells. Immunoprecipitation was performed with anti-FLAG or anti-HA antibodies. The control immunoblot with anti-FLAG showed that FLAG-Buffy (33 kDa) was precipitated (Figure 2, middle panel). Immunoblotting of the FLAG immunoprecipitates with anti-HA revealed the HA-Debcl protein (Figure 2, bottom panel), suggesting that the two proteins can co-immunoprecipitate. Therefore, like the pro- and anti-apoptotic members of the mammalian Bcl-2 family, Debcl and Buffy can physically interact.
Fig. 2. Buffy interacts with the pro-apoptotic Drosophila Bcl-2 homolog, Debcl. Flag-tagged Buffy was coexpressed with HA-tagged Debcl in 293T cells. (Top panel) Lysates blotted with HA antibody. (Middle panel) Lysates immunoprecipitated with an anti-FLAG antibody and blotted with anti-FLAG. (Bottom panel) Lysates immunoprecipitated with anti-FLAG and blotted with anti-HA.
Buffy protein colocalizes with mitotracker to the mitochondria
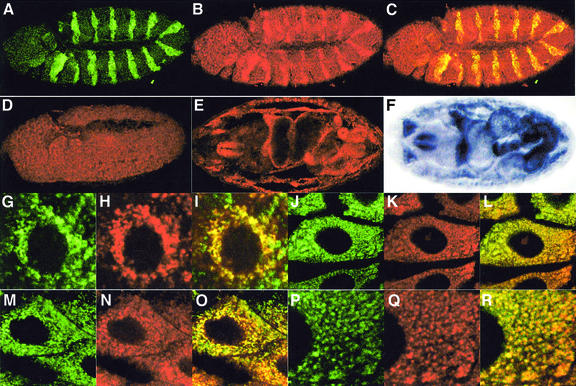
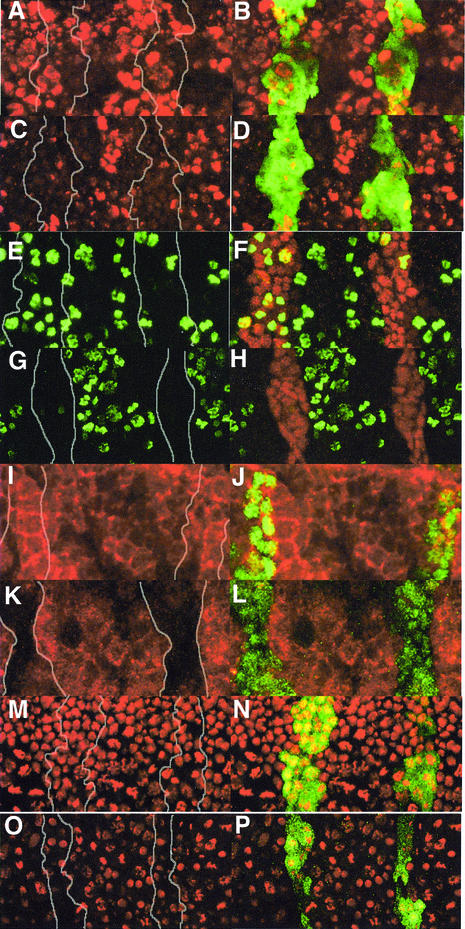
The subcellular distribution of Buffy protein was determined using a rat polyclonal Buffy antibody. We tested the specificity of this antibody using the en-GAL4 driver to ectopically express the upstream activator sequence (UAS)-buffy transgene and a UAS-GFP transgene to mark cells expressing engrailed (Figure 3A–C). Significantly, increased levels of anti-Buffy antibody staining were observed in the Engrailed (En) stripes compared with the levels of protein in adjacent cells (Figure 3B). Leaky expression of the UAS-buffy transgene was suggested by the finding that Buffy antibody staining was consistently higher across the entire embryo when compared with the level of endogenous protein from wild-type embryos (Figure 3D). Further evidence for leaky expression was the greater general protection from irradiation-induced cell death (see below). Although mutants were not available to verify the specificity of our antibodies further, we found a clear reduction in the level of staining for buffy double-stranded (ds) RNA ablation embryos (see below). In addition, the pattern of Buffy antibody staining in stage-16 wild-type embryos was similar to that observed for buffy mRNA expression (compare Figure 3E with F).
Fig. 3. Analysis of subcellular distribution of Buffy protein. (A–C) Stage 11 en-GAL4,UAS-Buffy,UAS-GFP/+ embryos with (A) anti-GFP antibody in green, (B) anti-Buffy antibody in red, and (C) merge. (D) en-GAL4,UAS-GFP/+ embryos with anti-Buffy antibody in red. Intensity of the confocal laser was equivalent in (B) and (D). Wild-type stage 16 embryos with (E) anti-Buffy antibody in red and (F) mRNA in situ for buffy. (G–I) Wild-type third instar brain lobe cells: (G) anti-Buffy antibody (green), (H) mitotracker (red), and (I) merge. (J–L) Wild-type larval midgut cells: (J) anti-Buffy antibody, (K) mitotracker, and (L) merge. (M–O) Wild-type larval midgut cells 2 h after treatment with 8 Gy ionizing radiation: (M) anti-Buffy antibody, (N) mitotracker, and (O) merge. (P–R) High power wild-type, unirradiated larval midgut cells: (P) anti-Buffy antibody, (Q) mitotracker, and (R) merge.
The pro-survival Bcl-2 proteins are localized to intracellular membranes, including mitochondria, endoplasmic reticulum and nuclear envelope (Janiak et al., 1994; reviewed in Cory and Adams, 2002). The Buffy C-terminus contains a putative hydrophobic membrane anchor, similar to the sequence found in many Bcl-2 family proteins (Brachmann et al., 2000; Colussi et al., 2000). To determine whether Buffy localized to mitochondria, we co-stained Drosophila tissues with the mitochondrial marker mitotracker and anti-Buffy antibody. Mitotracker has been used previously to show that the mitochondria of Drosophila larval brain are scattered throughout the entire cytoplasm and surround the nucleus (Iyengar et al., 2002). We reproduced this pattern of mitotracker staining in larval neuroblast cells (Figure 3H), and found co-localization with Buffy protein predominantly in mitotracker-positive regions (Figure 3I). Ionizing radiation has been used previously to induce apoptosis in Drosophila tissues (Ollmann et al., 2000). Similar Buffy staining was seen in γ-irradiated tissues compared with untreated ones, including eye discs, wing discs, salivary glands (data not shown) and midgut (Figure 3J–O, and high power in Figure 3P–R). Therefore, like Bcl-2, Buffy localizes to mitochondria in both normal and irradiated cells, unlike the pro-apoptotic Bax proteins that only become localized to the mitochondrial membrane following stress signals (reviewed in Cory and Adams, 2002).
Buffy is required for cell survival during embryogenesis
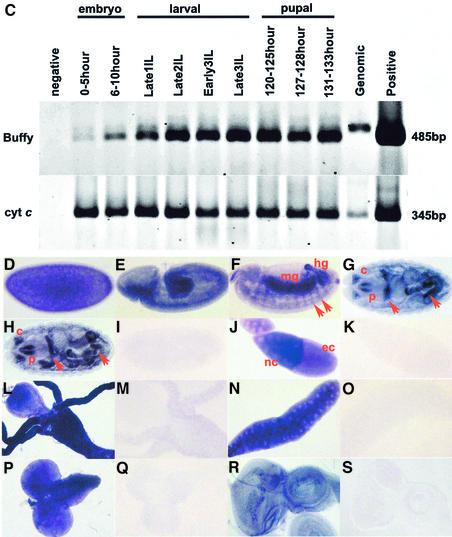
As no specific buffy mutants are currently available, we used RNAi to knock down Buffy expression, which has been used extensively in Drosophila to knock down gene function (reviewed in Sharp, 2001). buffy dsRNA was injected into pre-blastoderm embryos, which were allowed to develop for 6–7 h before TUNEL and staining with anti-Buffy antibody, to measure the efficiency of Buffy protein ablation. On average, a 7-fold increase in TUNEL cells was observed in buffy dsRNA-injected embryos (Figure 4). Wild-type, stage-11 embryos have small populations of apoptotic cells in the amnioserosa, brain lobes and developing central nervous system (CNS) (Abrams et al., 1993) [Figure 4A (TUNEL = 90 ± 12, n = 4) ], and ubiquitous staining for Buffy protein (Figure 4B). Control embryos injected with GFP dsRNA and aged to stage 11 have a similar low level of apoptosis [Figure 4C (TUNEL = 93 ± 23, n = 4) ] and ubiquitous staining for Buffy protein (Figure 4D). Reduction of Buffy protein, shown using the Buffy antibody (Figure 4F), correlated with increased levels of ectopic apoptosis [Figure 4E (TUNEL = 802 ± 115, n = 4)]. In a separate experiment, embryos were aged to between stages 14 and 16, following injection with either buffy dsRNA or buffer only. Antibody staining revealed that Buffy protein was barely detectable by stage 14–16 in embryos injected with buffy dsRNA (Figure 4L), compared with control embryos at stage 14, where epidermal and neural staining is observed (Figure 4H), and at stage 16 (Figure 4J). The older buffy RNAi embryos were fragile, with many disintegrating during collection; those remaining had three times the number of TUNEL-positive cells [Figure 4K (TUNEL = 1015 ± 149, n = 4)] compared with control embryos at stages 14 or 16 [Figure 4G and Figure 4I, respectively (TUNEL = 287 ± 79, n = 4)]. Reduced numbers of cells and very few surviving neural cells were also observed in similarly injected and aged embryos (data not shown). Thus, ablation of Buffy function results in cell death, indicating that Buffy is necessary for embryonic cell survival.
Fig. 4. RNAi using buffy dsRNA fragments. (A, C, E, G, I, K) Embryos stained for TUNEL. (B, D, F, H, J, L) Embryos stained for Buffy protein. (A and B) Wild-type stage 11 embryos. (C and D) Control embryos, injected with GFP dsRNA and aged to stage 11. (E and F) Embryos injected with buffy dsRNA fragments and aged to stage 11. (G–J) Embryos injected with buffer and aged at stage 14 (G and H) or stage 16 (I and J). (K and L) Embryos injected with buffy dsRNA fragments and aged between stages 14 and 16 (note that these embryos are difficult to stage due to impaired development).
Buffy inhibits developmental cell death and apoptosis induced by ionizing radiation
Apoptosis commences during stage 11 of Drosophila embryogenesis, and as development proceeds, TUNEL-labeled cells are observed throughout the embryo, particularly in cells of the nervous system (Abrams et al., 1993). In order to determine whether buffy overexpression inhibits developmental PCD, we overexpressed buffy in Drosophila embryos using the UAS-GAL4 system (Brand and Perrimon, 1993). Buffy protein was expressed using the En-GAL4 driver, which drives expression in the pair-rule striped pattern of the embryo (Kornberg et al., 1985) (Figure 5A and D). En stripes from En-GAL4,UAS-GFP,UAS-Buffy embryos [Figure 5E (2.3 ± 0.8, n = 14)] contained approximately half the number of TUNEL-positive cells when compared with control embryos [Figure 5B (4.86 ± 1.2, n = 14)]. Therefore, ectopic expression of Buffy can inhibit developmentally regulated PCD during Drosophila embryogenesis. Furthermore, ubiquitous expression of buffy using the Armadillo-GAL4 driver can result in additional neural cells, suggesting that Buffy overexpression can block the normal pattern of PCD in the developing peripheral nervous system (data not shown).
Fig. 5. Overexpression of Buffy with the engrailed (en)-GAL4 driver blocks developmental and irradiation-induced cell death in stage 13 embryos, and irradiation-induced cell death in third instar wing discs. (A) Control en-GAL4/+ embryo stained with TUNEL (blue) and anti-GFP antibody (green). (B) The embryo in (A) stained with TUNEL (positions of the En stripes are circled). (C) Irradiated control en-GAL4/+ embryo stained with anti-En (green) and TUNEL (blue) after 2.5 h recovery. (D) en-GAL4/+,UAS-GFP/+,UAS-buffy/+ stained with TUNEL (blue) and anti-GFP antibody (green). (E) The embryo in (D) showing the position of the En stripes. (F) Irradiated en-GAL4/+,UAS-buffy/+ embryo after 2.5 h of recovery, stained with anti-En (green) and TUNEL (blue). (G–J) Irradiated third instar larval wing discs after 4 h of recovery, stained with anti-GFP (green) and labeled with TUNEL (blue). (G and H) en-GAL4/+,UAS-GFP/+; (I and J) en-GAL4/+,UAS-buffy/+,UAS-GFP/+. (K) Irradiated en-GAL4/+,UAS-buffyΔN/+ embryo stained with anti-En (green) and TUNEL (blue).
Overexpression of Bcl-2 impairs the stress-induced apoptotic response of cells (Strasser et al., 1994; Cory and Adams, 2002). In order to determine whether buffy overexpression inhibits stress-induced apoptosis, we compared TUNEL from γ-irradiated en-GAL4,UAS-GFP,UAS-Buffy embryos with control embryos (Figure 5). Two UAS-buffy constructs were generated: a wild-type construct predicted to generate full-length protein and an N-terminal deletion construct (buffyΔN). The deletion eliminates 128 amino acids from the N-terminus, removing two putative α-helices that may be ancestral to the amphipathic α-helix from the BH4 domain of pro-survival Bcl-2 proteins (Figure 1A and B). As the BH4 domain is required for anti-apoptotic function of Bcl-2 (Hunter et al., 1996), by comparing the properties of buffyΔN with full-length buffy we sought to determine whether the extended N-terminal region of Buffy was important for cell survival.
Embryos expressing either full-length buffy (Figure 5F) or buffyΔN (Figure 5K) in the En pattern were protected from γ-irradiation-induced apoptosis (Figure 5B). TUNEL labeling within the En stripe was reduced 7-fold for full-length buffy (4.3 ± 1.7, n = 10) and 6-fold for buffyΔN (5.6 ± 1.5, n = 10), compared with wild type (32 ± 7, n = 9). There was also a reduced level of TUNEL in the inter-stripe region for full-length buffy (34 ± 8, n = 10) and buffyΔN (29 ± 10, n = 10) compared with wild type (64 ± 17, n = 9). This general reduction of TUNEL labeling, which was reproducible over three experiments, may be due to leaky expression of the UAS-buffy transgene (see above). To determine whether Buffy could inhibit stress-induced apoptosis in other tissues, we expressed Buffy using en-GAL4, which is also expressed in the posterior of third instar larval wing discs (Kornberg et al., 1985). Expression of two copies of UAS-buffy with en-GAL4 is embryonic lethal (data not shown), therefore wing discs expressing only one copy of full-length buffy were examined. Cells in the posterior compartment of the wing disc were protected from γ-irradiation-induced apoptosis, compared with the high level of cell death observed in the anterior compartment (Figure 5I and J), or with the extensive TUNEL observed in irradiated wild-type discs (Figure 5G). The observation that protection from apoptosis does not occur in the anterior compartment of the wing disc suggests that either: (i) leaky expression of the UAS-buffy transgene does not occur to the same degree in the wing discs as in the embryo; or (ii) that when only one copy of UAS-buffy is present, leaky expression does not occur at a level that provides protection from irradiation-induced cell death.
Increased levels of Buffy are therefore sufficient to inhibit the Drosophila apoptotic pathway that normally responds to DNA damaging agents such as ionizing radiation. Furthermore, since both full-length Buffy and BuffyΔN protected embryos from γ-irradiation-induced apoptosis, the region of the protein encompassing the three BH domains, and C-terminal membrane anchor, is sufficient for the anti-apoptotic function of Buffy.
Genetic analysis places Buffy downstream of Rpr, Grim and Hid, and upstream of the the apical caspase Dronc
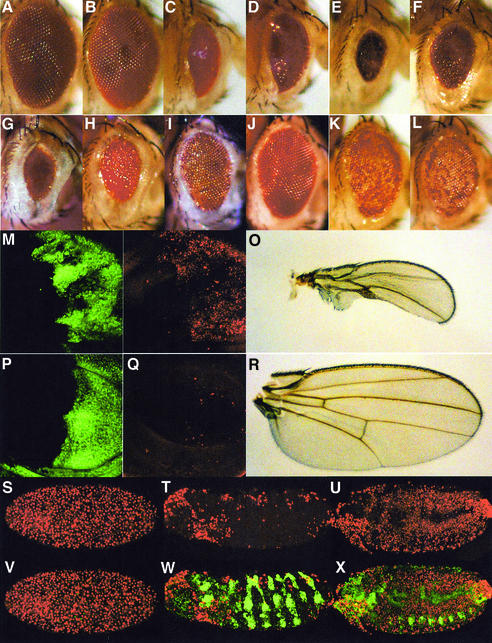
To examine genetic interactions between Buffy and other apoptotic pathway genes, we used Glass multimer reporter (GMR)-GAL4 to drive the UAS-buffy transgene in the posterior region of the third instar eye imaginal disc. Recombinants of the UAS-buffy transgene with GMR-GAL4 on the second chromosome, when heterozygous (GMR-GAL4:UAS-buffy/+), produce flies with eyes of wild-type appearance (compare Figure 6B with A). Similarly, ectopic expression of the Drosophila inhibitor of apoptosis, DIAP1, using the GMR driver results in normal-appearing adult eyes (data not shown). However, expression of diap1 can inhibit apoptotic phenotypes generated by overexpression of caspases (Hawkins et al., 2000; Meier et al., 2000; Quinn et al., 2000), rpr and hid (Goyal et al., 2000; Meier et al., 2000). GMR-diap1 also suppresses the GMR-GAL4/+;UAS-debcl/+ ablated eye phenotype (data not shown), consistent with the notion that Debcl induces apoptosis by functioning upstream of DIAP1-dependent caspase inhibition (Colussi et al., 2000).
Fig. 6. Genetic interactions between Buffy and cell death genes for Rpr, Grim, Hid, Debcl and Dronc. Adult eyes of the following genotypes: (A) GMR-GAL4/+;+/+; (B) GMR-GAL4/+,UAS-buffy/+;+; (C) GMR-GAL4/+;UAS-debcl/+; (D) GMR-GAL4/+,UAS-buffy/+;UAS-debcl/+; (E) GMR-rpr/+;+; (F) GMR-rpr/+,GMR-GAL4/+,UAS-buffy/+;+/+; (G) GMR-hid/+;+/+; (H) GMR-hid/+,GMR-GAL4/+,UAS-buffy/+;+/+; (I) GMR-grim/+;+/+; (J) GMR-grim/+,GMR-GAL4/+,UAS-buffy/+;+/+; (K) GMR-GAL4/+,UAS-dronc/+;+/+; and (L) GMR-GAL4/+,UAS-buffy/+,UAS-dronc/+;+/+. (M–P) Third instar larval wing discs: (M and N) en-GAL4/+,UAS-GFP/+;UAS-debcl/+; (O and P) en-GAL4/+,UAS-buffy/+,UAS-GFP/+;UAS-debcl/+. En-expressing cells stained using GFP (green) (M and O) and TUNEL (red) (N and P). (Q and R) Wings from adult females: (Q) en-GAL4/+,UAS-GFP/+;UAS-debcl/+ and (R) en-GAL4/+,UAS-buffy/+,UAS-GFP/+;UAS-debcl/+. (S–X) en-GAL4/+,UAS-GFP/+;th5/th5 (S and V), and en-GAL4/+,UAS-buffy/+,UAS-GFP/+;th5/th5 at stage 10 (T and W) and stage 14 (U and X). Embryos are labeled with TUNEL (red) (S, T and U) and stained with the anti-GFP antibody (green) (merged images V, W and X). Note, in (V), that cells expressing GFP have been destroyed.
The strong ablated eye phenotype from GMR-GAL4/+;UAS-debcl/+, shown in Figure 6C, could be partially suppressed by coexpression of buffy (Figure 6D). The extreme nature of the GMR-GAL4;UAS-Debcl/+ phenotype suggests a high level of Debcl protein expression, and thus the slight suppression by Buffy suggests that this UAS-buffy line is not expressed at high enough levels to sequester the excess Debcl protein. However, the en-GAL4-UAS-debcl ablated wing phenotype (Figure 6O) was suppressed by coexpression of UAS-buffy (Figure 6R). TUNEL labeling of third instar wing imaginal discs revealed that this suppression was due to Buffy inhibiting Debcl-induced apoptosis in the posterior compartment (compare Figure 6Q with M).
Ectopic expression of Rpr, Hid and Grim causes the Drosophila IAP homolog Diap1/Thread(th) to be sequestered and inactivated, thus resulting in ectopic cell death (reviewed in Hay, 2000). Overexpression of buffy with GMR-GAL4 partially suppressed the ablated eye phenotypes of rpr (Figure 6F), hid (Figure 6H) and grim (Figure 6J). Furthermore, the ectopic TUNEL observed in homozygous diap1 (th5) mutant embryos (Wang et al., 1999) (Figure 6S and V) was inhibited by overexpression of the UAS-buffy transgene with en-GAL4 (Figure 6T, U, W and X). Taken together, this suggests that Buffy acts downstream of Rpr, Grim, Hid and DIAP1 to block caspase-dependent cell death.
Expression of the UAS-dronc transgene with GMR-GAL4 results in a small, mottled eye phenotype as a consequence of ectopic cell death, particularly of pigment cells (Figure 6K) (Meier et al., 2000; Quinn et al., 2000). This phenotype was not modified by coexpression of the UAS-buffy transgene (Figure 6L), suggesting that Buffy acts upstream of caspase activation. Similarly, overexpression of the N-terminal deletion construct (UAS-buffyΔN) with GMR-GAL4 suppressed the Rpr, Grim, Hid and Debcl-ablated eye phenotypes, but did not alter the Dronc mottled eye phenotype (data not shown). Therefore, only the C-terminal portion of the Buffy protein is required for suppression.
Buffy induces a G1 phase delay when overexpressed in embryos
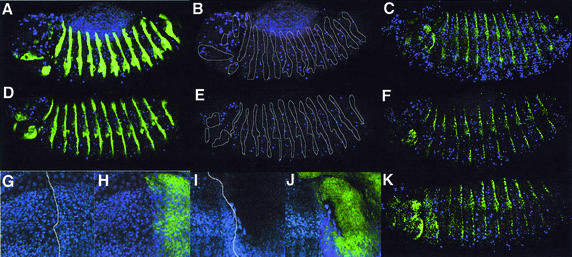
The mammalian Bcl-2 protein can inhibit cell cycle entry, independent of its anti-apoptotic function (Huang et al., 1997). Although the overall growth rate of proliferating cell cultures is not affected by ectopic Bcl-2, increased withdrawal from the cell cycle into G1 phase occurs (Vairo et al., 1996, 2000) and cell cycle re-entry is retarded (Linette et al., 1996; Mazel et al., 1996). Here we show that overexpression of the UAS-buffy transgene with the en-GAL4 driver results in inhibition of rapid embryonic cell cycles and an accumulation of cells in G1 (Figure 7). Although the cell cycle pattern is dynamic, generally there are comparable numbers of S-phase cells for the same sized region both inside and outside the En stripe in a normal stage-11 embryo (Quinn et al., 2001). The number of S-phase cells was clearly reduced, although not eliminated, in cells overexpressing two copies of the buffy transgene in the En-stripe (Figure 7C and D) (BrdU in the En stripe = 6.7 ± 1.1, n = 6) compared with the inter-stripe regions (BrdU in the interstripe = 37.7 ± 4.5, n = 6; the equivalently sized region to the En stripe was calculated by dividing total interstripe count by 3) and the control embryo (Figure 7A and B) (BrdU in the En stripe = 19.2 ± 1.5; n = 6, BrdU in the interstripe =33.3 ± 3.8, n = 6). Mitotic cells, visualized using the anti-phosphohistone H3 antibody (PH3), were scattered across the epithelium of stage-11 embryos (Quinn et al., 2001) (Figure 7E–H). Mitotic cells were almost eliminated within the buffy-expressing En stripe (PH3 in En stripe = 2.8 ± 1.8, n = 5), compared with cells between the stripe (Figure 7G and H) (PH3 between stripe = 23 ± 10, n = 5) and control embryos (Figure 7E and F) (PH3 in En stripe = 21 ± 3.9, n = 5; PH3 between stripe = 39 ± 11, n = 5).
Fig. 7. Cell cycle analysis of Drosophila embryos expressing the UAS-buffy transgene in En stripes. The En stripes shown are epidermal cells spanning the ventral region of thoracic segments 4 and 5 from stage 11 embryos. (a–d) S-phase cells labeled with BrdU (red) and with En marked with GFP [green in (B) and (D), or white lines in (A) and (C)]. (A and B) en-GAL4/en-GAL-4,UAS-GFP/UAS-GFP; (C and D) en-GAL4/enGAL4,UAS-GFP/+,UAS-buffy/+. (e–h) Mitotic cells of stage-11 embryos visualized using the anti-phosphohistone H3 antibody (PH3) in green, En antibody staining in red (F and H) or a white line (E and G). (E and F) en-GAL4/en-GAL4,UAS-GFP/UAS-GFP; (G and H) en-GAL4/enGAL4,UAS-GFP/+,UAS-buffy/+. (i–l) Cyclin B distribution in stage-11 embryos, Cyclin B (red) and En stripe marked with GFP (green) (J and L), or by a white line (I and K). (I and J) en-GAL4/en-GAL4,UAS-GFP/UAS-GFP; (K and L) en-GAL4/enGAL4,UAS-GFP/+,UAS-buffy/+. (m–p) DNA morphology analysis using propidium iodide (PI) in red and the En stripe marked by GFP in green (N and P) or by a white line (M and O). (M and N) en-GAL4/en-GAL4,UAS-GFP/UAS-GFP; (O and P) en-GAL4/en-GAL4,UAS-GFP/+,UAS-buffy/+.
That mitotic figures were almost eliminated, whilst some BrdU incorporation was observed, might suggest a G2 phase arrest, which would result in high levels of the G2–M cyclin, Cyclin B. However, staining with a Cyclin B antibody showed that Cyclin B was low in buffy-overexpressing cells when compared with neighboring regions (Figure 7K and L) and control embryos (Figure 7I and J). Therefore, high levels of Buffy do not appear to cause a G2 arrest or delay. Since there are decreased numbers of S-, G2- and M-phase cells, and the nuclei size is smaller (Figure 7O and P), arrest is consistent with a G1/early S phase arrest. Consistent with a cell cycle arrest when Buffy is overexpressed, the En stripe from an equivalent region of the embryo is often thinner and contains fewer cells compared with the control in stage 11/early stage 12 embryos (Figure 7O and P compared with M and N). The variability in the width of the En band is likely to be a consequence of the extremely rapid cycling of stage 11 embryos, combined with the fact the there will be a gradual accumulation of Buffy in the En stripes, because En only starts to be highly expressed at stage 11. Leaky expression of the UAS-buffy transgene (see above) does not appear to greatly affect cell cycle progression in the inter-stripe region of en-GAL4,UAS-buffy embryos, possibly because the level of Buffy protein required is higher than that needed to prevent apoptosis. Importantly, these results provide the first evidence within a whole animal that a member of the Bcl-2 family has a cell cycle inhibitory role.
Discussion
In this report, we have shown that Buffy functions in a similar manner to the pro-survival mammalian Bcl-2 proteins since it: (i) is required for cell survival; (ii) inhibits developmentally regulated apoptosis; (iii) inhibits γ-irradiation-induced apoptosis; (iv) binds the Drosophila pro-apoptotic Bcl-2 homolog Debcl, and can suppress Debcl-induced cell death; and (v) when overexpressed has an inhibitory effect on cell cycle progression.
In mammals there are multiple pro-survival Bcl-2 proteins that play a tissue-specific role in protecting cells from apoptosis. Therefore, targeted knockout of individual pro-survival Bcl-2 members does not result in the death of the entire organism, suggesting that other pro-survival Bcl-2 proteins can compensate. For example, knockout studies show that Bcl-2 is required for survival of stem cells from kidney and melanocytes and adult lymphocytes (Veis et al., 1993), whilst Bcl-XL is required for survival of neuronal and erythroid cells (Motoyama et al., 1995). In contrast, RNAi knockdown of buffy resulted in general embryonic cell death, suggesting that Buffy is a principle cell survival protein at this stage of Drosophila development. Indeed, database analysis shows no other Bcl-2-related proteins, apart from Buffy and Debcl.
Overexpression of Bcl-2 impairs the apoptotic response to DNA damaging agents such as ionizing radiation (Strasser et al., 1994). One of the reasons many tumors are resistant to chemotherapy and radiation therapy is that they mis-express Bcl-2 (reviewed in Cory and Adams, 2002). Here we have shown increased levels of Buffy are sufficient to inhibit the Drosophila apoptotic pathway that normally responds to DNA damaging agents.
Overexpression of mammalian Bcl-2 in Drosophila tissues has been shown to inhibit apoptosis, induced by either irradiation or Rpr overexpression (Gaumer et al., 2000; Brun et al., 2002). As we have found for Buffy, genetic analysis places the anti-apoptotic activity of Bcl-2 downstream of Rpr when expressed in flies (Brun et al., 2002). This is most likely a consequence of Bcl-2 protein binding to and sequestering Debcl, as has been shown previously (Colussi et al., 2000). In mammals, Bcl-2-mediated inhibition of apoptosis requires an α-helical domain within the N-terminal BH4 domain (Hunter et al., 1996). In contrast, Buffy’s N-terminus, and the putative α-helices therein, were not required for either inhibition of irradiation-induced apoptosis or suppression of Rpr-, Grim- and Hid-induced apoptosis. Thus, the C-terminal region containing the BH1, BH2, BH3 and membrane anchor is sufficient for Buffy’s cell survival function.
Certain factors controlling cell cycle progression are also sensitive to apoptotic stimuli (reviewed in Evan and Littlewood, 1998). Indeed, cell cycle factors may promote apoptosis under conditions unfavorable for proliferation, thus rendering cycling cells more vulnerable to apoptosis. Evidence that Bcl-2 plays a role in controlling cell cycle progression has been accumulating steadily (Linette et al., 1996; Mazel et al., 1996; O’Reilly et al., 1996; Vairo et al., 1996, 2000; Lind et al., 1999), and in this study we provide the first in vivo evidence that a member of the Bcl-2 family can result in cell cycle inhibition. However, in contrast to the serum-deprived G0 cells that have been used previously (Linette et al., 1996; Mazel et al., 1996; Vairo et al., 1996), Drosophila embryonic cells cycle normally prior to overexpression of Buffy.
The cell cycle delay that occurs as a consequence of Buffy overexpression is dose dependent. Expression of two copies of UAS-buffy via en-GAL4 is embryonic lethal, presumably as a consequence of the G1–S cell cycle arrest, which results in less cells in the En-stripes and insufficient cells to complete embryonic development. Two copies of en-GAL4,UAS-buffy (i.e. high level of Buffy protein) induce a cell cycle arrest and are embryonic lethal; however, flies expressing one copy (lower level of expression) are viable. Indeed, a low level of ubiquitous Buffy expression does not cause a cell cycle arrest, but results in the production of additional neural cells (L.Quinn and H.Richardson, unpublished data). Thus, the effect of Buffy overexpression is dose dependent; high levels of Buffy overexpression induce cell cycle arrest, which will ultimately result in fewer cells, whilst lower levels can inhibit developmental cell death and are associated with increased cell numbers.
Consistent with cell cycle arrest, Bcl-2 overexpression in mammalian cells correlates with increased levels of the CycE/Cdk2 inhibitor p27 (Brady et al., 1996; Linette et al., 1996), hyperphosphorylated and inactive retinoblastoma (RB) tumor suppressor (Mazel et al., 1996), and increased levels of RB-related protein p130 (Lind et al., 1999; Vairo et al., 2000). The inhibitory effect of Bcl-2 on the cell cycle is independent of p53, the cdk4/6 inhibitor p16 and RB, but requires p130 and p27 (O’Reilly et al., 1996; Vairo et al., 1996, 2000). The cell cycle inhibitory function of Bcl-2 can be separated from its cell survival function since the tyrosine residue, Y28, in the N-terminal BH4 domain is important for Bcl-2 inhibition of cell cycle re-entry, but is not required for cell survival (Huang et al., 1997). Although Buffy does not have an obvious BH4 domain, the cell cycle inhibitory function has been conserved between Buffy and Bcl-2. Thus, it will now be important to determine whether Buffy uses a similar mechanism to Bcl-2 to inhibit cell cycle progression.
Materials and methods
Cloning of Buffy cDNA
buffy was identified in a TBLASTN search using the Bcl-2 protein sequence (reported as 48A-E in Colussi et al., 2000). A 950-bp partial cDNA clone was isolated from a mixed stage Drosophila embryo cDNA library in λgt11 using a 450-bp probe derived from Drosophila genomic DNA using PCR. Sequencing of this clone confirmed that it encoded a Bcl-2 family member in the genomic region 48A-E and that it contained the predicted coding sequence for buffy. Further 5′ non-coding exon sequence was identified using 5′ RACE.
mRNA expression analysis
For RT–PCR, total RNA was prepared using RNAzol B (Tel-Test Inc.) and reverse transcribed using the First Strand cDNA Synthesis Kit (Amersham Pharmacia Biotech). PCR was performed using buffy-specific primers spanning an intron with forward sequence 5′-ATGCATCCCCGGATATACAAC-3′ and reverse sequence 5′-GCGGGATCCTTAGGAATTCGTAAATCGTTGGTA-3′ to yield a product of 465 bp. The cytochrome c control was amplified with the forward primer (5′-CCGGAATTCATGGGCGTTCCTGCTGGTGAT-3′) and reverse primer (5′-CCGGAATTCTTACTTGGTCGCCGACTTCAG-3′) to obtain a product of 345 bp. Since buffy mRNA is expressed at very low levels, and it is difficult to visualize using the standard immunological staining methods, the TSA™ system (New England Nuclear Life Science Products) was used in to amplify the in situ signal according the methods described in Colussi et al. (2000).
Buffy/Debcl co-immunoprecipitation experiments
The 900-bp coding region of buffy was PCR amplified using Pfu Turbo DNA Polymerase (Stratagene) with an in-frame N-terminal FLAG-tag, and cloned into mammalian expression vector pcDNA3 (Invitrogen). HA-tagged pcDNA3-Debcl has been described previously (Colussi et al., 2000). 293T cells were transfected with 2 µg of pcDNA3-HA Debcl or pcDNA3-FLAG Buffy, or co-transfected with 2 µg of each construct using FuGENE 6 Transfection Reagent (Roche). Twenty-four hours after transfection, cell lysates were prepared in lysis buffer (50 mM Tris–HCl pH 7.6, 150 mM NaCl, 0.1% NP-40) supplemented with Complete protease inhibitors (Roche). Immunoprecipitations were carried out as described previously (Colussi et al., 2000) using anti-Flag antibody (Sigma) and HA antibody (Roche).
Generation of Buffy antisera, antibody staining, BrdU, TUNEL and microscopy
To express Buffy in bacteria, the coding region of Buffy was PCR amplified from the cDNA clone and cloned into pGEX. Bacterially expressed Buffy protein was purified on a SDS polyacrylamide gel and used to inoculate rats. Rats were terminally bled after three boosts. Immunohistochemistry, including TUNEL and BrdU labeling of Drosophila tissues, was carried out as described previously (Quinn et al., 2000, 2001). Rat anti-Buffy antibody was detected using anti-rat-biotin conjugated secondary antibody followed by either streptavidin-lissamine rhodamine (Jackson) or Alexa-488 (Molecular Probes). TUNEL staining was carried out using the Roche in situ cell death kit, TRred. Other antibodies were: anti-BrdU monoclonal antibody (Becton Dickenson), anti-En monoclonal, rabbit anti-phosphohistone H3 (Santa Cruz) and rabbit anti-GFP (Molecular Probes). Staining with MitotrackerRed (Molecular Probes) was performed as described previously (Iyengar et al., 2002). All fluorescence-labeled samples were analyzed by confocal microscopy (Bio-Rad MRC1000), while AP colormetrically detected samples were analyzed on the Zeiss Axiophot Photophot using Nomarski optics.
RNA interference
buffy RNAi was pre-formed as described by Quinn et al. (2000). RNA transcripts were generated using the Ambion Megascript kit with linearized pOT2-buffy templates. Sense and antisense transcripts were purified, annealed and then dissolved in injection buffer (5 mM KCl in 0.1 mM phosphate buffer, pH 7.8) at 0.75 mg/ml. Between 250 and 300 pre-cellularized embryos were injected at 50% egg length with either buffy dsRNA or injection buffer alone, and aged until either stage 11–12 or stage 14–16.
Generation of transgenic flies and genetic interactions
PCR-generated, full-length buffy and buffyΔN-Flag fragments were cloned into pUAST (Brand and Perrimon, 1993) and transgenic flies were generated as previously described (Richardson et al., 1995). A UAS-buffy transgene on the second chromosome was used for all experiments. Recombinants of GMR-GAL4 and UAS-buffy on the second chromosome were used for genetic interactions. All general fly stocks were obtained from the Bloomington Stock Centre, except for en-GAL4,UAS-GFP (from Laura Johnston). All genetic interaction crosses with GMR-GAL4,UAS-buffy lines were carried out at 25°C, except those with GMR-hid and GMR-rpr, which were carried out at 18°C.
Acknowledgments
Acknowledgements
We thank David Glover for the anti-Dm Cyclin B antibody and Laura Johnston for the en-GAL4,UAS-GFP strain. This work was supported by the Wellcome Trust and the National Health and Medical Research Council (NHMRC). H.R. is a Wellcome Senior Research Fellow in Medical Science and S.K. is a NHMRC Principal Senior Research Fellow.
References
- Abrams J.M., White,K., Fessler,L.I. and Steller,H. (1993) Programmed cell death during Drosophila embryogenesis. Development, 117, 29–43. [DOI] [PubMed] [Google Scholar]
- Aravind L., Dixit,V.M. and Koonin,E.V. (2001) Apoptotic molecular machinery: vastly increased complexity in vertebrates revealed by genome comparisons. Science, 291, 1279–1284. [DOI] [PubMed] [Google Scholar]
- Baehrecke E.H. (2000) Steroid regulation of programmed cell death during Drosophila development. Cell Death Differ., 7, 1057–1062. [DOI] [PubMed] [Google Scholar]
- Baehrecke E.H. (2002) How death shapes life during development. Nat. Rev. Mol. Cell Biol., 3, 779–787. [DOI] [PubMed] [Google Scholar]
- Brachmann C.B., Jassim,O.W., Wachsmuth,B.D. and Cagan, R,L. (2000) The Drosophila bcl-2 family member dBorg-1 functions in the apoptotic response to UV-irradiation. Curr Biol., 10, 547–550. [DOI] [PubMed] [Google Scholar]
- Brady H.J.M., Gil-Gümez, Kirberg,J. and Berns,A.J.M. (1996) Baxα perturbs T cell development and affects cell cycle entry of T cells. EMBO J., 15, 6991–7001. [PMC free article] [PubMed] [Google Scholar]
- Brand A.H. and Perrimon,N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Brun S., Rincheval,V., Gaumer,S., Mignotte,B. and Guenal,I. (2002) reaper and bax initiate two different apoptotic pathways affecting mitochondria and antagonized by bcl-2 in Drosophila. Oncogene, 21, 6458–6470. [DOI] [PubMed] [Google Scholar]
- Buszczak M. and Cooley,L. (2000) Eggs to die for: cell death during Drosophila oogenesis. Cell Death Differ., 7, 1071–1074. [DOI] [PubMed] [Google Scholar]
- Christich A., Kauppila,S., Chen,P., Sogame,N., Ho,S.I. and Abrams,J.M. (2002) The damage-responsive Drosophila gene sickle encodes a novel IAP binding protein similar to but distinct from reaper, grim and hid. Curr. Biol., 12, 137–140. [DOI] [PubMed] [Google Scholar]
- Colussi P.A., Quinn,L.M., Huang,D.C.S., Coombe,M., Read,S.H., Richardson,H. and Kumar,S. (2000) Debcl a proapoptotic Bcl-2 homologue, is a component of the Drosophila melanogaster cell death machinery. J. Cell Biol., 148, 703–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradt B. and Horvitz,H.R. (1998) The C.elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell, 93, 519–529. [DOI] [PubMed] [Google Scholar]
- Cory S. and Adams,J.M. (2002) The Bcl-2 family: regulators of the cellular life-or-death switch. Nat. Rev. Cancer, 2, 647–656. [DOI] [PubMed] [Google Scholar]
- Cryns V. and Yuan,J. (1998) Proteases to die for. Genes Dev., 12, 1551–1570. [DOI] [PubMed] [Google Scholar]
- Deveraux Q.L. and Reed,J.C. (1999) IAP family proteins-suppressors of apoptosis. Genes Dev., 13, 239–252. [DOI] [PubMed] [Google Scholar]
- Du C., Fang,M., Li,Y., Li,L. and Wang,X. (2000) Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell, 102, 33–42. [DOI] [PubMed] [Google Scholar]
- Evan G. and Littlewood,T. (1998) A matter of life and death. Science, 281, 1317–1321. [DOI] [PubMed] [Google Scholar]
- Gaumer S., Guénal,I., Brun,S., Théodore,L. and Mignotte,B. (2000) Bcl-2 and Bax mammalian regulators of apoptosis are functional in Drosophila. Cell Death Differ., 7, 804–814. [DOI] [PubMed] [Google Scholar]
- Goyal L., MacCall,K., Agapite,J., Hartwieg,E. and Steller,H. (2000) Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J., 19, 589–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins C.J., Yoo,S.J., Peterson,E.P., Wang,S.L., Vernooy,S.Y. and Hay,B.A. (2000) The Drosophila caspase DRONC is a glutamate/aspartate protease whose activity is regulated by DIAP1, HID and GRIM. J. Biol. Chem., 275, 27084–27093. [DOI] [PubMed] [Google Scholar]
- Hay B.A. (2000) Understanding IAP function and regulation: a view from Drosophila. Cell Death Differ., 7, 1045–1056. [DOI] [PubMed] [Google Scholar]
- Hengartner M.O. (2001) Apoptosis. DNA destroyers. Nature, 412, 27–29. [DOI] [PubMed] [Google Scholar]
- Horvitz H.R. (1999) Genetic control of programmed cell death in the nematode Caenorhabditis elegans. Cancer Res., 59, 1701S–1706S. [PubMed] [Google Scholar]
- Huang D.C.S., O’Reilly,L.A., Strasser,A. and Cory,S. (1997) The anti-apoptosis function of Bcl-2 can be genetically separated from its inhibitory effect on cell cycle entry. EMBO J., 16, 4628–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter J.J., Bond,B.L. and Parslow,T.G. (1996) Functional dissection of the human Bcl-2 protein: sequence requirements for inhibition of apoptosis. Mol. Cell. Biol., 16, 877–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaki T., Kanuka,H., Inohara,N., Sawamoto,K., Nunez,G., Okano,H. and Miura,M. (2000) Drob-1, a Drosophila member of the Bcl-2/CED-9 family that promotes cell death. Proc. Natl Acad. Sci. USA, 97, 662–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar B., Luo,N., Farr,C.L., Kaguni,L.S. and Campos,A.-R. (2002) The accessory subunit of DNA polymerase γ is essential for mitochondrial DNA maintenance and development in Drosophila melanogaster. Proc. Natl Acad. Sci. USA, 99, 4483–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janiak F., Leber,B. and Andrews,B.W. (1994) Assembly of Bcl-2 into microsomal and outer mitochondrial membranes. J. Biol. Chem., 269, 9842–9849. [PubMed] [Google Scholar]
- Jiang C., Baehrecke,E.H. and Thummel,C.S. (1997) Steroid regulated programmed cell death during Drosophila metamorphosis. Development, 124, 4673–4683. [DOI] [PubMed] [Google Scholar]
- Kornberg T., Siden,I., O’Farrell,P. and Simon,M. (1985) The engrailed locus of Drosophila: in situ localization of transcripts reveals compartment-specific expression. Cell, 40, 45–53. [DOI] [PubMed] [Google Scholar]
- Lind E.F., Wayne,J., Wang,Q.-Z., Staeva,T., Stolzer,A. and Petrie,H.T. (1999) Bcl-2 induced changes in E2F regulatory complexes reveal the potential for integrated cell cycle and cell death functions. J. Immunol., 162, 5374–5379. [PubMed] [Google Scholar]
- Linette G.P., Li,Y., Roth,K. and Korsmeyer,S.J. (1996) Cross talk between cell death and cell cycle progression: BCL-2 regulates NFAT-mediated activation. Proc. Natl Acad. Sci. USA, 93, 9545–9552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazel S., Burtrum,D. and Petrie,H.T. (1996) Regulation of cell division cycle progression by bcl-2 expression: a potential mechanism for inhibition of programmed cell death. J. Exp. Med., 183, 2219–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall K. and Steller,H. (1997) Facing death in the fly: genetic analysis of cell death in Drosophila. Trends Genet., 13, 222–226. [DOI] [PubMed] [Google Scholar]
- Meier P., Silke,J., Leevers,S.J. and Evan,G.I. (2000) The Drosophila caspase DRONC is regulated by DIAP1. EMBO J., 19, 598–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller D.T. and Cagan,R.L. (1998) Local induction of patterning and programmed cell death in the developing Drosophila retina. Development, 125, 2327–2335. [DOI] [PubMed] [Google Scholar]
- Motoyama N. et al. (1995) Massive cell death of immature hematopoietic cells and neurons in Bcl-x deficient mice. Science, 267, 1506–1510. [DOI] [PubMed] [Google Scholar]
- Ollmann M. et al. (2000) Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell, 101, 91–101. [DOI] [PubMed] [Google Scholar]
- O’Reilly L.A., Huang,D.C.S. and Strasser,A. (1996) The cell death inhibitor Bcl-2 and its homologues influence control of cell cycle entry. EMBO J., 15, 6979–6990. [PMC free article] [PubMed] [Google Scholar]
- Quinn L.M., Dorstyn,L., Colussi,P.A., Chen,P., Coombe,M., Abrams,J., Kumar,S. and Richardson,H. (2000) An essential role for the caspase Dronc in developmentally programmed cell death in Drosophila. J. Biol. Chem., 275, 40416–40424. [DOI] [PubMed] [Google Scholar]
- Quinn L.M., Herr,A., McGarry,T. and Richardson,H. (2001) The Drosophila Geminin homologue: roles for Geminin in limiting DNA replication, in anaphase and in neurogenesis. Genes Dev., 15, 2741–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson H. and Kumar,S. (2002) Death to flies: Drosophila as a model system to study programmed cell death. J. Immunol. Methods, 265, 21–38. [DOI] [PubMed] [Google Scholar]
- Richardson H., O’Keefe,L.V., Marty,T. and Saint,R. (1995) Ectopic cyclin E expression induces premature entry into S-phase and disrupts pattern formation in the Drosophila eye imaginal discs. Development, 121, 3371–3379. [DOI] [PubMed] [Google Scholar]
- Sharp P.A. (2001) RNA interference—2001. Genes Dev., 15, 485–490. [DOI] [PubMed] [Google Scholar]
- Srinivasula S.M. et al. (2002) sickle, a novel Drosophila death gene in the reaper/hid/grim region, encodes an IAP-inhibitory protein. Curr. Biol., 12, 125–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser A., Harris,A.W., Jacks,T. and Cory,S. (1994) DNA damage can induce apoptosis in proliferating lymphoid cells via p53-independent mechanisms inhibitable by Bcl-2. Cell, 79, 329–339. [DOI] [PubMed] [Google Scholar]
- Truman J.W., Talbot,W.S., Fahrbach,S.E. and Hogness,D.S. (1994) Ecdysone receptor expression in the CNS correlates with stage-specific responses to ecdysteroids during Drosophila and Manduca development. Development, 120, 219–234. [DOI] [PubMed] [Google Scholar]
- Vairo G., Innes,K.M. and Adams,J.M. (1996) Bcl-2 has a cell cycle inhibitory function separable from its enhancement of cell survival. Oncogene, 13, 1511–1519. [PubMed] [Google Scholar]
- Vairo G., Soos,T.J., Upton,T.M., Zalvide,J., DeCaprio,J.A., Ewen,M.E., Koff,A. and Adams,J.M. (2000) Bcl-2 retards cell cycle entry through p27Kip1, pRB relative p130 and altered E2F reulation. Mol. Cell. Biol., 20, 4745–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veis D.J., Sorenson,C.M., Shutter,J.R. and Korsmeyer,S.J. (1993) Bcl-2 deficient mice demonstrate fulminant lymphoid apoptosis, polycystic kidneys and hypopigmented hair. Cell, 75, 229–240. [DOI] [PubMed] [Google Scholar]
- Verhagen A.M. et al. (2000) Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell, 102, 43–53. [DOI] [PubMed] [Google Scholar]
- Wang S.L., Hawkins,C.J., Yoo,S.J., Muller,H.A. and Hay,B.A. (1999) The Drosophila Caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell, 98, 453–463. [DOI] [PubMed] [Google Scholar]
- Wing J.P., Karres,J.S., Ogdahl,J.L., Zhou,L., Schwartz,L.M. and Nambu,J.R. (2002) Drosophila sickle is a novel grim-reaper cell death activator. Curr. Biol., 12, 131–135. [DOI] [PubMed] [Google Scholar]
- Zou H., Henzel,W.J., Liu,X., Lutschg,A. and Wang,X. (1997) Apaf-1, a human protein homologous to C.elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell, 90, 405–413. [DOI] [PubMed] [Google Scholar]