Abstract
Human immunodeficiency virus type 1 (HIV-1), like other lentiviruses, can infect non-dividing cells. This property depends on the active nuclear import of its intracellular reverse transcription complex (RTC). We have studied nuclear import of purified HIV-1 RTCs in primary macrophages and found that importin 7, an import receptor for ribosomal proteins and histone H1, is involved in the process. Nuclear import of RTCs requires, in addition, energy and the com ponents of the Ran system. Depletion of importin 7 from cultured cells by small interfering RNA inhibits HIV-1 infection. These results provide a new insight into the molecular mechanism for HIV-1 nuclear import and reveal potential targets for therapeutic intervention.
Keywords: HIV-1/importins/nuclear import/reverse transcription complex/small interfering RNA
Introduction
Since it was first described (Gottlieb et al., 1981), the AIDS epidemic has killed some 24 million people and an estimated 40 million are currently infected with the human immunodeficiency virus (HIV) (UNAIDS, 2002). HIV mainly infects cells of the immune system expressing CD4, such as T-helper lymphocytes and macrophages (Freed and Martin, 2001). Studies of sexually transmitted HIV infection in animal models and in patients have indicated that initial virus infection of resident macrophages is crucial for subsequent virus spread to lymphoid organs and T-helper lymphocytes (Cohen and Fauci, 2001). The ability of HIV-1 to infect non-dividing cells such as mature macrophages depends on the active nuclear import of the intracellular complex that mediates reverse transcription and integration of the viral genome into host chromosomes (hereafter called reverse transcription complex, or RTC) (Bukrinsky et al., 1992). The HIV-1 RTC shows biophysical properties typical of a large nucleo protein complex and contains the viral proteins reverse transcriptase (RT), integrase (IN), nucleocapsid (NC, p7Gag), Vpr and small amounts of matrix (MA, p17Gag) in addition to the viral nucleic acids (Farnet and Haseltine, 1991; Bukrinsky et al., 1993; Miller et al., 1997; Fassati and Goff, 2001). MA, Vpr and IN possess nuclear localization signals (NLSs) and have been implicated in the nuclear import of RTCs, although their precise role is controversial (reviewed by Goff, 2001). Structural elements in the viral genome also appear to stimulate RTC nuclear import. HIV-1 mutants lacking the central polypurine tract (cPPT), a second site of initiation of plus strand DNA synthesis present in all lentiviruses, have a reduced ability to access the nucleus (Zennou et al., 2000). This reduction is small (2- to 5-fold) and dependent on the HIV-1 strain used (Dvorin et al., 2002; Limon et al., 2002). Alternatively, HIV-1 may gain access to the nucleus by breaking through limited areas of the nuclear envelope (de Noronha et al., 2001). While there is considerable knowledge on the cellular factors involved in nuclear export of the HIV-1 genome (Malim and Cullen, 1991; reviewed by Cullen, 2002), little is known about the cellular factors that mediate its nuclear import.
Nuclear import of proteins is mediated mainly by a large superfamily of factors related to importin β (impβ)/karyopherin β1 that shuttle continuously between the nucleus and the cytoplasm (reviewed in Görlich and Kutay, 1999). Members of this superfamily share the ability to bind to three components; the small GTPase Ran at the N-terminus, their cargo and nucleoporins, the latter being constituents of the nuclear pore complex (NPC) (Rout et al. 2000). The first step in nuclear import involves binding of the nuclear import receptor to its cargo in the cytoplasm, followed by interaction of the complex with phenylalanine-rich repeat regions of the nucleoporins. The receptor–cargo complex translocates through the NPC channel into the nucleoplasm where nuclear import receptors dissociate from the cargo and shuttle back into the cytoplasm. The cycle is controlled by a RanGTP gradient across the nuclear envelope. Low levels of RanGTP in the cytoplasm allow the binding of importins to their cargoes, whereas high RanGTP levels in the nucleus induce their dissociation (Rexach and Blobel, 1995; Bischoff et al., 2002). The RanGTP gradient is maintained by RCC1, the guanine nucleotide exchange factor that stimulates the conversion of RanGDP into RanGTP in the nucleus, RanGAP1 that, in combination with either RanBP1 or RanBP2, stimulates RanGTP hydrolysis in the cytoplasm (Bischoff et al., 2002), and NTF2 that imports cytoplasmic RanGDP into the nucleus (Ribbeck et al., 1998). These proteins, together with Ran, constitute the Ran system.
Many soluble factors involved in the nuclear import of cellular proteins were discovered and/or characterized using the nuclear import assay (Adam et al., 1990). In this assay, cells are treated with digitonin to permeabilize selectively the plasma membrane, leaving intact the nuclear envelope. By this procedure, cytosolic contents are washed out and nuclear import is reconstituted by the addition of a fluorescent-labelled substrate, cytosolic extracts or purified transport receptors, the components of the Ran system and an energy-regenerating system. Nuclear accumulation of the substrate is then analysed by fluorescence microscopy (Adam and Gerace, 1991; Görlich et al., 1994). Here, we have performed nuclear import assays in primary macrophages using purified HIV-1 RTCs as substrate and found that imp7 is a mediator of HIV-1 nuclear import. The biological relevance of this finding has been confirmed by small interfering RNA (siRNA)-mediated depletion of imp7 in cultured cells.
Results
Purified HIV-1 RTCs can be visualized by confocal microscopy
To purify intracellular viral complexes, we have modified a protocol used previously to yield functional HIV-1 and murine leukaemia virus (MLV) RTCs (Fassati and Goff, 1999, 2001). Briefly, recombinant HIV-1 virus pseudotyped with the vesicular stomatitis virus glycoprotein (VSV-G) was prepared by transient transfection and purified. Cytosolic extracts were prepared from acutely infected HeLa cells by Dounce homogenization in hypotonic buffer. RTCs were purified from the extracts by velocity sedimentation through a 5–20% linear sucrose gradient, followed by density fractionation in a 20–70% linear sucrose gradient and dialysis in a large pore cellulose membrane (see Materials and methods). Uninfected cells were processed in an identical way in parallel and used as negative controls.
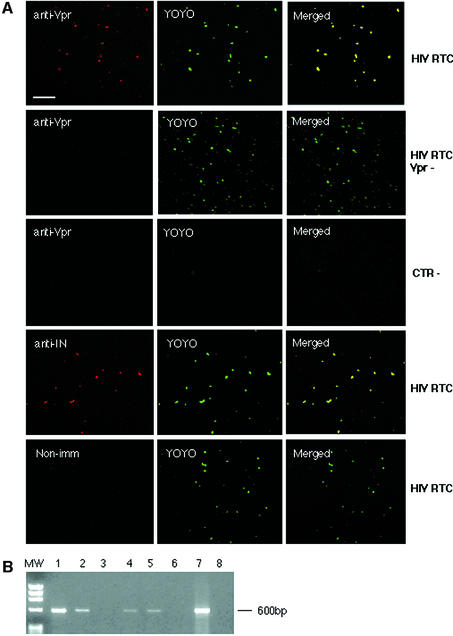
To examine the degree of RTC purification, purified fractions were analysed by confocal microscopy. Samples were adsorbed onto plastic slides, fixed and labelled with an anti-Vpr or an anti-IN rabbit antiserum and YOYO-1, a sensitive nucleic acids dye. Confocal microscopy detected co-localization of viral proteins and nucleic acids in >90% of fluorescent particles (Figure 1A). No co-localized particles were seen in samples containing mutant RTCs lacking Vpr, when rabbit non-immune sera was used or in control samples purified from uninfected cells. YOYO-1-only labelled particles were also rare in uninfected controls (Figure 1A). Overall, free nucleic acids represented <10% of green fluorescent dots in the RTC-containing samples. These data demonstrate that RTCs were highly purified and could be visualized specifically by confocal microscopy.
Fig. 1. Analyses of purified HIV-1 RTCs by confocal microscopy and endogenous reverse transcription assay. (A) Purified RTCs were adsorbed onto a plastic tissue culture dish, fixed and labelled with the nucleic acid dye YOYO-1 and anti-Vpr or anti-IN antibodies. Images were acquired sequentially and merged using the Confocal Assistant software. Mutant RTCs lacking Vpr (RTC Vpr–), samples from uninfected cells (CTR–) and non- immune rabbit sera were used as controls. Scale bar, 15 µm. (B) Endogenous RT assay on the equilibrium density fractions containing the peak of the viral DNA. Samples were incubated in the presence or absence of exogenous dNTPs and then subjected to PCR with primers specific for the (+) strand DNA (expected band size is 600 bp). MW, DNA molecular weight markers; 1, RTC fraction incubated in the presence of exogenous dNTPs; 2, same fraction without dNTPs; 3, fraction from uninfected cells; 4–6, same conditions as in 1–3 but RTCs were extracted in 160 mM KCl buffer after the first extraction in hypotonic buffer; 7, positive control; 8, no DNA.
To test the activity of purified RTCs, aliquots from the 1.34 g/ml density fractions were subjected to an endogenous RT assay. This assay examines the integrity of the RT machinery and its ability to synthesize full-length viral DNA. Samples were incubated in the presence or absence of exogenous dNTPs and analysed by semi-quantitative PCR with primers specific for the (+) strand DNA, a late reverse transcription product (Figure 1B). Contamination with plasmid DNA in fractions containing RTCs was excluded by PCR amplification of a sequence unique to the plasmid but absent in the reverse-transcribed viral genome (not shown). Addition of dNTPs consistently stimulated the synthesis of the (+) strand DNA. No viral DNA was found in samples from uninfected cells (Figure 1B). To test if most RTCs were extracted from the infected cells by Dounce homogenization in hypotonic buffer, an additional extraction step in 160 mM KCl was performed. Only small amounts of viral DNA were recovered by this second extraction, and RTCs were unable to reverse-transcribe (Figure 1B). Thus, purified RTCs extracted in hypotonic buffer are competent for reverse transcription in vitro.
HIV-1 RTCs accumulate at the nuclear envelope and in the nucleoli of digitonin-permeabilized cells
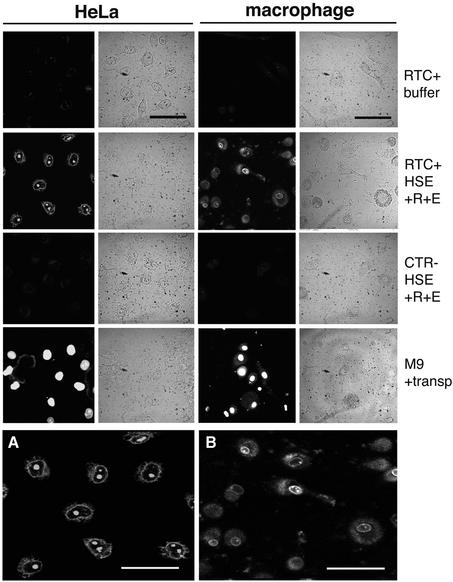
The nuclear import assay requires the substrate of interest to be fluorescent. Thus, the 1.34 g/ml density fraction containing purified RTCs was incubated with YOYO-1 and dialysed extensively to eliminate excess dye. The efficiency of the reaction was monitored by confocal microscopy, and labelled RTCs were added to digitonin-permeabilized HeLa cells or primary human macrophages. Primary macrophages were chosen for their direct relevance to HIV-1 replication in vivo. Following incubation at 37°C, cells were fixed and analysed by confocal microscopy (Figure 2). Labelled RTCs accumulated mainly at the nuclear periphery and in the nucleoli of both HeLa cells and macrophages in the presence of the constituents of the Ran system (Ran mix), an energy-regenerating system (energy mix) and 60S high-speed cytosolic extract (HSE). The binding of RTCs to the nuclear envelope was confirmed in separate nuclear import experiments, in which RTCs were found to co-localize with a fluorescent-labelled mutant impβ that binds irreversibly to the NPC (not shown) (Kutay et al., 1997). Nuclear import was undetectable using YOYO-1-labelled fractions from uninfected cells or in the absence of Ran mix, energy mix and HSE (Figure 2). Some background nuclear import was found in HeLa cells when HSE was not added, presumably because enough import receptors and Ran remained available after permeabilization. This phenomenon was less prominent in macrophages. We tested a number of different conditions for the nuclear import assay and compared the behaviour of RTCs with that of a GST–green fluorescent protein (GFP) fusion construct bearing the ‘classic’ SV40 large T antigen NLS (GFP–nls) (Kalderon et al., 1984). Incubation of cells at 25°C instead of 37°C resulted in poor import of RTCs but had no effect on GFP–nls. Similarly to the classic NLS, RTC import was stimulated by addition of Ran and energy mix.
Fig. 2. Nuclear import assay with labelled RTCs in HeLa cells and primary human macrophages. Following permeabilization with digitonin, cells were incubated with 103 RTCs/cell (RTC+) or an equal volume of sample from uninfected cells (CTR–) or 1 µM fluorescent-labelled M9-bearing nucleoplasmin core (M9) plus transportin (transp, 1 µM). Incubation was performed for 15 min at 37°C in the presence of energy mix (E), Ran mix (R) and HeLa high-speed cytosolic exctracts (HSE, 0.5 mg/ml final concentration) or buffer. Fluorescent and transmission images of the same fields are shown. Scale bars, 25 µm. (A) Enlarged image corresponding to the HeLa RTC HSE panel. (B) Enlarged image corresponding to the macrophage RTC HSE panel. Scale bars, 25 µm.
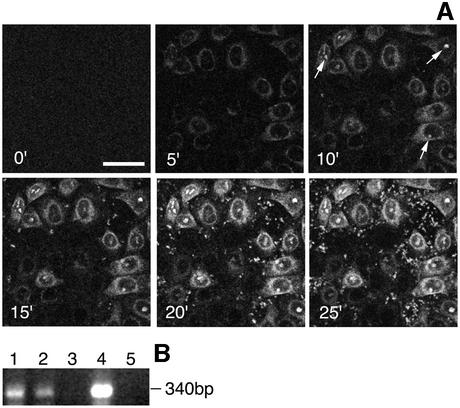
Time-lapse microscopy was used to analyse the kinetics of RTC nuclear accumulation in permeabilized HeLa cells (Figure 3A). Nuclear import assays of labelled RTCs were performed at 37°C in the presence of HSE, energy mix and Ran mix, and images were acquired every 30 s. After 3–5 min incubation, RTCs accumulated at the nuclear envelope. Five minutes later, they started accumulating in the nucleoli, and import reached a peak in ∼20 min (Figure 3A). Large-scale import assays were also performed using unlabelled RTCs. Cells were washed extensively following the import assay and viral DNA present in the permeabilized cells was extracted (Hirt, 1967) and analysed by PCR. The (+) strand DNA was clearly detected and the signal was stronger in samples incubated with HSE, energy and Ran mix compared with samples incubated with energy and Ran mix only, suggesting that RTCs had not been degraded (Figure 3B).
Fig. 3. Time-lapse microscopy of RTC nuclear import in permeabilized HeLa cells. (A) Permeabilized cells were placed on a heated plate (37°C) and incubated in the presence of 0.5 mg/ml HeLa HSE, 103 labelled RTCs/cell, 1× energy mix and Ran mix. Images were acquired every 30 s using a Zeiss Axiovert S100 fluorescent microscope over a 25 min period, and images were processed using the Meta Imaging series 4.5 (Universal Imaging Corp.). Arrows point to fluorescent nucleoli; scale bar, 20 µm. (B) PCR analysis of viral DNA in HeLa cells after the nuclear import assay with purified RTCs. Following large-scale nuclear import assays, Hirt DNA was extracted (Hirt, 1967) and analysed by PCR with primers specific for the (+) strand DNA, a late reverse transcription product. 1, cells incubated in the presence of HSE, Ran mix and energy mix; 2, cells incubated in the absence of HSE; 3, cells incubated with the fraction from uninfected cells; 4, cells infected with the HIV-1 vector; 5, uninfected cells.
Imp7 and the imp7–impβ heterodimer mediate nuclear import of RTCs
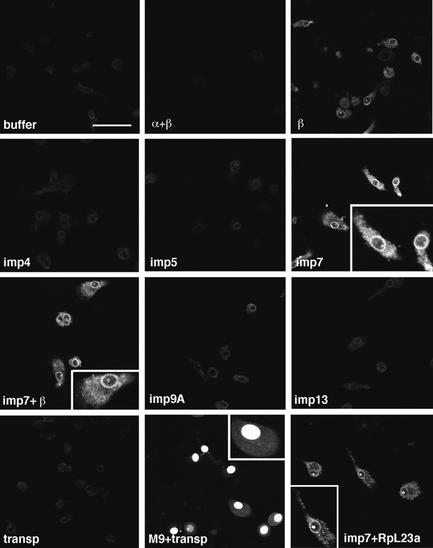
Addition of cell extracts to permeabilized cells stimulated nuclear import of RTCs, suggesting that they contained active factors. A growing number of impβ-related nuclear import receptors is emerging, each one displaying import activity for one or more cell proteins. Some import receptors can also function as heterodimers, e.g. impβ and imp7 form a heterodimer that mediates nuclear import of histone H1 (Görlich et al., 1997; Jäkel et al., 1999). It is therefore conceivable that one or more of these known factors could mediate HIV-1 RTC nuclear import. We tested this hypothesis by performing the nuclear import assay on primary macrophages in the presence of labelled RTCs, energy mix, Ran mix and all known nuclear import receptors. Labelled fusion proteins bearing the M9 nuclear import domain, recognized by transportin (Siomi et al., 1997), and the ribosomal protein (Rp) L23a, recognized by imp7 among other importins (Jäkel and Görlich, 1998), were used as positive controls in the assay. Remarkably, imp7, alone or in combination with impβ, was the most active receptor in stimulating RTC nuclear import (Figure 4). As expected, rpL23a was imported into the nucleoli efficiently in the presence of imp7, and the M9-bearing protein was imported into the nuclei efficiently in the presence of transportin. Similar results were obtained in HeLa cells (not shown).
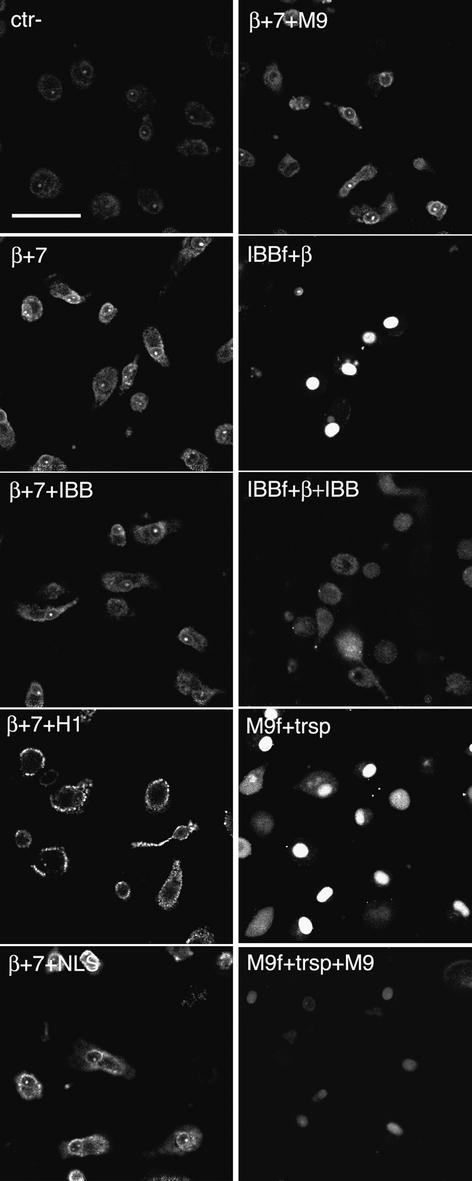
Fig. 4. Imp7 and the impβ–imp7 heterodimer stimulate nuclear import of RTCs. Primary human macrophages were permeabilized with digitonin and incubated at 37°C for 15 min in the presence of 103 labelled RTCs/cell, 1× energy mix, 1× Ran mix and the following import receptors: 0.5 µM each impα + impβ (α+β), 0.5 µM impβ (β), 0.5 µM imp4, imp5, imp7, imp9A, imp13, transportin (transp) and 0.5 µM imp7 + 0.25 µM impβ (imp7+β). MBP fused to the M9 peptide (M9) and the ribosomal protein L23a (RpL23a) were used as positive controls at 1 µM each + 1 µM specific import receptor. Cells were fixed and analysed by confocal microscopy. Scale bar, 30 µm.
To elucidate the specific role of imp7 and impβ in RTC nuclear import, we carried out import assays in primary macrophages in the presence of the two importins and a panel of specific competitors (Figure 5). RTC nuclear accumulation was unaffected by competition with a ‘classical’ NLS peptide at a concentration sufficient to inhibit NLS-dependent nuclear import completely (see Supplementary figure 2 available at The EMBO Journal Online) or by the addition of similar amounts of M9 domain, that specifically inhibits transportin-mediated nuclear import. Competition with the impβ-binding domain (IBB) from impα, which specifically binds to impβ, slightly reduced the accumulation of RTCs at the nuclear envelope, whereas addition of histone H1, which binds to the imp7–impβ heterodimer, abolished RTC nuclear import. These results indicate that imp7 is the major determinant and that impβ has a more limited activity in RTC nuclear import.
Fig. 5. Histone H1 inhibits RTC nuclear import in primary macrophages. Primary human macrophages were permeabilized with digitonin and incubated at 37°C for 15 min with 103 labelled RTCs/cell, 1× energy mix, 0.75× Ran mix, 0.75 µM each impβ and imp7 (β+7) or without importins (Ctr–) and 1.5 µM of the following recombinant factors as indicated: MBP–IBB (IBB), MBP–M9 (M9), histone H1 (H1) and BSA–NLS (NLS). Control samples were incubated in the same conditions with 0.5 µM fluorescently labelled GST–IBB (IBBf) + 1 µM impβ or labelled MBP–M9 (M9f) + 1 µM transportin with or without their specific competitors (IBB and M9, respectively). Cells were fixed and analysed by confocal microscopy. Scale bar, 25 µm.
The Ran system is required for imp7-mediated nuclear import of RTCs
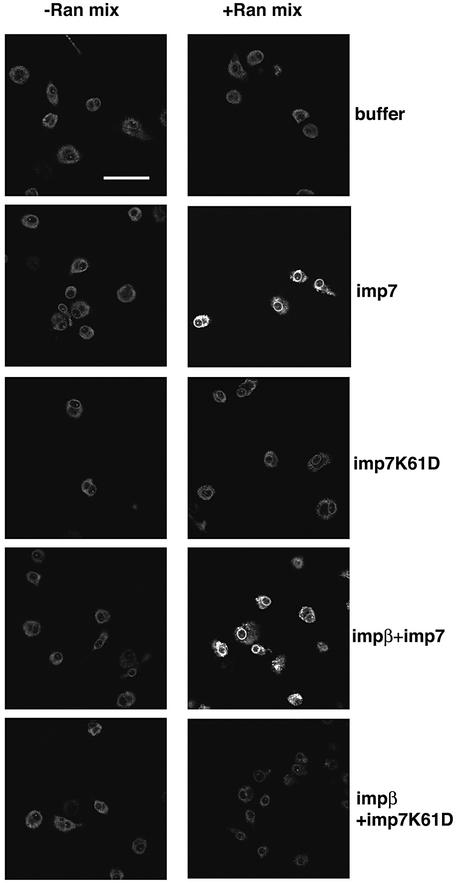
The role of imp7, impβ and the Ran system in the nuclear import of RTCs was examined further. Nuclear import assays of RTCs were performed with or without the components of the Ran system and different combinations of imp7, impβ and imp7 K61D, a mutant with reduced affinity for Ran (Jäkel et al., 1999). Imp7 alone or in combination with impβ did not induce RTC nuclear import significantly in the absence of Ran mix, but significant import was observed upon addition of this mix (Figure 6). The mutant imp7 K61D did not induce RTC nuclear import even in the presence of Ran mix. Interestingly, the mutant imp7 inhibited RTC import when mixed with impβ (Figure 6). Thus, impβ alone was insufficient to sustain significant RTC nuclear import, and the presence of a functional imp7 was necessary.
Fig. 6. Effect of the Ran system and the mutant imp7 K61D on nuclear import of RTCs. Primary human macrophages were treated as in Figure 3 and incubated at 37°C for 15 min with 103 labelled RTCs/cell, 1 µM import receptors with or without energy and Ran mixes. The mutant imp7 K61D is unable to bind Ran. Samples were fixed and analysed by confocal microscopy. Scale bar, 25 µm.
Interestingly, we have found that imp7 and the imp7–impβ heterodimer together with the components of the Ran system were also able to stimulate nuclear import of purified IN, a protein that remains bound to the viral nucleic acids and is part of the RTC (see Supplementary data).
Imp7 depletion inhibits HIV-1 infection
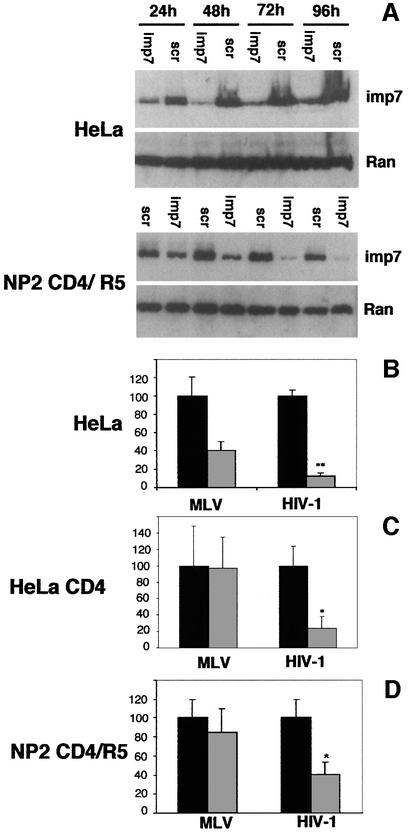
We tested if imp7 could be depleted in cells using siRNA (Elbashir et al., 2001). Primary HIV-1 isolates use predominantly the CCR5 chemokine co-receptor for infection (Deng et al., 1996); thus we tested siRNA in NP2 cells stably expressing CD4 and CCR5, which can be infected by primary HIV-1 isolates (Soda et al., 1999). We also tested siRNA in normal HeLa cells and HeLa cells stably expressing CD4 that can be infected by HIV-1 vectors. The system was optimized until imp7 was reduced to ∼10–15% of endogenous levels. Below that level, toxic effects appeared in treated cells. Briefly, HeLa and HeLa CD4 cells were transfected once and NP2 cells were transfected twice, at a 24 h interval, with an siRNA homologous to nucleotides 1392–1414 of human imp7 mRNA. Levels of imp7 were examined 24–96 h post-transfection by western blot on total cell lysates (Figure 7A). Depletion of imp7 was detected 48 h post-transfection in HeLa cells and 72 h after the first transfection in NP2 cells, while Ran levels remained unaffected, and so did imp7 levels when cells were transfected with a scramble siRNA, demonstrating the specificity of siRNA (Figure 7A). HeLa cells were more susceptible to imp7 siRNA depletion, presumably because they were transfected with higher efficiency, and grew slightly more slowly than control cells transfected with a scramble siRNA (not shown).
Fig. 7. Imp7 depletion inhibits HIV-1 infection. (A) Western blot showing specific depletion of imp7 by siRNA in HeLa cells and NP2 cells expressing CD4 and CCR5. Cells were transfected with 60 pmol siRNA duplex and analysed by western blot 24–96 h post-transfection. Imp7, anti-importin 7 siRNA; scr, scramble siRNA. Levels of Ran are shown as loading controls. (B) HeLa cells transfected with imp7 siRNA (grey bars) or with a scramble siRNA (black bars) were infected at an m.o.i. of 0.01 with HIV-1 or MLV vectors pseudotyped with VSV-G. (C) HeLa CD4 cells transfected with imp7 siRNA (grey bars) or scramble siRNA (black bars) were infected with the HIV-1 vector harbouring the gp120 envelope or an MLV vector pseudotyped with VSV-G. (D) NP2 cells, similarly transfected with imp7 siRNA (grey bars) or scramble siRNA (black bars), were infected at the same m.o.i. with SF-162 HIV-1 primary isolate or with amphotropic MLV. Infected cells were counted 48 h post-infection. Values are expressed as mean number of infected cells ± SD. HIV titres in imp7-depleted cells were reduced compared with both scramble siRNA-treated cells and MLV-infected cells (paired Student t-test, P < 0.004).
We next examined if depletion of imp7 had an effect on HIV-1 infection. HeLa cells were infected 48 h post-siRNA transfection with a VSV-G pseudotyped HIV-1 vector expressing GFP, and positive cells were scored 2 days after infection. HeLa CD4 cells were similarly infected with the same HIV-1 vector harbouring the gp120 envelope glycoprotein. NP2 cells were infected 48 h after the second siRNA transfection with a CCR5-tropic primary HIV-1 isolate and infected cells were scored 2 days later by in situ immunocytochemistry with an anti p24Gag monoclonal antibody. The number of HeLa and HeLa CD4 cells infected with the HIV-1 vector was reduced 5- to 10-fold in cultures treated with imp7 siRNA compared with those transfected with a scramble siRNA or infected in parallel with MLV (Figure 7B). Similar results were obtained in NP2 cells infected with a primary HIV-1 isolate. HIV-1 restriction in siRNA-treated cells was dose dependent; it was maximal at an m.o.i. of 0.01 and disappeared at an m.o.i. ≥1. Some decrease in the number of MLV-infected cells was found in HeLa cell cultures transfected with imp7 siRNA, presumably because of their slower growth. Indeed, MLV can only infect cells that undergo mitosis (Roe et al., 1993). It is also possible that imp7 is required for steps other than nuclear import in the MLV life cycle. In any case, the efficiency of infection of both the HIV-1 vector and the primary HIV-1 isolate was always significantly lower than that of MLV in these cells (paired Student-t test, P < 0.004). To test for the specificity of the imp7 activity on HIV-1 infection, we have knocked-down a second importin, imp9, which is also involved in nuclear import of ribosomal proteins (Jäckel et al., 2002). HeLa cells were transfected once with an siRNA homologous to nucleotides 527–547 of human imp9 mRNA. Depletion of imp9 reached a peak 72 h post-transfection and was as efficient as for imp7 (not shown). HIV-1 vector infection was modestly reduced (from 2 to 38% reduction in three independent experiments) in imp9 knock-down cells compared with controls, but the difference was not statistically significant (not shown). These results indicate that downregulation of imp7 inhibits HIV-1 infection in vivo, although a modest effect due to cell toxicity cannot be ruled out.
Discussion
The nuclear import assay as a tool to study nuclear import of HIV-1 RTCs
The nuclear import assay has been employed to study import of cellular proteins, large macromolecules and, more recently, influenza virus, hepatitis B virus and adenoviruses (O’Neill et al., 1995; Kann et al., 1999; Sapphire et al., 2000; Trotman et al., 2001). Here we have used this assay to study nuclear import of HIV-1. The combined use of purified intracellular RTCs and primary human macrophages provided the most physiologically relevant model to study nuclear import of HIV-1. Purified RTCs were competent to reverse-transcribe their genome in vitro and were visualized specifically by confocal microscopy. The degree of purification was high and allowed the use of YOYO-1-labelled RTCs as substrate in the nuclear import assay. For some aspects, RTCs behaved like molecules bearing a ‘classic’ NLS: their nuclear import was stimulated by the addition of Ran mix and energy mix and required passage through the NPC. However, differences between RTCs and classic NLS-bearing molecules were also apparent. In the case of RTCs, import intermediates accumulated very prominently at NPCs, suggesting that passage of these large complexes through NPCs is slower or less efficient than passage of smaller substrates. This would also explain why nuclear import of NLS-bearing molecules is still efficient at 25°C whereas RTC import is not.
Several lines of evidence indicate that fluorescence accumulation at NPCs and nucleoli of permeabilized cells was not due to diffusion of YOYO-1 or HIV DNA degradation, but rather recapitulated physiological events. First, import was inefficient at 0°C and required energy, consistent with previous reports showing that HIV-1 nuclear import in infected cells requires ATP (Bukrinsky et al., 1992). Secondly, labelled RTCs co-localized with a mutant impβ that binds irreversibly to the NPC and was inhibited by excess import receptors. Thirdly, HIV DNA could be detected by PCR in permeabilized cells following the nuclear import assay. Fourthly, time-lapse microscopy indicated that RTCs accumulate at the nuclear envelope first and then in the nucleoli, reaching a peak after 20 min. Such selective labelling of intracellular structures argues against the passive diffusion of YOYO-1, a dye able to bind to both DNA and RNA.
Imp7 is a mediator of HIV-1 nuclear import
Our results indicate that imp7 is involved in HIV-1 RTC nuclear import. Addition of imp7 stimulated RTC nuclear import in both HeLa cells and primary macrophages in a Ran- and energy-dependent way. Addition of specific competitors to the nuclear import assay performed in the presence of both imp7 and impβ as well as experiments performed in the presence of the mutant imp7 K61D showed that imp7 is the major player while impβ seems to have a weaker activity. Consistently, depletion of imp7 from HeLa, HeLa CD4 and NP2 CCR5/CD4 cells resulted in restriction to HIV-1 infection. This effect was observed using an HIV-1 vector with the wild-type gp120 or the VSV-G envelope or by using a primary HIV-1 isolate. Depletion of imp9 as a control had only a modest effect, further supporting the effect of imp7 depletion on infection by HIV-1. Restriction to HIV-1 infection in imp7-depleted cells was observed only at a low m.o.i. The residual 10–15% of imp7 in treated cells may explain such a phenotype. Alternatively, additional cellular and viral factors may be involved in RTC nuclear import.
Imp7 is mildly hydrophobic, belongs to the impβ superfamily and is involved in nuclear import and chaperoning of basic, nucleic acid-binding proteins such as ribosomal proteins and histone H1 (Jäkel and Görlich, 1998; Jäkel et al., 1999, 2002). Thus, imp7 would be well suited to interact with basic proteins present in the RTC. Indeed, we have found that one of such proteins, IN, binds to and is imported by imp7. Interestingly, the impβ–imp7 heterodimer has also been implicated in nuclear import of adenovirus type 2 DNA (Trotman et al., 2001), suggesting that imp7 could have a more general role in mediating nuclear import of viral nucleic acids.
A model for HIV-1 nuclear import
Based on the findings reported here, we propose that imp7 is likely to interact with basic proteins bound to the viral nucleic acids, such as IN. Once associated to the RTC, imp7 may facilitate passage of the large, hydrophilic DNA molecule through the NPC channel that acts as a mildly hydrophobic selective phase (Ribbeck and Görlich, 2002). We predict that the large intracellular RTC has to undergo substantial structural rearrangements to cross the NPC channel and that other cellular factors may be required for this process. In this regard, it is tempting to draw a parallel with the export of mRNA, also organized in a large ribonucleoprotein complex (mRNP) (Izaurralde, 2002). Export of such mRNPs through the NPC requires several export factors and dynamic rearrangements of RNA secondary structure and RNA–protein interactions (Cullen, 2002; Izaurralde, 2002). Interestingly, Dbp5, a DEAD-box ATP-dependent RNA helicase, recently has been implicated in facilitating the late steps of mRNP export (reviewed by Izaurralde, 2002). Perhaps a similar mechanism exists for HIV-1 RTC nuclear import and might explain the different requirements of this process compared with nuclear import of simple, one-protein substrates. The system described here will help to elucidate further the complex mechanisms involved in HIV-1 nuclear import and may be used to screen for compounds that interfere with this process.
Materials and methods
Cell culture
HeLa and 293T cells were grown in Dulbecco’s modified Eagle’s medium (Gibco Laboratories, Paisley, UK) supplemented with 10% fetal calf serum (Helena Bioscience, Newcastle, UK) and 2 mM glutamine at 37°C in a humidified atmosphere containing 5% CO2. Human peripheral blood mononuclear cells were isolated from buffy coats (50 ml) by Ficoll gradients. Monocytes were selected by magnetic sorting in the presence of CD14 microbeads (6 µl/107 cells) according to the manufacturer’s instructions (Moltenyi Biotech, Bergisch Gladbach, Germany). Monocytes were washed and plated at the desired density onto 5 mm diameter poly-l-lysine-coated glass-bottomed microwell dishes (Matek Corp., Ashland, MA) and grown in RPMI medium (Gibco) plus 10% heat-inactivated autologous serum for 1 week, until spontaneous differentiation into mature macrophages occurred (Neil et al. 2001). For production of viral stocks, see Supplementary data.
RTCs purification and labelling
Hypotonic cytoplasmic extracts were prepared from infected and uninfected cells as described previously (Fassati and Goff, 1999, 2001). Cytoplasmic extracts were loaded on a 5–20% continuous sucrose gradient in 50 mM Na phosphate buffer pH 7.4 containing 1 mM dithiothreitol (DTT), 20 µg/ml aprotinin and 20 µg/ml leupeptin, and centrifuged at 23 000 r.p.m. for 1 h at 4°C in a Sorvall AH-650 rotor. Fractions (0.4 ml each) were collected by puncturing the bottom of the tube, and the position of the viral DNA was monitored by PCR with primers specific for the strong stop or (+) strand DNA as previously described (Fassati and Goff, 2001). The fraction density was measured by weighing 100 µl, and the S value was calculated (McEwen, 1967). Calibration of the system was performed by independently running 32S-labelled poliovirus, intact MLV and naked viral DNA through identical sucrose gradients. Fractions of the sedimentation velocity gradient containing the peak of the viral DNA (sedimenting at ∼350S) were diluted in a 20% sucrose solution in 50 mM Na phosphate buffer pH 7.4 containing 1 mM DTT, 20 µg/ml aprotinin and 20 µg/ml leupeptin, and mixed with a cold 70% sucrose solution in D2O to obtain a continuous 20–70% gradient using a gradient maker (Biocomp, Frederikton, NB, Canada). Gradients were centrifuged at 35 000 r.p.m. at 4°C for 18 h in a Sorvall AH-650 rotor and divided into 12 fractions by puncturing the bottom of the tube. The density was calculated by weighing 100 µl of each fraction, and the position of the viral DNA was monitored by PCR as before. The 1.34 g/ml density fraction containing the peak of the viral DNA was diluted 1:1 in import buffer (20 mM HEPES pH 7.3, 110 mM potassium acetate, 5 mM magnesium acetate, 0.5 mM EGTA, 250 mM sucrose) and incubated in the dark for 1 h at room temperature in the presence of YOYO-1 (1:10 000) (Molecular Probes, Eugene, OR). Samples were loaded onto 1 ml of cellulose float-A-lyzer, 300 kDa cut-off (Spectrum Laboratories, Cheshire, UK), and dialysed against large volumes of import buffer at 4°C.
Preparation of high-speed cytosolic extracts (HSEs)
The pellet from ∼1010 HeLa cells was washed once in phosphate-buffered saline (PBS) and resuspended in 5 vols of hypotonic buffer on ice. The supenatant was centrifuged at 3300 g for 10 min at 4°C, resuspended in 5 vols of hypotonic buffer and incubated on ice for 10 min with gentle stirring. Digitonin was added to a final concentration of 40 µg/ml while stirring, and samples were centrifuged at 3300 g for 15 min at 4°C. The supernatant was centrifuged at 7500 g for 20 min at 4°C and then ultracentrifuged at 100 000 g for 4.5 h at 4°C. The supernatant was passed through a glass wool filter and through a 0.45 µm filter. Samples were then de-salted, and buffer exchanged to import buffer using HiTrap columns (Amersham).
PCR
PCR was performed in a final volume of 50 µl containing 1× PCR buffer, 100 µM each dNTP, 2.5 mM MgCl2, 5 U of Taq polymerase (Perkin Elmer) and 30 pmol of each primer. Primer sequences for pHR′ and pHRSINcPPT vector plasmids were as follows: strong stop forward primer, 5′-GGCTAACTAGGGAACCCACTG-3′; reverse complementary primer, 5′-CTGCTAGAGATTTTCCACACTGAC-3′; positive strand forward primer, 5′-AGGGCTAATTCACTCCCAACGAAG-3′; reverse complementary primer, 5′-GCCGTGCGCGCTTCAGCAAGC-3′. Cycle parameters were: 94°C for 3 min for the first cycle, and 94°C for 1 min, 55°C for 30 s, 68°C for 1 min for 27–35 cycles. RTC copy number was quantitated by limiting dilution PCR in the same conditions as described above using 2-fold serial dilutions of the RTC template and the pHR′SINcPPT plasmid. After 25–30 cycles, PCR bands were quantified by the Kodak digital science 1D image analysis software, and the linear region of the reaction was then selected to calculate RTC copy number, assuming that 1 fg of a 7 kb plasmid DNA corresponds to 1.3 × 102 molecules. Approximately 108 viral DNA copies/ml were obtained following the purification protocol.
Endogenous RT assay
Endogenous RT reactions were carried out in 30 µl of buffer (20 mM Tris–HCl pH 8.1, 15 mM NaCl, 6 mM MgCl2, 1 mM DTT, 2 mM each dNTP). A 5 µl aliquot from the density equilibrium fractions was added to the buffer and incubated for 4–6 h at 37°C. The products of reverse transcription were detected by PCR using 5 µl of the endogenous reaction as template with primers specific for the (+) strand DNA.
Immunochemistry
Rabbit antiserum to HIV-1 Vpr (amino acids 1–46) from Dr Jeffrey Kopp was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIH; antiserum R5 and R4 to HIV-1 IN peptide 1 and peptide 2, respectively (from Dr Ranjbar), were obtained through the Centralised Facility for AIDS Reagents, UK. Aliquots of the density fractions (10 µl) were diluted in 2 vols of PBS containing 8 µg/ml polybrene and incubated on tissue culture plastic slides for 1 h at 37°C in a tissue culture incubator. Samples were fixed with 4% paraformaldehyde in PBS for 20 min at room temperature and incubated with anti-Vpr (1:250) or anti-IN (1:300) antibodies for 1 h at room temperature, followed by incubation with tetramethylrhodamine isothiocyanate (TRITC)-conjugated secondary antibody (1:300) (Dako, Cambridgeshire, UK) and YOYO-1 (Molecular Probes, Eugene, OR), diluted 1:10 000 in PBS, for 1 h at room temperature. Samples were mounted with Vectashield mounting medium and analysed on a Bio-Rad MRC 1024 confocal microscope equipped with a krypton–argon laser. Images were acquired sequentially and merged using Lasersharp Confocal Assistant software (Bio-Rad).
Western blot
Antibody C-20 against Ran was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-imp7 and anti-imp9 antibodies were raised against a C-terminal peptide in rabbits. Antibodies against impα, impβ and transportin have been described previously (Jäkel and Gorlich, 1998). Anti-rabbit and anti-goat IgG horseradish peroxidase (HRP)-conjugated antibodies were purchased from Jackson Laboratories (Bar Harbor, MN) and from Sigma, respectively. After SDS–PAGE, the proteins were transferred to a PVDF membrane (Bio-Rad, Hercules, CA) and probed with the primary antibodies. HRP-conjugated secondary antibodies were used diluted 1:3000 in 10% non-fat milk. Chemiluminescence (ECL, Amersham) was used to develop the blots as described by the manufacturer. Autoradiography films were exposed for different periods of time to ensure linearity of the signal.
Recombinant protein expression
Expression of nuclear transport factors has been described previously (Görlich et al., 1996; Izaurralde et al., 1997; Kutay et al., 1997; Jakel and Görlich, 1998; Mingot et al., 2001; Ribbeck and Görlich, 2002). Recombinant HIV-1 IN (from Dr Pemberton) was obtained through the NIBSC Centralised Facility for AIDS Reagents, UK. The His tag was cleaved by incubating 1.8 mg of IN with 0.54 U of factor Xa (Sigma) for 16 h on ice in 50 mM Tris–HCl pH 7.8, 0.5 M NaCl. Cleavage of the His tag was confirmed by western blot using anti-His antibody (Sigma) and anti-IN antibody. Purified IN was labelled with stoichiometric amounts of Alexa 594 (Molecular Probes).
Nuclear import assay
Cells (∼50% confluent in 5 mm diameter microdishes) were washed in import buffer and permeabilized for 5 min on ice in import buffer containing 40 µg/ml digitonin (Fluka). After one wash in import buffer, cells were incubated with a mixture of 0.3 µM each IBB, BIB and M3 domains fused to the maltose-binding protein (MBP) for 5 min on ice to adsorb endogenous transport receptors, and washed three times in import buffer. IN (0.5 µM final) was pre-incubated on ice for 45 min in the presence of the transport receptors. Cells were incubated for 15 min at 37°C (25°C for IN) in a tissue culture incubator in the presence of ∼103 labelled RTCs/cell, 0.5 mg/ml HSE or recombinant import factors (concentration indicated in figure legends), 2 µM Ran (GDP form), 0.2 µM NTF2, 0.2 µM RanBP1, 0.2 µM RanGAP1 (Ran mix) and an energy-regenerating system (1 mM ATP, 1 mM GTP, 2 mM creatine phosphate, 40 U/ml creatine phosphokinase). Samples were washed three times in import buffer, fixed on ice for 5 min with 2% paraformaldehyde in import buffer and analysed directly by confocal microscopy.
SiRNA
Twenty-one-nucleotide duplexes with symmetric two-nucleotide 3′ (2-deoxy) thymidine overhangs, corresponding to imp7 mRNA nucleotides 1392–1414 and to imp9 mRNA nucleotides 527–547, and a scramble siRNA were synthesized by Dharmacon Research (Lafayette, CO). RNA sequences: imp7 sense, 5′-GAUGGAGCCCUGCAUAUGAdTdT; imp7 complement, 5′-dTdTCUACCUCGGGACGUAUACU-3′; imp9 sense, 5′-GUUACAGACACACAGAUGCdTdT; imp9 complement, 5′-dTdTC AAUGUCUGUGUGUCUACG-3′; scramble sense, 5′-CAGUCGCGUU UGCGACUGGdTdT; scramble complement, 5′-dTdTGUCAGCGC AAACGCUGACC. HeLa cells were plated at 2 × 104 cells/well in 24-well plates and transfected the next day with 60 pmol siRNA duplex and 3 µl/well oligofectamine (Invitrogen, Paisley, UK) following the manufacturer’s instructions. Cells were infected 48 h post-transfection with serial dilutions of a HIV vector (VSV-G pseudotyped or harbouring the gp120 envelope) or an MLV (VSV-G pseudotyped) vector expressing GFP. GFP-positive cells were counted 48 h post-transfection. NP2 cells were plated at 4 × 104 cells/well in 24-well plates and were transfected twice at a 24 h interval. At 24 h after the second transfection, cells were split 1:2 (imp7 siRNA) or 1:6 (scramble siRNA) and infected 24 h later with serial dilutions of the primary HIV-1 isolate SF-162 or amphotropic MLV. At the time of infection, cell density was the same for samples transfected with scramble siRNA and imp7 siRNA. Infected cells were identified by in situ immunocytochemistry using anti-p24 (HIV-1) or anti-p30 (MLV) monoclonal antibodies as previously described (Clapham et al. 1992).
Supplemenary data
Supplementary data are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Robin Weiss and Mary Collins for helpful discussions throughout the course of this work and for critical review of the manuscript, all members of the Görlich lab for their assistance, Didier Trono and Adrian Thrasher for the HIV-1 vectors, and Stuart Neil for the SF-162 HIV-1 primary isolate. This work was supported by a Wellcome Trust Career Development Fellowship (to A.F.) and by the Alfried Krupp von Bohlen und Halbach-Stiftung (to D.G.).
References
- Adam S.A. and Gerace,L. (1991) Cytosolic proteins that specifically bind nuclear location signals are receptors for nuclear import. Cell, 66, 837–847. [DOI] [PubMed] [Google Scholar]
- Adam S.A., Sterne Marr R. and Gerace,L. (1990) Nuclear import in permeabilized mammalian cells requires soluble cytoplasmic factors. J. Cell Biol., 111, 807–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff F.R., Scheffzek,K. and Postingl,H. (2002) How Ran is regulated. In Weis,K. (ed.), Nuclear Transport. Springer-Verlag, Berlin, Germany, pp. 49–66. [Google Scholar]
- Bukrinsky M.I., Sharova,N., Dempsey,M.P., Stanwick,T.L., Bukrinkaya,A.G., Haggerty,S. and Stevenson,M. (1992) Active nuclear import of human immunodeficiency virus type 1 preintegration complexes. Proc. Natl Acad. Sci. USA, 89, 6580–6584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukrinsky M.I., Sharova,N., McDonald,T.L., Pushkarskaya,T., Tarpley,W.G. and Stevenson M. (1993) Association of integrase, matrix and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl Acad. Sci. USA, 90, 6125–6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham P.R., McKnight,A. and Weiss,R.A. (1992) Human immunodeficiency virus type-2 infection and fusion of CD4-negative human cell lines: induction and enhancement by soluble CD4. J. Virol., 66, 3531–3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen O.J. and Fauci,A.S. (2001) Pathogenesis and medical aspects of HIV-1 infection. In Knipe,D.M. and Howley,P.M. (eds), Fields Virology. Lippincot Williams & Wilkins, Philadelphia, PA, pp. 2043–2094. [Google Scholar]
- Cullen B.R. (2002) Using retroviruses to study the nuclear export of mRNA. In Weis,K. (ed.), Nuclear Transport. Springer-Verlag, Berlin, Germany, pp. 151–168. [DOI] [PubMed] [Google Scholar]
- Demiason C., Parsley,K., Brouns,G., Scherr,M., Battmer,K., Kinnon,C., Grez,M. and Thrasher,A.J. (2002) High-level transduction and gene expression in hematopoietic repopulating cells using a HIV-1 based lentiviral vector containing an internal spleen focus forming virus promoter. Hum. Gene Ther., 13, 803–813. [DOI] [PubMed] [Google Scholar]
- Deng H. et al. (1996) Identification of a major co-receptor for primary isolates of HIV-1. Nature, 381, 661–666. [DOI] [PubMed] [Google Scholar]
- de Noronha C.M., Sherman,M.P., Lin,H.W., Cavrois,M.V., Moir,R.D., Goldman,R.D. and Greene,W.C. (2001) Dynamic disruptions in nuclear envelope architecture and integrity induced by HIV-1 Vpr. Science, 294, 1105–1108. [DOI] [PubMed] [Google Scholar]
- Dvorin J.D., Bell,P., Maul,G.G., Yamashita,M., Emerman,M. and Malim,M.H. (2002) Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J. Virol., 76, 12087–12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Harborth,J., Lendeckel,W., Yalcin,A., Weber,K. and Tuschl,T. (2001) Duplexes of 21-nucleotide RNAs mediate interference in cultured mammalian cells. Nature, 411, 494–498. [DOI] [PubMed] [Google Scholar]
- Engelman A. and Craigie,R. (1992) Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol., 66, 6361–6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnet C.M. and Haseltine,W.A. (1991) Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J. Virol., 65, 1910–1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassati A. and Goff,S.P. (1999) Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J. Virol., 73, 8919–8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fassati A. and Goff,S.P. (2001) Characterization of intracellular reverse transcription complexes of human immunodeficiency type-1. J. Virol., 75, 3626–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed E.O. and Martin,M.A. (2001) HIVs and their replication. In Knipe,D.M. and Howley,P.M. (eds). Fields Virology. Lippincot Williams & Wilkins, Philadelphia, PA, pp. 1971–2041. [Google Scholar]
- Gallay P., Hope,T., Chin,D., Trono,D. (1997) HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl Acad. Sci. USA, 94, 9825–9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff S.P. (2001) Intracellular trafficking of retroviral genomes during the early phase of infection: viral exploitation of cellular pathways. J. Gene Med., 3, 517–528. [DOI] [PubMed] [Google Scholar]
- Görlich D. and Kutay,U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell. Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Görlich D., Prehn,S., Laskey, R A. and Hartmann,E. (1994) Isolation of a protein that is essential for the first step of nuclear protein import. Cell, 79, 767–778. [DOI] [PubMed] [Google Scholar]
- Görlich D., Henklein,P., Laskey,R.A. and Hartmann,E. (1996) A 41 amino acid motif in importin α confers binding to importin β and hence transit into the nucleus. EMBO J., 15, 1810–1817. [PMC free article] [PubMed] [Google Scholar]
- Görlich D., Dabrowski,M., Bischoff,F.R., Kutay,U., Bork,P., Hartmann,E., Prehn,S. and Izaurralde,E. (1997) A novel class of RanGTP binding proteins. J. Cell Biol., 138, 65–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb M.S., Schroff R., Schanker H.M., Weisman,J.D., Fan,P.T, Wolf,R.A. and Saxon A. (1981) Pneumocystis carinii pneumonia and mucosal candidiasis in previously healthy homosexual men: evidence of a new acquired cellular immunodeficiency. N. Engl. J. Med., 305, 1425–1431. [DOI] [PubMed] [Google Scholar]
- Henderson B.R. and Percipalle,P. (1997) Interactions between Rev and nuclear import and export factors: the rev nuclear localisation signal mediates specific binding to human importin B. J. Mol. Biol., 274, 693–707. [DOI] [PubMed] [Google Scholar]
- Hirt B. (1967) Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol., 26, 365–369. [DOI] [PubMed] [Google Scholar]
- Izaurralde E. (2002) Nuclear export of messenger RNA. In Weis,K. (ed.) Nuclear Transport. Springer-Verlag, Berlin, Germany, pp. 133–150. [DOI] [PubMed] [Google Scholar]
- Izaurralde E., Kutay,U., von Kobbe,C., Mattaj,I.W. and Görlich,D. (1997) The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J., 16, 6535–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäkel S. and Görlich,D. (1998) Importin β, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J., 17, 4491–4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäkel S., Albig,W., Kutay,U., Bischoff,F.R., Schwamborn,K., Doenecke,D. and Görlich,D. (1999) The importin β/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J., 18, 2411–2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jäkel S., Mingot J-M., Schwarzmaier,P., Hartmann,E. and Görlich,D. (2002) Importins fulfil a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J., 21, 377–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalderon D., Roberts,B.L., Richardson,W.D. and Smith,A.E. (1984) A short amino acid sequence able to specify nuclear location. Cell, 39, 499–509. [DOI] [PubMed] [Google Scholar]
- Kann M., Sodeik,B., Vlachou,A., Gerlich,W.H. and Helenius,A. (1999) Phosphorylation dependent binding of hepatitis B virus core particles to the nuclear pore complex. J. Cell Biol., 145, 45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U., Izaurralde,E., Bischoff,F.R., Mattaj,I. and Görlich,D. (1997) Dominant-negative mutants of importinβ block multiple pathways of import and export through the nuclear pore complex. EMBO J., 16, 1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limon A., Nakajima,N., Lu,R., Ghory,H.Z. and Engelman,A. (2002) Wild-type levels of nuclear localization and human immunodeficiency virus type 1 replication in the absence of the central DNA flap. J. Virol., 76, 12078–12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malim M.H. and Cullen,B.R. (1991) HIV-1 structural gene expression requires the binding of multiple Rev monomers to the viral RRE: implications for HIV-1 latency. Cell, 65, 241–248. [DOI] [PubMed] [Google Scholar]
- McEwen C.R. (1967) Tables for estimating sedimentation through linear concentration gradients of sucrose solution. Anal. Biochem., 20, 114–149. [DOI] [PubMed] [Google Scholar]
- Miller M.D., Farnet,C.M. and Bushman,F.D. (1997) Human immunodeficiency virus type-1 preintegration complexes: studies of organization and composition. J. Virol., 71, 5382–5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingot J.-M., Kostka,S., Kraft,R., Hartmann,E. and Görlich,D. (2001) Importin 13: a novel mediator of nuclear import and export. EMBO J., 20, 3685–3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L., Blomer,U., Gallay,P., Ory,D., Mulligan,R., Gage,F.H., Verma,I.M. and Trono.D. (1996) In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science, 272, 263–267. [DOI] [PubMed] [Google Scholar]
- Neil S., Martin,F., Ikeda,Y. and Collins,M. (2001) Postentry restriction to human immunodeficiency virus-based vector transduction in human monocytes. J. Virol., 75, 5448–5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill R., Jaskunas,R., Blobel,G., Palese,P. and Moroianu,J. (1995) Nuclear import of influenza virus RNA can be mediated by viral nucleoprotein and transport factors required for protein import. J. Biol. Chem., 270, 22701–22704. [DOI] [PubMed] [Google Scholar]
- Rexach M. and Blobel,G. (1995) Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors and nucleoporins. Cell, 83, 683–692. [DOI] [PubMed] [Google Scholar]
- Ribbeck K. and Görlich,D. (2002) The permeability barrier of nuclear pore complexes appears to operate via hydrophobic exclusion. EMBO J., 21, 2664–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K. Lipowsky,G., Kent,H.M., Stewart,M. and Görlich,D. (1998) NTF2 mediates nuclear import of Ran. EMBO J., 17, 6587–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roe T., Reynolds,G., Yu,G. and Brown,P.O. (1993) Integration of murine leukemia virus DNA depends on mitosis. EMBO J., 12, 2099–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M.P., Aitchinson,J.D., Suprapto,A., Hjertaas,K., Zhao,Y. and Chait,B.T. (2000) The yeast nuclear pore complex: composition, architecture and transport mechanism. J. Cell Biol., 148, 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapphire A.C.S., Guan,T., Schirmer,E.C., Nemerow,G.R. and Gerace,L. (2000) Nuclear import of adenovirus DNA in vitro involves the nuclear protein import pathway and hsc70. J. Biol. Chem., 275, 4298–4304. [DOI] [PubMed] [Google Scholar]
- Siomi M.C., Eder,P.S., Kataoka,N., Wan,L., Liu,Q. and Dreyfuss,G. (1997) Transportin-mediated nuclear import of heterogeneous nuclear RNP proteins. J. Cell Biol., 138, 1181–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soda Y., Shimizu,N., Jinno,A., Liu,H.Y., Kanabe,K., Kitamura,T. and Hoshino,H. (1999) Establishment of a new system for determination of coreceptor usages of HIV based on the human glioma NP-2 cell line. Biochem. Biophys. Res. Commun., 258, 313–321. [DOI] [PubMed] [Google Scholar]
- Trotman L.C., Mosberger,N., Fornerod,M., Stidwill,R.P. and Greber,U.F. (2001) Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nature Cell Biol., 3, 1092–1100. [DOI] [PubMed] [Google Scholar]
- UNAIDS 2002) Report on the global HIV/AIDS epidemic. http://www.unaids.org/epidemic_update/report_july02/english/embargo.htm
- Zennou V., Petit,C., Guetard,U., Nerhbass,U., Montagnier,L. and Charneau,P. (2000) HIV-1 genome nuclear import is mediated by a central DNA flap. Cell, 101, 173–185. [DOI] [PubMed] [Google Scholar]
- Zufferey R., Nagy,D., Mandel,R.J., Naldini,L. and Trono,D. (1997) Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat. Biotechnol., 15, 871–875. [DOI] [PubMed] [Google Scholar]