Abstract
In this review, we discuss the structural and functional diversity of protein–protein interactions (PPIs) based primarily on protein families for which three-dimensional structural data are available. PPIs play diverse roles in biology and differ based on the composition, affinity and whether the association is permanent or transient. In vivo, the protomer’s localization, concentration and local environment can affect the interaction between protomers and are vital to control the composition and oligomeric state of protein complexes. Since a change in quaternary state is often coupled with biological function or activity, transient PPIs are important biological regulators. Structural characteristics of different types of PPIs are discussed and related to their physiological function, specificity and evolution.
Keywords: complexes/protein–protein interactions/protein structures/transient interactions
Types of protein–protein interactions
Homo- and hetero-oligomeric complexes
Protein–protein interactions (PPIs) occur between identical or non-identical chains (i.e. homo- or hetero-oligomers; Figure 1). Oligomers of identical or homologous protein units can be organized in an isologous or heterologous way (Monod et al., 1965) with structural symmetry (Goodsell and Olson, 2000). An isologous association involves the same surface on both monomers (e.g. Arc repressor and lysin; Figure 1A and C), related by a 2-fold symmetry axis. In contrast to an isologous association that can only further oligomerize using a different interface (e.g. form a dimer of dimers with three 2-fold axes of symmetry), heterologous assemblies use different interfaces that, without a closed (cyclic) symmetry, can lead to infinite aggregation.
Fig. 1. Examples of different types of protein–protein interactions as described in the text: (A) obligate homodimer, P22 Arc repressor; (B) obligate heterodimer, human cathepsin D that consists of a non-homologous light (red) and heavy (green) chain; (C) non-obligate homodimer, sperm lysin; (D) non-obligate heterodimer, RhoA (green) and RhoGAP (red) signalling complex; (E) non-obligate permanent heterodimer, thrombin (red) and rodniin inhibitor (green); (F) non-obligate transient heterotrimer, bovine G protein, i.e. the interaction between Gα (green) and Gβγ (red, orange) is transient.
Non-obligate and obligate complexes
As well as composition, two different types of complexes can be distinguished on the basis of whether a complex is obligate or non-obligate. In an obligate PPI, the protomers are not found as stable structures on their own in vivo. Such complexes are generally also functionally obligate; for example, the Arc repressor dimer (Figure 1A) is essential for DNA binding. Many of the hetero-oligomeric structures in the Protein Data Bank involve non-obligate interactions of protomers that exist independently, such as intracellular signalling complexes (e.g. RhoA–RhoGAP; Figure 1D) and antibody–antigen, receptor–ligand and enzyme–inhibitor (e.g. thrombin–rodniin; Figure 1E) complexes. The components of such protein–protein complexes are often initially not co-localized and thus need to be independently stable. However, some homo-oligomers, which by definition are co-localized, can also form non-obligate assemblies (e.g. sperm lysin; Figure 1C).
Transient and permanent complexes
PPIs can also be distinguished based on the lifetime of the complex. In contrast to a permanent interaction that is usually very stable and thus only exists in its complexed form, a transient interaction associates and dissociates in vivo. We distinguish weak transient interactions that feature a dynamic oligomeric equilibrium in solution, where the interaction is broken and formed continuously (e.g. lysin; Figure 1C), and strong transient associations that require a molecular trigger to shift the oligomeric equilibrium. For example, the heterotrimeric G protein (Figure 1F) dissociates into the Gα and Gβγ subunits upon guanosine triphosphate (GTP) binding, but forms a stable trimer with guanosine diphosphate (GDP) bound. Structurally or functionally obligate interactions are usually permanent, whereas non-obligate interactions may be transient or permanent.
It is important to note that many PPIs do not fall into distinct types. Rather, a continuum exists between non-obligate and obligate interactions, and the stability of all complexes very much depends on the physiological conditions and environment (see below). An interaction may be mainly transient in vivo but become permanent under certain cellular conditions. Folding data, as well as data on the dynamics of the assembly at different physiological conditions or environments, are often not available. However, the subcellular location of subunits and the function of the protein will often suggest the biologically relevant type of interaction; for example, interactions in intracellular signalling are expected to be transient, since their function requires a ready association and dissociation.
Control of oligomeric state
Ultimately, all interactions and complexes are driven by the concentration of the components and the free energy of the complex, relative to alternative states. PPIs can be controlled either by altering the local concentration of the protein components or by influencing the binding affinity, determined by the physicochemical and geometrical interface properties (Figure 2A). We identify three types of control: (i) Encounter. The association of two proteins or protomers relies on an encounter of the interacting surfaces, requiring co-localization in time and space. Such encounters may occur upon co-expression or localization within a compartment (e.g. intracellular signalling complexes) or between components that usually reside in different compartments. For encounters of these proteins from different locations, (directed) diffusion or (vascular) transport is essential. (ii) Local concentration. Control mechanisms that alter the effective local concentration include gene-expression or secretion levels, protein degradation, temporary storage, the local molecular environment and diffusion or viscosity. Clearly, the anchoring of (one or both) proteins in a membrane (e.g. transmembrane protein oligomerization) or other structural complex, as well as localization by adjacent domains in multidomain proteins, can help to increase the local concentration. (iii) Local physicochemical environment. The mutual affinity of components of a complex can be altered by the presence of an effector molecule (e.g. ATP, Ca2+) or a change in physiological conditions. Typically, the concentration of ions, chemicals or proteins, changes in pH and temperature, or covalent modifications such as phosphorylation (addition of PO4–) all affect binding.
Fig. 2. Illustration of (A) the control of protein oligomerization and (B) the relation between different types of protein–protein interactions (PPIs), their binding affinity and the localization of their protomers. The triggers that control the transient oligomerization are given in red in (B). *Large conformational changes are usually associated with these transient PPIs.
Protomers involved in strong obligate interactions are often expressed simultaneously and are thus co-localized upon synthesis (Figure 2B). This is obviously true for homo-multimers but may also occur in hetero-complexes; for example, the genes that encode the non-identical subunits of cathepsin D (Figure 1B) have the same promotor (Faust et al., 1985). Most non-obligate interactions perform a regulatory role, and the control mechanisms are vital to their biological function. The control mechanism of the PPI is often related to the affinity of the complex it is regulating. PPIs such as receptor–ligand, enzyme–inhibitor and antibody–antigen interactions that are regulated by localization generally have a high affinity towards each other, and the association once made is often permanent and irreversible (Figure 2B). The thrombin–rodniin complex (Figure 1E), for example, has a dissociation constant in the nanomolar range. Such very strong interactions are usually only perturbed by proteolysis. In contrast, physicochemically regulated transient interactions need the ability to change the affinity between protomers, sometimes by orders of magnitude; for example, the Gα–Gβγ subunit assembly of the heterotrimeric G protein (Figure 1F), controlled by GTP/GDP exchange, exhibits a 1000-fold change in affinity. Such regulatory switches permit the effective control of dynamic protein networks in biology.
Structural characteristics of protein–protein interfaces
Given these distinct functional types of interactions, we can explore the nature of the structural interfaces involved to see whether the type of PPI can be identified from knowledge of the structure. Structural data is available for various protein or protomer complexes, and the structure of the interfaces has been assessed by parameters such as the size of the contact area, the polarity of the interface, protrusion and flatness (Chothia and Janin, 1975; Miller et al., 1987; Argos, 1988; Janin et al., 1988; Jones and Thornton, 1995). Apart from a possible symmetry restraint in homo-oligomers, we do not expect to see differences between homo- and hetero-oligomers. A few general rules can be extracted from structural studies on PPIs so far: (i) The interfaces in obligate complexes, such as most homodimers, are generally larger and more hydrophobic than non-obligate associations (Jones and Thornton, 1996; LoConte et al., 1999) (Figure 3). Such homodimers can co-fold their co-expressed protomers and form stable structurally obligate complexes with large, intertwined and hydrophobic interfaces. Proteins that form complexes but can also exist on their own (i.e. non-obligate complexes) exhibit a more polar interface, presumably to meet the requirements for independent protomer folding and solubility. (ii) Complexes with interfaces larger than ∼1000 Å2 are likely to undergo conformational changes upon complexation (LoConte et al., 1999; Nooren and Thornton, 2003). This includes intertwined homodimers, non-obligate encounter complexes that form an induced-fit permanent complex such as the thrombin–rodniin complex (contact area of 1737 Å2; Figure 1E) and ‘strong’ transient complexes such as the heterotrimeric G protein of which the Gα and Gβγ subunits dissociate upon GTP/GDP exchange (contact area of 1159 Å2; Figure 1F). (iii) The trigger that changes the oligomeric state of a transient protein complex can be related to the stability of the complex (Nooren and Thornton, 2003). Oligomerization that is controlled by concentration or environmental triggers such as pH or temperature usually have only a weak effect on the oligomeric equilibrium state and show small and planar interfaces (denoted by the green ellipse in Figure 3). In contrast, more powerful molecular triggers such as GTP/GDP exchange or phosphorylation may induce rather drastic physicochemical or geometrical changes that can cause even strong complexes to dissociate. This may involve large conformational changes, and interfaces may be larger and less polar. (iv) Overall, the binding energy ΔG between protomers does not appear to be correlated with the size of the interface or other interface parameters such as the planarity and polarity for most PPIs. However, for complexes of co-expressed, co-localized protomers (e.g. homodimers), a weak correlation between the stability of the complex and size of the interface and hydrophobicity could be demonstrated (Brooijmans et al., 2002; Nooren and Thornton, 2003).
Fig. 3. Contact area and polarity of the interfaces of various non- obligate and obligate complexes. Obligate complexes with a small and hydrophobic interface include coiled-coil proteins. The ellipse denotes the contact area–polarity space of weak transient interactions.
In general, although some structural differences can be found between non-obligate and obligate complexes, it remains difficult to couple distinct structural patterns with different types of PPIs. A continuum exists between non-obligate and obligate and transient and permanent interactions, and current structural characterization parameters appear inadequate to differentiate between different affinities or specificities of diverse PPIs.
Specificity of PPIs
A protein generally resides in a crowded environment with many potential binding partners with different surface properties. Most proteins are very specific in their choice of partner, although some are multispecific, having multiple (competing) binding partners on coinciding or overlapping interfaces. Most often, protein complexes such as hormone–receptor and antibody–antigen complexes that are formed between protomers that are initially not co-localized, and functionally relevant interactions such as enzyme–inhibitor assemblies, are highly specific. Specificity clearly derives from the complementarity of shape and chemistry that determine the free energy of binding, but localization also has a role to play.
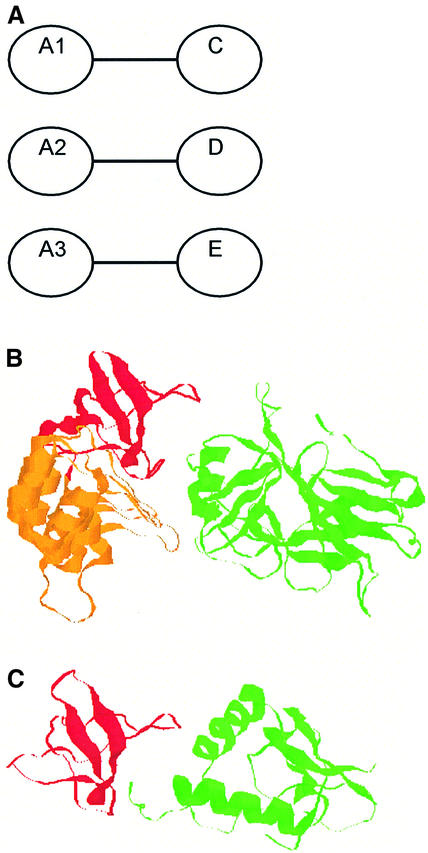
Within a family of proteins, paralogues will have frequently evolved different specificities, within a generic class of target ligands (e.g. nucleic acids, sugars). For PPIs, we can distinguish multispecificity between two homologous families of proteins (Figure 4) or between a homologous family and a set of non-homologous protein ligands (Figure 5). Multispecific binding between two protein families (i.e. multiple As and Bs; Figure 4A) is very common in regulatory pathways or networks such as in extracellular and intracellular cell signalling (e.g. Cdc42–Cdc42GAP and RhoA–RhoGAP; Figure 1D). When the As and Bs are homologous or identical, homo- and hetero-oligomerization can occur with coinciding interfaces (e.g. the HU histones; Figure 4B). Similarly, the NFκB transcription factor forms homologous homodimers and heterodimers that each select different signalling pathways. In contrast to the structurally obligate HU dimers, the interaction between the dimerization domains of NFκB is non-obligate and features a rather small and flat interface (Figure 4C). The immunoglobin and trypsin-like proteinases are examples of large homologous families with multiple target ligands that are not necessarily homologous (Figure 5A). However, the members of the protein family often recognize a specific pattern or surface patch on the target protein. For example, the SH2 and SH3 domains bind to proteins with phosphotyrosine and proline-rich sequences, respectively. Two structures of complexes between homologous SH3 domains and different non-homologous partners at the same interface are shown in Figure 5B.
Fig. 4. (A) Illustration of multispecific oligomerization between two families of homologous proteins (A1–A3 and B1–B3). The lines between the proteins denote interactions with differing affinities. Protein contacts are exchanged between homologous members of a family (e.g. A1–B1 and A1–B2), depending on protomer co-localization and concentration, and their mutual affinities. Examples are given for multispecific oligomerization within one protein family (i.e. where A∼B): (B) Bacillus stearothermophilus HU homodimer and Escherichia coli IHF heterodimer (the α and β chain are depicted in red and orange, respectively), including the target DNA; (C) mouse NFκB P50–P50 homodimer and P50–P65 heterodimer (the P50 and P65 protomers are depicted in red and orange, respectively) of the dimerization and DNA-binding domain, including the bound operator DNA.
Fig. 5. (A) Illustration of homologous monospecific oligomerization between one family (A1–A3) and three different ligands (C, D and E). Each member of the family has a specific binding partner. An example is given for the SH3-substrate heterodimers: (B) p53bp2, including the ankyrin repeat (orange) and the SH3 domain (red), with p53 (green), and (C) Fyn kinase SH3 domain (red) with HIV-1 Nef (green).
In the competition between binding partners, the control of oligomerization—i.e. the co-localization (in time and space), the local concentration of competing proteins and the affinity for the target protein (possibly regulated by molecular triggers; see also Figure 2)—are clearly important. A change of composition of a multispecific, permanent obligate interaction (e.g. HU; Figure 4B) would need to occur at the gene-expression level. Transient oligomerization allows for a change in binding partners or the composition of the complex at any time. Competition between proteins that form ‘weak’ transient complexes (e.g. NFκB; Figure 4C) requires a change in the local concentration of components. In contrast, a change in the composition of a strong transient interaction such as that between the Gα and Gβγ subunits of the heterotrimeric G protein (Figure 1F) requires the right GTP/GDP ratio and concentrations for GTP to bind and cause dissociation.
Evolution of PPIs
The structure and affinity of a PPI is tuned to its biological function and the physiological environment and control mechanism (Figure 2). PPIs presumably evolve to optimize ‘functional’ efficacy. This does not necessarily involve strong interactions. Clearly, weak transient interactions that are efficiently controlled are also very important in cellular processes. Obligate complexes may simply reflect the need for stability or the evolution of a function that requires both protomers; for example, symmetric DNA-binding modules, or intersubunit active sites with catalytic residues on different subunits (i.e. ∼1/6 of oligomeric enzymes; G.Bartlett, private communication), that would be inactive as separate proteins. Whereas some oligomerizations are obligate from a functional perspective, others may be incidental to function (e.g. oligomerization of cytokines whose primary function lies in receptor binding as a monomer). Proteins bump into each other all the time. They may evolve an interaction with no functional reason, which survives because there is no selective pressure to reject it from the evolutionary path. Often, the functional rationale of oligomerization is not clear, which may suggest a happenstance oligomerization.
The evolution of protein complexes is often obscure. It may be related to the folding of the oligomer. In two-state folding complexes (Xu et al., 1998), folding of the individual protomers and oligomerization occur concurrently. In contrast, in non-obligate interactions, each protomer folds independently and the interaction site has presumably evolved on the surface of the stable monomer. Some oligomers may evolve through domain swapping that involves a rearrangement of domains where interdomain interactions are replaced by intermonomer interactions (Bennett et al., 1994). Varying oligomeric states or structures within a homologous protein family can give further clues to the evolution of the family.
Variation in oligomeric state in homologous protein families
Within a family of homologous proteins (i.e. paralogues and orthologues), the stability of the oligomer will adapt to the different conditions in which they perform their function (i.e. different organisms, tissues or subcellular locations with differing metabolism or physiology). Also, during evolution, the function of a protein might have changed with a change in oligomeric state (e.g. the homologous monomeric methionine aminopeptidase and the homodimeric creatinase; Hoeffken et al., 1988). In at least 23 (out of 167) homologous CATH (hierarchical classification scheme of protein domain structures according to Class, Architecture, Topology and Homologous superfamily) superfamilies of enzymes, a variation of quaternary state or structure was found between members (Todd et al., 2001).
Although it is often unclear how particular families have evolved a variation of quaternary structure or oligomeric stability, it may occur in different ways: (i) Sequence drifting, leading to a change in affinity and specificity that can alter the preferred oligomeric state, structure or composition. For example, different interfaces as well as different quaternary states (i.e. dimers and tetramers; Clore and Gronenborn, 1995) are found in the homologous family of IL-8-like cytokines. Dimers are found with two different interfaces, involving completely different residues, consistent with the evolution of a PPI interface on the surface of the monomer. (ii) Gene duplication and fusion (i.e. covalent linkage of monomers). For example, a monomeric dihaemic cytochrome c4 from Pseudomonas stutzeri mimics the homologous dimeric cytochrome c4 from Pseudomonas nautica (Brown et al., 1999). (iii) Change in fragment or domain organization, related to the physiological domain swapping equilibrium (see above). (iv) Gene duplication and cross-over (i.e. fragment insert or deletion that mimics the oligomerization site). For example, the structure of the monomeric Lactobacillus leichmannii B12 dependent ribonucleotide reductase features an insert of a fragment that mimics the intersubunit allosteric site found in homologous dimeric ribonucleotide reductases (Sintchak et al., 2002).
Notably, variations in oligomeric state such as domain swapping dimers and mixtures of monomers and weak dimers may reflect or relate to a dynamic, transient oligomeric equilibrium of the protein in vivo. For a conserved oligomeric state, the residues at the interface are preferentially conserved compared with the rest of the surface (Valdar and Thornton, 2001). However, in large families that have members with varying oligomeric states or structures, these residues are found to be less conserved, as expected (Nooren and Thornton, 2003).
Concluding remarks
With more data, we are starting to understand the underlying evolutionary, functional and structural principles of PPIs, revealing their great diversity. Current proteomics studies have recently allowed the identification of PPIs on a large scale (Ho et al., 2002; Gavin et al., 2002). The protein networks found underline the multispecificity and dynamics of PPIs involving transient interactions, since many proteins were shown to be involved in more than one multiprotein complex or binary interaction. These multicomponent transient complexes have yet to be characterized in terms of their detailed structures or energetics. We can expect the full range of interactions, from rigid to dynamic, weak to strong, obligate and non-obligate. The functional rationale for many of these complexes is not known, and ultimately the optimization of function during evolution will be the key determinant of the observed character of each complex. Much work over the next few years will enormously expand our knowledge of protein complexes and hopefully improve our ability to model and predict their structures and their energetics and functions.
References
- Argos P. (1988) An investigation of protein subunit and domain interfaces. Protein Eng., 2, 101–113. [DOI] [PubMed] [Google Scholar]
- Bennett M.J., Choe,S. and Eisenberg,D. (1994) Domain swapping: entangling alliances between proteins. Proc. Natl Acad. Sci. USA, 91, 3127–3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooijmans N., Sharp,K.A. and Kuntz,I.D. (2002) Stability of macromolecular complexes. Proteins, 48, 645–653. [DOI] [PubMed] [Google Scholar]
- Brown K., Nurizzo,D., Besson,S., Shepard,W., Moura,J., Moura,I., Tegoni,M. and Cambillau,C. (1999) MAD structure of Pseudomonas nautica dimeric cytochrome c552 mimicks the c4 Dihemic cytochrome domain association. J. Mol. Biol., 289, 1017–1028. [DOI] [PubMed] [Google Scholar]
- Chothia C. and Janin,J. (1975) Principles of protein–protein recognition. Nature, 256, 705–708. [DOI] [PubMed] [Google Scholar]
- Clore G.M. and Gronenborn,A.M. (1995) Three-dimensional structures of α and β chemokines. FASEB J., 9, 57–62. [DOI] [PubMed] [Google Scholar]
- Faust P.L., Kornfeld,S. and Chirgwin,J.M. (1985) Cloning and sequence analysis of cDNA for human cathepsin D. Proc. Natl Acad. Sci. USA, 82, 4910–4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A.C. et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature, 415, 141–147. [DOI] [PubMed] [Google Scholar]
- Goodsell D.S. and Olson,A.J. (2000) Structural symmetry and protein function. Annu. Rev. Biophys. Biomol. Struct., 29, 105–153. [DOI] [PubMed] [Google Scholar]
- Ho Y. et al. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature, 415, 180–183. [DOI] [PubMed] [Google Scholar]
- Hoeffken H.W., Siegward,H.K., Bartlett,P.A. and Huber,R. (1988) Crystal structure determination, refinement and molecular model of creatine amidinohydrolase from Pseudomonas putida. J. Mol. Biol., 204, 417–433. [DOI] [PubMed] [Google Scholar]
- Janin J., Miller,S. and Chothia,C. (1988) Surface, subunit interfaces and interior of oligomeric proteins. J. Mol. Biol., 204, 155–164. [DOI] [PubMed] [Google Scholar]
- Jones S. and Thornton,J.M. (1995) Protein–protein interactions: a review of protein dimer structures. Prog. Biophys. Mol. Biol., 63, 31–65. [DOI] [PubMed] [Google Scholar]
- Jones S. and Thornton,J.M. (1996) Principles of protein–protein interactions. Proc. Natl Acad. Sci. USA, 93, 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoConte L., Chothia,C. and Janin,J. (1999) The atomic structure of protein–protein recognition sites. J. Mol. Biol., 285, 2177–2198. [DOI] [PubMed] [Google Scholar]
- Miller S., Lesk,A.M., Janin,J. and Chothia,C. (1987) The accessible surface area and stability of oligomeric proteins. Nature, 328, 834–836. [DOI] [PubMed] [Google Scholar]
- Monod J. Wyman,J. and Changeux,J. (1965) On the nature of allosteric transitions: a plausible model. J. Mol. Biol., 12, 88–118. [DOI] [PubMed] [Google Scholar]
- Nooren I.M. and Thornton,J.M. (2003). Structural characterisation and functional significance of transient protein–protein interactions. J. Mol. Biol., 325, 991–1018. [DOI] [PubMed] [Google Scholar]
- Sintchak M.D., Arjara,G., Kellogg,B.A., Stubbe,J. and Drennan,C.L. (2002) The crystal structure of class II ribonucleotide reductase reveals how an allosterically regulated monomer mimics a dimer. Nat. Struct. Biol., 9, 293–300. [DOI] [PubMed] [Google Scholar]
- Todd A.E., Orengo,C.A. and Thornton,J.M. (2001) Evolution of function in protein superfamilies, from a structural perspective. J. Mol. Biol., 307, 1113–1143. [DOI] [PubMed] [Google Scholar]
- Valdar W.S. and Thornton,J.M. (2001) Protein–protein interfaces: analysis of amino acid conservation in homodimers. Proteins, 42, 108–124. [PubMed] [Google Scholar]
- Xu D., Tsai,C.J. and Nussinov,R. (1998) Mechanism and evolution of protein dimerisation. Protein Sci., 7, 533–544. [DOI] [PMC free article] [PubMed] [Google Scholar]