Abstract
We have analyzed in vivo how model signal sequences are inserted and oriented in the membrane during cotranslational integration into the endoplasmic reticulum. The results are incompatible with the current models of retention of positive flanking charges or loop insertion of the polypeptide into the translocon. Instead they indicate that these N-terminal signals initially insert head-on with a cytoplasmic C-terminus before they invert their orientation to translocate the C-terminus. The rate of inversion increases with more positive N-terminal charge and is reduced with increasing hydrophobicity of the signal. Inversion may proceed for up to ∼50 s, when it is terminated by a signal-independent process. These findings provide a mechanism for the topogenic effects of flanking charges as well as of signal hydrophobicity.
Keywords: endoplasmic reticulum/protein topology/protein translocation/signal sequence
Introduction
Sorting of proteins to the mammalian endoplasmic reticulum (ER), for secretion or for insertion into the membrane, is mediated by a signal sequence that is characterized by a stretch of 7–25 mainly apolar residues (Walter and Johnson, 1994). The signal is first recognized by the signal recognition particle (SRP), which associates with the ribosome–nascent chain complex, slows down translation, and targets the complex to the ER membrane by binding to the SRP receptor (SR) (Keenan et al., 2001). This process is regulated by three GTPases: the 54 kDa subunit of SRP (SRP54) and the two subunits of SR (Connolly and Gilmore, 1993; Miller et al., 1993; Powers and Walter, 1995; Rapiejko and Gilmore, 1997; Song et al., 2000). Upon GTP hydrolysis, the ribosome engages with the translocon, which is composed of 3–4 Sec61 complexes (Hartmann et al., 1994; Hanein et al., 1996; Wang and Dobberstein, 1999), and resumes translation. SRP is then released, and the signal is transferred to the translocon by an unknown mechanism. Site-specific photocrosslinking has shown that the hydrophobic segment of the signal contacts a defined site in Sec61α as well as lipids, indicating that the signal sequence is specifically situated at the interface between the channel and the surrounding lipids (High et al., 1993; Jungnickel and Rapoport, 1995; Martoglio et al., 1995; Mothes et al., 1998).
In addition to their function in targeting, signal sequences play an important role in protein topogenesis by orienting themselves in the translocon and the membrane (Spiess, 1995). Cleavable signals of secretory and type I membrane proteins ultimately expose the C-terminal cleavage site to signal peptidase on the lumenal surface of the ER membrane, the N-terminus thus pointing towards the cytosol. The same orientation is attained by uncleaved signal–anchor sequences of type II membrane proteins (e.g. many glycosyltransferases), anchoring the polypeptide in an Ncyt/Cexo orientation in the bilayer. In both cases, the polypeptide transiently forms a loop while protein synthesis is still ongoing. Reverse signal–anchor sequences, in contrast, acquire the opposite Nexo/Ccyt orientation, as is the case, for example, in the cytochromes P450.
The most prominent determinant of signal orientation is the distribution of charged amino acids on either end of the hydrophobic sequence. The more positively charged flanking sequence is usually found on the cytosolic side of the membrane in both prokaryotic and eukaryotic cells (the positive-inside and charge difference rules; von Heijne, 1986; Hartmann et al., 1989). The effect of the flanking charges on the orientation of the signal is most likely due to electrostatic interactions with charges at or near the translocon.
Although several studies have shown that mutations of charged residues flanking a signal sequence affect its orientation, an asymmetric distribution of charges is often not sufficient to generate a uniform topology of the mutant proteins (e.g. Beltzer et al., 1991; Parks and Lamb, 1991, 1993; Andrews et al., 1992). It is thus clear that additional factors contribute to topogenesis. One such factor is the folding of sequences N-terminal to an internal signal, which may sterically prevent translocation of the N-terminus irrespective of the charge distribution (Denzer et al., 1995). Another one is the hydrophobicity of the apolar core of the signal itself. The more hydrophobic this sequence, the higher the tendency for translocation of the N-terminus into the ER lumen (Sakaguchi et al., 1992; Wahlberg and Spiess, 1997; Eusebio et al., 1998; Harley et al., 1998; Rösch et al., 2000). With a sufficently hydrophobic core, even a signal with a more positive N- than C-terminal flanking region may insert in an Nexo/Ccyt orientation. How the hydrophobicity of the signal exerts its topogenic effect is not obvious. It has been proposed that a hydrophobicity gradient within the apolar core of the signal affects orientation, rather than the overall hydrophobicity (Harley et al., 1998). The more hydrophobic end of a signal has been found to be more efficiently translocated, which may suggest a signal binding site that is more hydrophobic towards the lumenal surface of the translocon.
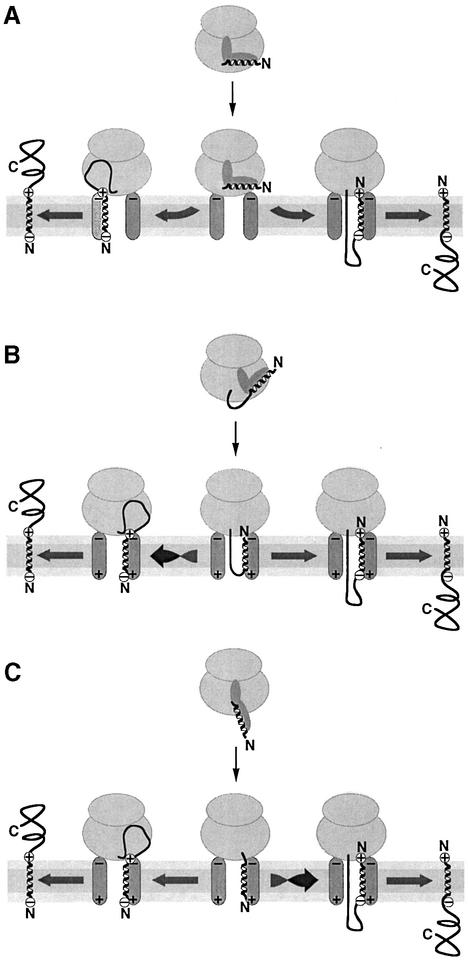
There are three basic models of how an N-terminal signal sequence may enter the translocon and achieve its correct orientation based on its flanking charges. In a first model, the signal is oriented by retention of the more positively charged flanking sequence at the cytosolic surface of the ER by interaction with negative charges at or near the translocon (Figure 1A). The other end of the signal enters the channel and is translocated. In Escherichia coli, anionic lipids, which influence topogenesis, have been suggested to prevent membrane passage of positive charges near the apolar signal sequence (van Klompenburg et al., 1997). Similarly, it has also been proposed for the mammalian ER that positive charges are retained on the cytosolic side (Parks and Lamb, 1991). A consequence of the retention model is that the signal acquires its final orientation early in the integration process, simultaneously with its initial insertion into the translocon.
Fig. 1. Potential mechanisms of signal orientation in the translocon. (A) Retention of the more positive flanking sequence of the signal by interaction with negative charges on the cytosolic side at or near the translocon. (B) Loop insertion of the polypeptide followed by inversion of reverse signal–anchors with a more negative N-terminus. (C) Head-on insertion with subsequent inversion of cleavable signals and type II signal–anchors with a more positive N-terminal flanking sequence. For simplicity, the SRP receptor has been omitted. The signal sequence is schematically drawn as a helix; yet, its conformation is not known during targeting and reorientation.
According to a second model (Figure 1B), the signal is initially transferred into the translocon in an Ncyt/Cexo orientation, with the nascent polypeptide forming a loop through the translocation pore. This could be a consequence of how the signal is presented to the translocon by SRP upon docking to the ER membrane. The initial signal orientation already corresponds to the final disposition of cleavable signals and type II signal–anchors (Shaw et al., 1988). Reverse signal–anchors will subsequently invert their orientation under the influence of a local electric potential acting on the flanking charges that are more positive on the C-terminal end. There is evidence that dynamic reorientation of signal and transmembrane sequences can indeed occur in the translocation apparatus (Goder et al., 1999; Nilsson et al., 2000).
A third model (Figure 1C) predicts initial insertion of the signal in the opposite, Nexo/Ccyt orientation, corresponding to the final topology of reverse signal–anchor sequences. Cleavable signals and type II signal–anchors then invert due to the mismatched flanking charges and the C-terminus translocates across the membrane.
Here, we present experiments using model proteins expressed in transfected COS cells that address the mechanism by which the signal orients itself in the translocon and how the flanking charges and the hydrophobicity of the signal sequence affect topogenesis. We provide evidence that signal orientation takes a maximum of ∼50 s to be committed. This is incompatible with a mechanism of retention of the more positive end of a signal on the cytosolic face of the membrane. Instead, our findings indicate that the signal first translocates its N-terminus and only inverts its orientation if the N-terminus is sufficiently positively charged (Figure 1C). Inversion is inhibited by increasing hydrophobicity of the signal, thus explaining the topogenic effect of the hydrophobic signal core.
Results
Polypeptide length affects signal orientation
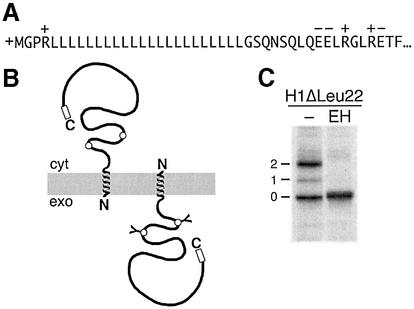
Signal orientation by retention of the more positive flanking sequence (Figure 1A) should be determined in the early phase of the insertion/translocation process and be independent of the length of the protein. Alternatively, if the preferred orientation is only reached after dynamic reorientation of the signal in the translocon, the process may take considerably longer and may not have arrived at a final topology when translation is completed. We therefore tested whether topogenesis depends on the length of the polypeptide using a series of diagnostic constructs with a signal sequence that generates mixed topologies. The construct H1ΔLeu22 (Figure 2) is derived from subunit H1 of the human asialoglycoprotein receptor, a typical type II single-spanning membrane protein (Wahlberg and Spiess, 1997). The N-terminal hydrophilic portion preceding the signal–anchor segment was truncated to only four residues with two positive charges (the α-amino group of the polypeptide and an arginine). The apolar core of the signal was replaced by an oligo-leucine sequence of 22 residues followed by the natural C-terminal portion of H1 with 230 amino acids, including two sites for N-linked glycosylation in the ER lumen. Despite the positively charged N-terminus, only ∼55% of the proteins produced in transfected COS-1 cells inserted in an Ncyt/Cexo orientation as indicated by glycosylation (Figure 2C). The rest was not glycosylated, indicating that the C-terminal portion of the protein remained in the cytosol, but anchored in the membrane because it could not be extracted with saponin (Wahlberg and Spiess, 1997; see also below). This intermediate insertion pattern lends itself to test changes in either direction upon extending or shortening the C-terminal portion of the protein.
Fig. 2. A model protein to study membrane protein topogenesis. (A) The chimeric model protein H1ΔLeu22 consists of an N-terminal signal–anchor sequence with a 230-residue C-terminal domain. It inserts in both orientations as schematically depicted (B). The C-terminal domain contains two diagnostic glycosylation sites (circles) that are core-glycosylated upon translocation into the ER lumen. (C) Upon labeling of transfected COS-1 cells with [35S]methionine for 40 min, followed by immunoprecipitation using an antibody against the very C-terminal sequence (open box), SDS–gel electrophoresis and autoradiography, the glycosylated and endoglycosidase H (EH) sensitive Ncyt/Cexo forms are easily distinguished from the unglycosylated Nexo/Ccyt forms. The number of attached glycans is indicated. Cyt, cytoplasm; exo, exoplasm.
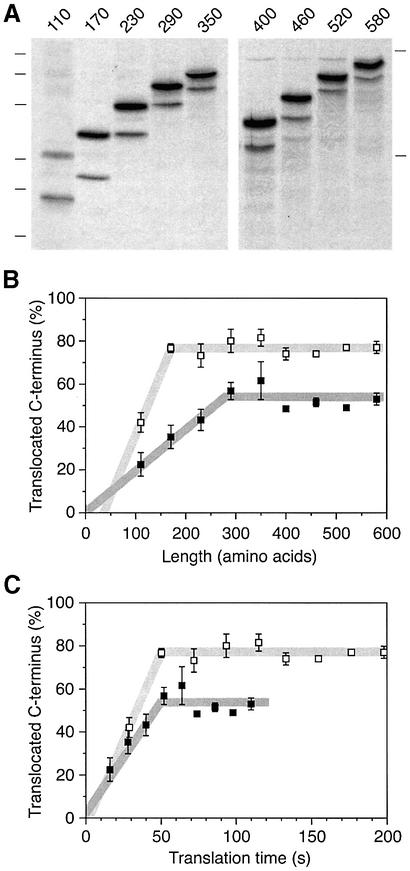
Constructs were prepared by deleting sequences from the C-terminus or by duplicating sequences to extend the C-terminal domain (Figure 3A). The N-terminal sequences of all constructs including the signal–anchor sequence and the following 96 amino acids are identical. These constructs were expressed in COS-1 cells and labeled with [35S]methionine for 40 min. The products were immunoprecipitated and analyzed by SDS–gel electrophoresis and autoradiography. As the C-terminal sequence was shortened from 230 to 170 and to 110 amino acids, the ratio of glycosylated to unglycosylated forms shifted in favor of the unglycosylated species (Figure 3B and C). In contrast, lengthening the C-terminal sequence to 290 residues resulted in an increase of the glycosylated fraction. Additional extension, however, did not further change the ratio of the two forms significantly.
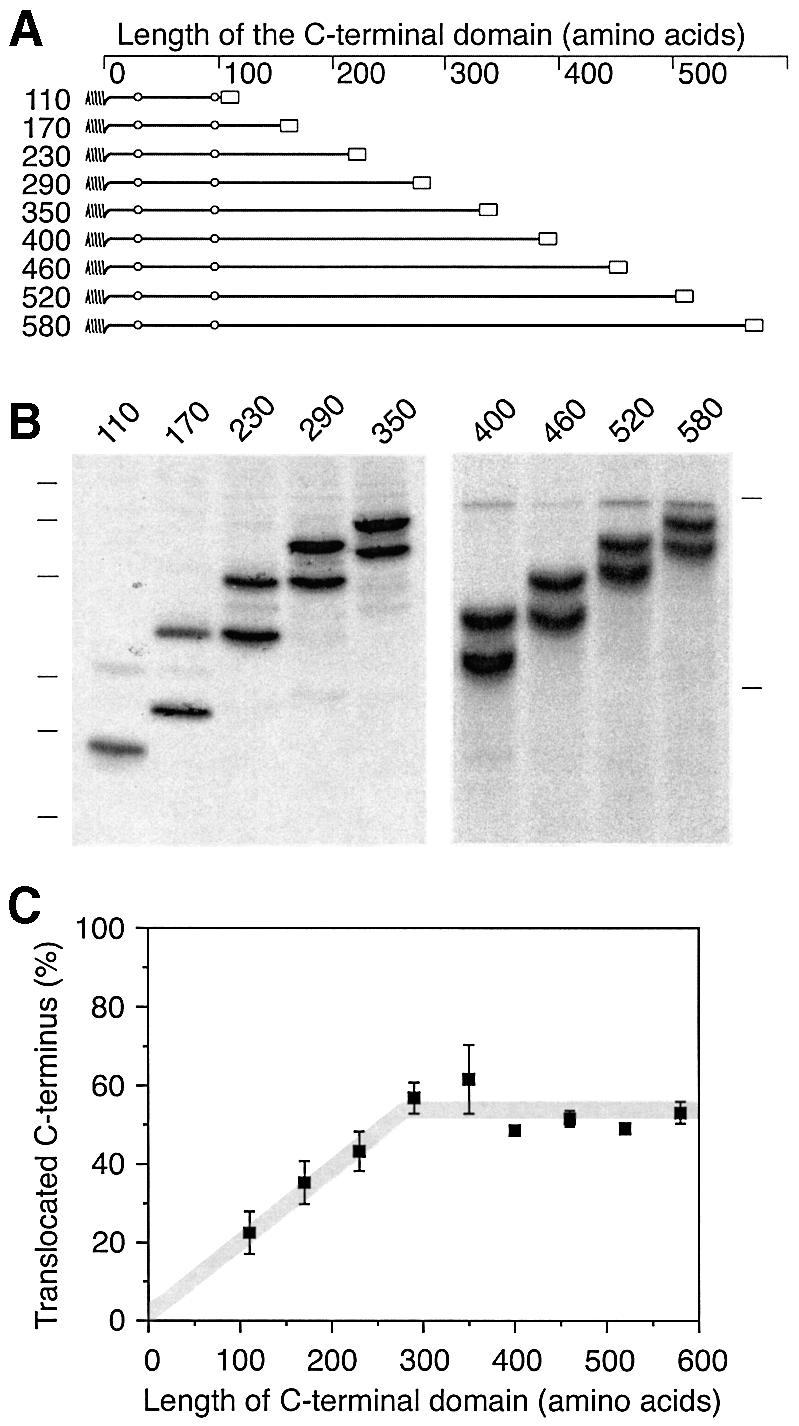
Fig. 3. Signal orientation depends on the size of the protein. (A) Schematic representation of the constructs H1ΔLeu22[#] with different C-terminal domain lengths. All constructs share the signal and the following 96 amino acids of H1ΔLeu22 as well as the C-terminal epitope for immunoprecipitation (open box). (B) The constructs were expressed in COS-1 cells, labeled with [35S]methionine for 40 min, immunoprecipitated and analyzed by SDS–gel electrophoresis (14 and 10% polyacrylamide on the left and the right gel, respectively) and phosphorimaging. The positions of molecular weight markers of 15, 20, 26, 37, 50 and 64 kDa (left) and 61 and 84 kDa (right) are indicated. (C) The fraction of polypeptides with a glycosylated and thus translocated C-terminus was quantified using phosphorimager analysis. The average and standard deviation of three experiments (including the one shown in B) was plotted versus the length of the C-terminal domain.
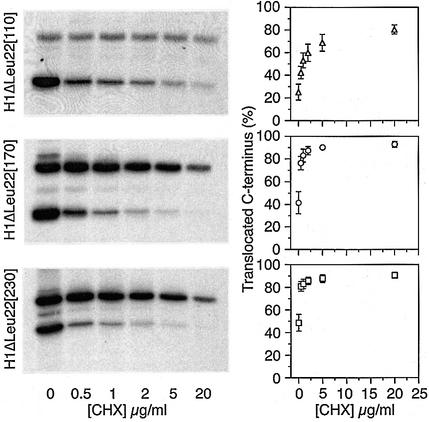
To test whether the observed change in the glycosylation pattern correctly reflects a change in protein topology, we analyzed the three constructs which differed most, H1ΔLeu22[110], [170] and [230], with respect to membrane integration and their orientation in the membrane. Cells expressing these constructs were labeled and subjected to alkaline extraction to distinguish between integral membrane proteins and soluble or only peripherally membrane-associated proteins. As a soluble control protein we also expressed H20–, a secretory protein composed of the C-terminal domain of H1 preceded by the cleavable signal sequence of influenza hemagglutinin (Goder et al., 1999). Upon pelleting the membranes, all products were sedimented except for H20– (Figure 4A), indicating that all H1ΔLeu22[#] polypeptides were stably integrated into the ER membrane independently of protein size.
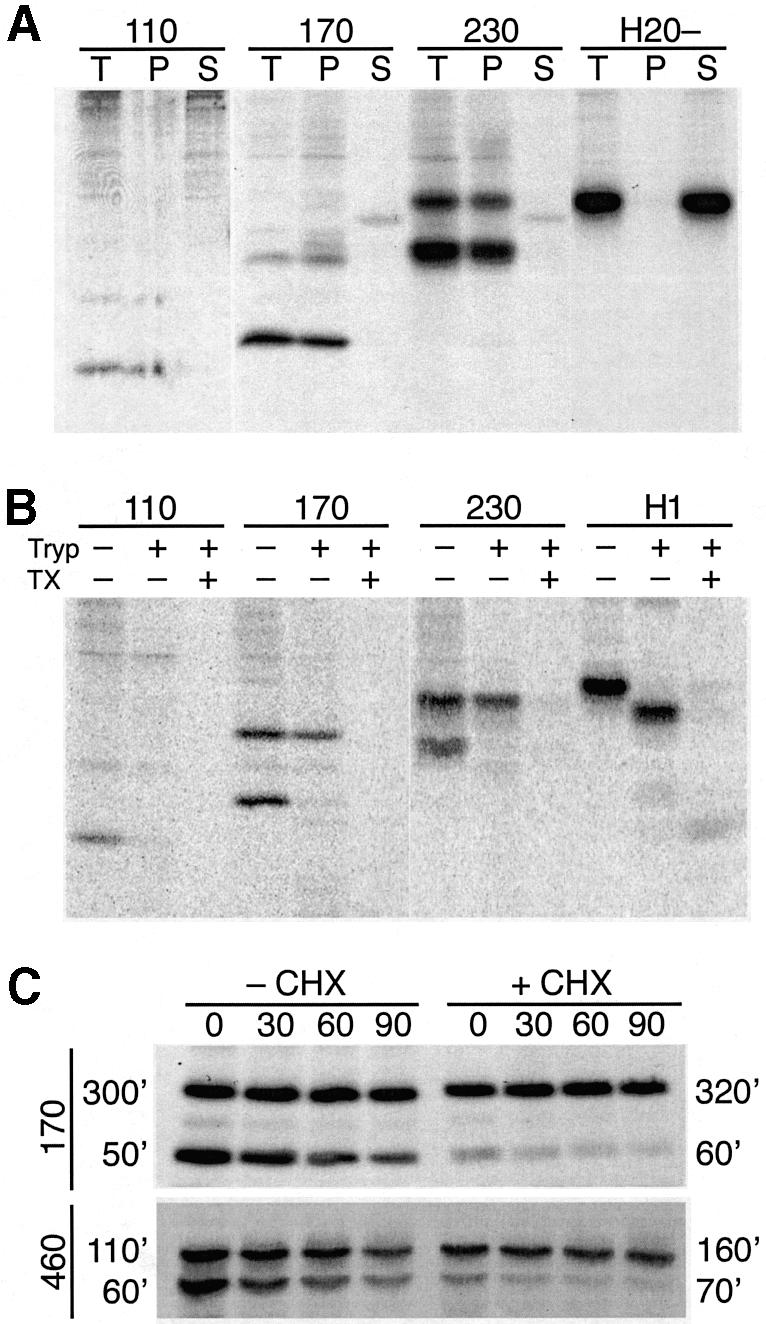
Fig. 4. Glycosylation state reflects protein topology. (A) Alkaline extraction: COS-1 cells expressing H1ΔLeu22[110], [170], [230] or the secretory protein H20– were labeled with [35S]methionine, and subjected to alkaline extraction. After centrifugation, the pellet (P) and the supernatant (S), as well as an equal aliquot of the total starting material (T) were analyzed by immunoprecipitation, SDS–gel electrophoresis, and autoradiography. (B) Protease protection: COS-1 cells expressing H1ΔLeu22[110], [170], [230] or wild-type H1 were labeled with [35S]methionine, broken by swelling and scraping, and incubated with or without trypsin (Tryp) and with or without Triton X-100 (TX). Trypsin was then inhibited and the proteins were analyzed by immunoprecipitation, SDS–gel electrophoresis, and autoradiography. Since the broken cells were incubated at 4°C in the absence of protease inhibitors, some unglycosylated polypeptides were lost even in the absence of added trypsin, most likely by released lysosomal proteases. (C) Protein stability: COS-1 cells expressing H1ΔLeu22[170] or [460] were pulse-labeled for 40 min with [35S]methionine and chased with excess unlabeled methionine for 0–90 min. Both pulse and chase were performed in the presence (+CHX) or absence (–CHX) of 1 µg/ml cycloheximide. The proteins were analyzed by immunoprecipitation, SDS–gel electrophoresis, and autoradiography. Upon quantitation of the glycosylated and unglycosylated products, their half-lives were calculated as indicated in minutes. The half-lives were used to correct the apparent fraction of C-terminally translocated polypeptides for degradation during the labeling period. Correction changed the values for H1ΔLeu22[170] from 36 to 31% and for H1ΔLeu22[460] from 51 to 48% in the absence of cycloheximide, and from 77 to 74% and from 73 to 71%, respectively, in the presence of cycloheximide. Protein degradation thus did not significantly distort the results.
To test the localization of the products, the plasma membrane of labeled cells was broken by swelling and scraping, and the broken cells were incubated at 4°C with or without trypsin (Figure 4B). Wild-type H1, which was analyzed as a control protein, shifted its position in SDS–gel electrophoresis upon trypsin treatment due to digestion of its 40-amino acid cytoplasmic domain. Whereas the glycosylated forms of all three H1ΔLeu22[#] constructs were resistant to protease, the unglycosylated ones were efficiently digested. Upon permeabilization of membranes by detergent, all products were sensitive. The C-terminal portion of the glycosylated polypeptides was therefore translocated across the ER membrane, whereas that of the unglycosylated ones was exposed to the cytosol.
These control experiments confirm that the glycosylation state of the proteins analyzed in Figure 3 directly reflects their orientation in the membrane. The topologies generated by the same signal sequence thus depend on the length of the polypeptide, with predominantly N-terminal translocation for very short proteins and reaching a maximum of ∼55% C-terminal translocation for polypeptides of ∼300 amino acids or more. This is incompatible with a retention mechanism of signal–anchor orientation.
Topology depends on the time of translation rather than on the length of the polypeptide
These results may be interpreted as a time course of reorientation of the signal sequence in the translocon which is interrupted upon termination of translation. Alternatively, the observed topologies may depend directly on polypeptide size. To distinguish between a topogenic effect of protein size per se and a change in signal orientation with time, we tested the effect of extending the time of translation on the resulting topologies of the same constructs. To achieve this, we reduced the translation rate with low concentrations of the reversible elongation inhibitor cycloheximide. Cells expressing the three shortest constructs with C-terminal domains of 110, 170 or 230 residues were labeled with [35S]methionine for 40 min in the presence of different cycloheximide concentrations. As expected, the total amount of protein synthesized was decreased in relation to the reduced translation rate. In addition, the apparent ratio of topologies shifted in favor of C-terminal translocation (Figure 5).
Fig. 5. Topology depends on translation time. COS-1 cells expressing H1ΔLeu22[110] (triangles), H1ΔLeu22[170] (circles) or H1ΔLeu22 [230] (rectangles) were labeled with [35S]methionine in the presence of different concentrations of cycloheximide. The labeled proteins were immunoprecipitated and analyzed by gel electrophoresis and autoradiography. Protein oriention was quantified by phosphorimager analysis and plotted as a function of cycloheximide concentration. The average with standard deviation of three determinations is shown.
To exclude the possibility that cycloheximide treatment affected the relative stability of the two products, a pulse–chase analysis was performed for H1ΔLeu22[170] and H1ΔLeu22[460], i.e. a short and a long construct, in the presence or absence of 1 µg/ml cycloheximide (Figure 4C). As previously observed (Wahlberg and Spiess, 1997), the unglycosylated Nexo/Ccyt forms of the proteins were less stable than the glycosylated Ncyt/Cexo forms. This results in a slight underestimation of the Nexo/Ccyt forms. In addition, the half-lives varied somewhat with protein size and with the presence or absence of cycloheximide. However, when the observed ratios of glycosylated to unglycosylated forms are corrected for degradation during the labeling time, the percentage of polypeptides with a translocated C-terminus is reduced by only 2–5%. The observed shift towards the glycosylated forms with increasing protein size and cycloheximide concentration is thus not an artefact of altered protein stabilities, but reflects a shift towards Ncyt/Cexo orientation of the proteins.
In Figure 6, we determined the topologies of all nine constructs in the presence of 1 µg/ml cycloheximide, which reduces translation rate by a factor of 1.8 (as determined from the amount of labeled protein produced; see also Goder et al., 2000). Every construct inserted with increased Ncyt/Cexo orientation (Figure 6B, open symbols). Whereas at normal translation rate, a maximum was reached at ∼55% with a C-terminal sequence of 290 amino acids, at reduced translation rate it was reached at ∼75% already with a sequence of 170 residues. In Figure 6C, the same data were plotted as a function of the translation time from the moment when the signal has just emerged from the ribosome to insert into the translocon (i.e. with 30 residues following the hydrophobic core of the signal still hidden within the ribosome; Matlack and Walter, 1995; Morgan et al., 2000) until the ribosome has reached the stop codon. The calculation is based on a translation rate of ∼5 amino acids/s in the absence of cycloheximide, as determined for cultured cell lines (Hershey, 1991). This presentation reveals that under both conditions the maximal amount of Ncyt/Cexo insertion was reached at the same time, after ∼50 s. Because the rate of inversion of the protein was higher at slow translation, a higher level of Ncyt/Cexo insertion was attained at this moment. The rate of inversion may be higher at low translation rate, because the growing polypeptides are on average shorter during the first 50 s and thus may more easily move through the translocation pore as the signal reorients itself.
Fig. 6. Signal inversion stops after ∼50 s. H1ΔLeu22[#] constructs were expressed in COS-1 cells in the presence of 1 µg/ml cycloheximide and analyzed as in Figure 3. (A) Protein orientation was quantified by phosphorimager analysis and plotted with open squares either as a function of the length of the C-terminal domain (B) or as a function of translation time (C). The elongation rate in the presence of 1 µg/ml cycloheximide was reduced by a factor of 1.8, as determined from the reduction in the total signal. For comparison, protein orientation determined in the absence of cycloheximide (from Figure 3C) is shown with filled squares. The average with standard deviation of three determinations is shown.
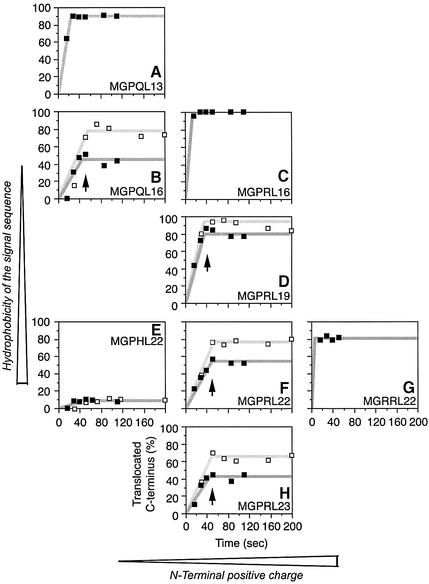
Flanking charges and hydrophobicity determine the rate of signal inversion
The force driving inversion of the signal sequence is likely to be electrostatic, acting on the flanking charges. To test this hypothesis, we analyzed the behavior of charge mutants of the constructs used above. Upon mutation of the N-terminal arginine to histidine, thus reducing the N-terminal positive charge by nearly one unit, inversion was almost completely abolished and the proteins inserted with a cytosolic C-terminus irrespective of the length of the polypeptides and of the presence or absence of cycloheximide (Figure 7E). In contrast, addition of a second arginine at the N-terminus yielded 80% C-terminal translocation even for the shortest construct at normal translation speed (Figure 7G), consistent with rapid inversion of the signal. Why inversion is incomplete is not clear.
Fig. 7. Flanking charges and hydrophobicity determine the rate of signal inversion. Starting from the construct series H1ΔLeu22[#] (panel F), further series of proteins with increasing or decreasing hydrophobicity of the signal sequence and/or with higher or lower N-terminal positive charge were analyzed. Protein orientation was plotted versus translation time as in Figure 6C. The N-terminal amino acid sequence and the length of the oligo- leucine segment are indicated. Experiments were performed in the absence (filled squares) or presence of 1 µg/ml cycloheximide (open squares). The arrows mark the time at which signal inversion stops. Each value represents the average of at least two individual determinations.
To test how the hydrophobicity of the signal affects the insertion process, we generated an additional construct series in which the oligo-leucine core of the signal was either extended or shortened. Addition of a single leucine to a total length of Leu23 (Figure 7H) appeared to slightly reduce the inversion rate both in the absence and in the presence of cycloheximide. On the contrary, a shortened hydrophobic core of only 19 leucines (Figure 7D) caused an increased rate of inversion. With 16 leucines (Figure 7C), inversion was too quick to allow detection of any molecules with an Nexo/Ccyt orientation. However, in combination with a reduced charge at the N-terminus (Figure 7B) the kinetics of topogenesis slowed down again and were similar to that of the constructs shown in Figure 7H, i.e. of proteins with a longer apolar core but also higher N-terminal positive charge. Further shortening the signal to 13 leucines (Figure 7A) accelerated reorientation once more, as expected.
It is interesting to note that for none of the constructs analyzed here topology was observed to change after ∼50 s. For the constructs analyzed in Figure 7B, D, F and H, the change in topologies stopped at roughly the same time of ∼40–50 s. Both flanking charges and hydrophobicity affected the rate of signal inversion, but did not affect the time when topogenesis stopped. This time was also not affected by slowing down translation rate with cycloheximide. It thus appears to be a function of the translocation machinery.
Discussion
The results shown here are not consistent with the mechanism of cytosolic retention of the positive end of signal sequences (Figure 1A). Our model signals, which generate two populations with Ncyt/Cexo or Nexo/Ccyt orientation, change their integration patterns significantly with the length of the protein sequence following the signal between 100–350 amino acids. Signal orientation is thus not determined early in the insertion process, but changes over time from complete Nexo/Ccyt to an increasing fraction of Ncyt/Cexo orientation, as predicted by the model illustrated in Figure 1C. The rate of inversion mainly depends on the flanking charges and on the hydrophobicity/length of the core of the signal sequence.
Our results support the following scenario for the insertion and topogenesis of ER proteins with N-terminal signal sequences. As the signal emerges from the ribosome and is recognized by SRP, the hydrophobic core contacts the M domain of SRP54 (Römisch et al., 1990; Zopf et al., 1990; Lutcke et al., 1992). Conserved methionines form a hydrophobic groove which can accommodate diverse apolar signals (Keenan et al., 1998; Batey et al., 2000). Upon binding to SR, SRP54 changes its position relative to the ribosome (as was shown by crosslinking experiments; Pool et al., 2002), which may allow the ribosome to dock to the translocon. At the same time, SRP54 may connect to a hydrophobic surface in the translocation pore, thereby allowing the signal to slide into the translocon head-on without having to dissociate from the apolar environment. As a result, the initial orientation of the signal in the translocon is Nexo/Ccyt, which was observed as the predominant orientation of the shortest model proteins such as H1ΔLeu22[110]. As the polypeptide grows longer, the signal inverts its orientation driven by a local electrical potential acting on the flanking charges. If translation is slowed down with cycloheximide, short constructs gain time for signal inversion and end up with higher fractions of Ncyt/Cexo products.
The rate of inversion depends first of all on the flanking charges. It is increased by a more positive and decreased by a less positive N-terminal charge. Reverse signal–anchors with a more positive C-terminus do not invert at all. Importantly, also the hydrophobic core of the signal affects the rate of reorientation. The longer and the more hydrophobic the signal, the more slowly it inverts in the translocon. If the signal needs to dissociate from the apolar binding site in the translocation pore to reorient itself, this process will be slower the more hydrophobic the signal is. If inversion can occur in a hydrophobic environment, the length of the helix and its rigidity, which parallel hydrophobicity, may sterically hinder a rapid turnaround. In either case, our model can explain how increasing hydrophobicity of the signal favors N-terminal translocation, namely by impeding signal inversion. A strongly hydrophobic core of the reverse signal–anchor may therefore prevent inversion by flanking charges that violate the charge rules. This seems to be the case in microsomal epoxide hydrolase: despite flanking charges of 0 at the N-terminal and –2 at the C-terminal end of the signal, it inserts exclusively with a Nexo/Ccyt orientation. However, when the hydrophobic core is shortened, Ncyt/Cexo products are obtained as well (Eusebio et al., 1998), consistent with an increased inversion rate due to the reduced hydrophobicity of the signal.
The signals used in our study are mostly longer and certainly more hydrophobic than typical natural signal sequences. As a result, natural C-terminus translocating signals will generally invert much more rapidly than the model signals analyzed here and cannot be ‘caught’ in an Nexo/Ccyt orientation. We have analyzed truncated forms of γ-glutamyl-cleaving enzyme, a type II membrane protein with a rather hydrophobic signal and two positive charges at the N-terminus, but have been unable to detect an Nexo/Ccyt population (data not shown). The small size of most cleaved signals and a high frequency of ∼25% of hydrophilic amino acids (Thr, Ser, Gly, Asn, Gln, His, Pro) in signal–anchor sequences guarantees rapid inversion and efficient translocation of the C-terminus even for small proteins. It is also conceivable that less hydrophobic signals with a more positive N- than C-terminus might be affected by the local potential even before they have fully entered the translocon and thus might insert as a loop from the start. In Nexo/Ccyt proteins, appropriate flanking charges (more positive C- than N-terminal sequences) or a strongly hydrophobic core of the reverse signal–anchor prevent inversion.
The proposed model implies that, depending on the properties of the signal sequence, rather long polypeptide segments following the signal may be synthesized before translocation across the membrane is triggered by signal reorientation. This is reminiscent of the situation observed in translocational pausing of certain proteins (Chuck and Lingappa, 1992). Whereas in the latter case delayed translocation is caused by specific pause-transfer sequences in the polypeptide to be transported through the membrane, in our model proteins it is the result of the relatively slow orientation of the signal sequence.
It is intriguing that signal inversion can only be observed during the first ∼50 s after insertion into the translocon. If it takes longer than this period for the protein to be completed, either because of the length of the protein or because of reduced translation rate, the resulting ratio of topologies does not change further; topogenesis appears to come to a halt. The time when this occurs is not significantly affected by alterations in the signal, neither of the flanking charges nor of the apolar core. It is thus a property of the translocation machinery to commit the substrate to its current orientation ∼50 s after engagement of the protein with the translocon. This could happen for example by expelling the signal into the lipid bilayer, possibly to purge the translocon of the signal sequence. The topologies do not approach the final distribution asymptotically with time, but appear to become fixed rather synchronously. The molecular mechanism which triggers this phenomenon is currently unclear.
Materials and methods
DNA constructs
All constructs were made by polymerase chain reaction (PCR) using Vent polymerase (New England Biolabs). The final constructs were subcloned into the expression vector pECE (Ellis et al., 1986) and verified by sequencing. A series of cDNA constructs were generated encoding the signal sequence of H1ΔLeu22 (Wahlberg and Spiess, 1997) followed by 110–580 residues and ending with the C-terminal 14 residues of H1, which constitute the epitope of an anti-peptide antiserum. First, a KpnI site was introduced in front of the 14 last codons of the coding sequence of H1ΔLeu22 to generate H1ΔLeu22[230]. The sequence upstream of the KpnI site was truncated by PCR and ligated to the epitope sequence to yield H1ΔLeu22[110] and [170]. The sequence encoding the C-terminal 130 residues of H1ΔLeu22 was amplified with a 5′ KpnI site and ligated to the KpnI site of H1ΔLeu22[170] and [230] to produce H1ΔLeu22[290] and [350]. For further extension, a BglII site was introduced in front of the stop codon of the constructs encoding H1ΔLeu22, [170], [230], [290] and [350] and ligated to the BamHI site of the sequence BamHI–EcoRI encoding the entire C-terminal domain of 230 amino acids of H1(nog1nog2) producing H1ΔLeu22[400], [460], [520] and [580], respectively. In the DNA of H1(nog1nog2) the two glycosylations sites had been destroyed by mutation of the respective asparagine codons to glutamine. All constructs therefore contain two glycosylation sites to rule out differences due to a potential topogenic effect of glycosylation (Goder et al., 1999).
Constructs with different signal sequences were produced by fusing the 5′ end of the cDNAs of H1ΔQLeu13, H1ΔQLeu16, H1ΔLeu16 and H1ΔLeu19 from Wahlberg and Spiess (1997) up to the BstXI site (corresponding to the position of residue 85 of the C-terminal domain) to the 3′ fragments from the BstXI site of the constructs encoding H1ΔLeu22[110]–[580]. To generate H1ΔLeu23[110]–[580], the sequence of the signal of H1ΔLeu22 was extended by one leucine codon and fused to the segments encoding the C-terminal domains of H1ΔLeu22[110]–[580]. To introduce N-terminal charge mutations, the codon for Arg4 in H1ΔLeu22[110]–[580] was exchanged for that of a histidine or the codon for Pro3 for that of an arginine. H20– was described previously (Goder et al., 1999).
In vivo expression and labeling
Cell culture reagents were from Life Technologies, Inc. COS-1 cells were grown in modified Eagle’s minimal essential medium supplemented with 10% fetal calf serum, 2 mM l-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin at 37°C with 7.5% CO2. Transient transfection was performed with lipofectin (Life Technologies, Inc.) according to the manufacturer’s instructions in 6-well clusters. The cells were processed the second day after transfection. For in vivo labeling, transfected cells were incubated for 30 min in methionine-free medium, labeled for 40 min at 37°C with 100 µCi/ml [35S]methionine, transferred to 4°C and washed twice with phosphate-buffered saline (PBS). To release soluble proteins, cells were incubated with 0.1% saponin for 20 min at 4°C in the presence of protease inhibitors, washed once with PBS, and finally lysed and immunoprecipitated using a rabbit anti-serum directed against a synthetic peptide corresponding to residues 277–287 near the C-terminus of the ASGP receptor H1 (anti-H1C). The immune complexes were isolated with protein A–Sepharose (Pharmacia) and analyzed by SDS–polyacrylamide gel electrophoresis and autoradiography. For deglycosylation, the immune complexes were released from protein A–Sepharose by boiling in 50 mM Na citrate pH 6, 1% SDS and incubated with 1 mU endo-β-d-N-acetyl glucosaminidase H for 5 h at 37°C, before gel electrophoresis. Quantitation was performed using a phosphorimager (Molecular Dynamics Inc.).
For experiments at reduced elongation rate, 0.5–20 µg/ml cycloheximide were added to the labeling solution. The reduction of translation rate at 1 µg/ml cycloheximide was determined for the H1ΔLeu22[#] constructs from the total [35S]methionine signal corrected for cell numbers based on protein content in the saponin extract (as measured using the bicinchoninic acid method; Pierce).
Alkaline extraction with pelleting was performed as previously described (Wessels et al., 1991). To reduce the viscosity of the sample, the cells suspended in alkaline solution were pipetted up and down through a 25G needle to shear the DNA before loading onto the sucrose cushion. For the protease protection assay, labeled cells were incubated at 4°C with swelling buffer (15 mM HEPES/KOH pH 7.2, 15 mM KCl) and scraped with a rubber policeman. Aliquots were incubated without protease or with 100 µg/ml trypsin in the presence or absence of 0.5% Triton X-100 for 30 min at 4°C. Trypsin was then inhibited by addition of 500 µg/ml soybean trypsin inhibitor before immunoprecipitation and analysis by SDS–gel electrophoresis and autoradiography.
Acknowledgments
Acknowledgements
We thank Drs Natasha Kralli and Stephen Helliwell for critically reading the manuscript, and John Groffen for the cDNA of γ-glutamyl-cleaving enzyme. This work was supported by grant 31-061579.00 from the Swiss National Science Foundation.
References
- Andrews D.W., Young,J.C., Mirels,L.F. and Czarnota,G.J. (1992) The role of the N-region in signal sequence and signal–anchor function. J. Biol. Chem., 267, 7761–7769. [PubMed] [Google Scholar]
- Batey R.T., Rambo,R.P., Lucast,L., Rha,B. and Doudna,J.A. (2000) Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science, 287, 1232–1239. [DOI] [PubMed] [Google Scholar]
- Beltzer J.P., Fiedler,K., Fuhrer,C., Geffen,I., Handschin,C., Wessels,H.P. and Spiess,M. (1991) Charged residues are major determinants of the transmembrane orientation of a signal–anchor sequence. J. Biol. Chem., 266, 973–978. [PubMed] [Google Scholar]
- Chuck S.L. and Lingappa,V.R. (1992) Pause transfer: a topogenic sequence in apolipoprotein B mediates stopping and restarting of translocation. Cell, 68, 9–21. [DOI] [PubMed] [Google Scholar]
- Connolly T. and Gilmore,R. (1993) GTP hydrolysis by complexes of the signal recognition particle and the signal recognition particle receptor. J. Cell Biol., 123, 799–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denzer A.J., Nabholz,C.E. and Spiess,M. (1995) Transmembrane orientation of signal–anchor proteins is affected by the folding state but not the size of the N-terminal domain. EMBO J., 14, 6311–6317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis L., Clauser,E., Morgan,D.O., Edery,M., Roth,R.A. and Rutter,W.J. (1986) Replacement of insulin receptor tyrosine residues 1162 and 1163 compromises insulin-stimulated kinase activity and uptake of 2-deoxyglucose. Cell, 45, 721–732. [DOI] [PubMed] [Google Scholar]
- Eusebio A., Friedberg,T. and Spiess,M. (1998) The role of the hydrophobic domain in orienting natural signal sequences within the ER membrane. Exp. Cell Res., 241, 181–185. [DOI] [PubMed] [Google Scholar]
- Goder V., Bieri,C. and Spiess,M. (1999) Glycosylation can influence topogenesis of membrane proteins and reveals dynamic reorientation of nascent polypeptides within the translocon. J. Cell Biol., 147, 257–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goder V., Crottet,P. and Spiess,M. (2000) In vivo kinetics of protein targeting to the endoplasmic reticulum determined by site-specific phosphorylation. EMBO J., 19, 6704–6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanein D., Matlack,K.E., Jungnickel,B., Plath,K., Kalies,K.U., Miller,K.R., Rapoport,T.A. and Akey,C.W. (1996) Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell, 87, 721–732. [DOI] [PubMed] [Google Scholar]
- Harley C.A., Holt,J.A., Turner,R. and Tipper,D.J. (1998) Transmembrane protein insertion orientation in yeast depends on the charge difference across transmembrane segments, their total hydrophobicity and its distribution. J. Biol. Chem., 273, 24963–24971. [DOI] [PubMed] [Google Scholar]
- Hartmann E., Rapoport,T.A. and Lodish,H.F. (1989) Predicting the orientation of eukaryotic membrane-spanning proteins. Proc. Natl Acad. Sci. USA, 86, 5786–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann E., Sommer,T., Prehn,S., Gorlich,D., Jentsch,S. and Rapoport,T.A. (1994) Evolutionary conservation of components of the protein translocation complex. Nature, 367, 654–657. [DOI] [PubMed] [Google Scholar]
- Hershey J.W.B. (1991) Translational control in mammalian cells. Annu. Rev. Biochem., 60, 717–755. [DOI] [PubMed] [Google Scholar]
- High S., Andersen,S.S., Gorlich,D., Hartmann,E., Prehn,S., Rapoport,T.A. and Dobberstein,B. (1993) Sec61p is adjacent to nascent type I and type II signal–anchor proteins during their membrane insertion. J. Cell Biol., 121, 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungnickel B. and Rapoport,T.A. (1995) A post-targeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell, 82, 261–270. [DOI] [PubMed] [Google Scholar]
- Keenan R.J., Freymann,D.M., Walter,P. and Stroud,R.M. (1998) Crystal structure of the signal sequence binding subunit of the signal recognition particle. Cell, 94, 181–191. [DOI] [PubMed] [Google Scholar]
- Keenan R.J., Freymann,D.M., Stroud,R.M. and Walter,P. (2001) The signal recognition particle. Annu. Rev. Biochem., 70, 755–775. [DOI] [PubMed] [Google Scholar]
- Lutcke H., High,S., Romisch,K., Ashford,A.J. and Dobberstein,B. (1992) The methionine-rich domain of the 54 kDa subunit of signal recognition particle is sufficient for the interaction with signal sequences. EMBO J., 11, 1543–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martoglio B., Hofmann,M.W., Brunner,J. and Dobberstein,B. (1995) The protein-conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell, 81, 207–214. [DOI] [PubMed] [Google Scholar]
- Matlack K.E. and Walter,P. (1995) The 70 carboxyl-terminal amino acids of nascent secretory proteins are protected from proteolysis by the ribosome and the protein translocation apparatus of the endoplasmic reticulum membrane. J. Biol. Chem., 270, 6170–6180. [DOI] [PubMed] [Google Scholar]
- Miller J.D., Wilhelm,H., Gierasch,L., Gilmore,R. and Walter,P. (1993) GTP binding and hydrolysis by the signal recognition particle during initiation of protein translocation. Nature, 366, 351–354. [DOI] [PubMed] [Google Scholar]
- Morgan D.G., Menetret,J.F., Radermacher,M., Neuhof,A., Akey,I.V., Rapoport,T.A. and Akey,C.W. (2000) A comparison of the yeast and rabbit 80 S ribosome reveals the topology of the nascent chain exit tunnel, inter-subunit bridges and mammalian rRNA expansion segments. J. Mol. Biol., 301, 301–321. [DOI] [PubMed] [Google Scholar]
- Mothes W., Jungnickel,B., Brunner,J. and Rapoport,T.A. (1998) Signal sequence recognition in cotranslational translocation by protein components of the endoplasmic reticulum membrane. J. Cell Biol., 142, 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson I., Witt,S., Kiefer,H., Mingarro,I. and von Heijne,G. (2000) Distant downstream sequence determinants can control N-tail translocation during protein insertion into the endoplasmic reticulum membrane. J. Biol. Chem., 275, 6207–6213. [DOI] [PubMed] [Google Scholar]
- Parks G.D. and Lamb,R.A. (1991) Topology of eukaryotic type-II membrane proteins—importance of N-terminal positively charged residues flanking the hydrophobic domain. Cell, 64, 777–787. [DOI] [PubMed] [Google Scholar]
- Parks G.D. and Lamb,R.A. (1993) Role of NH2-terminal positively charged residues in establishing membrane protein topology. J. Biol. Chem., 268, 19101–19109. [PubMed] [Google Scholar]
- Pool M.R., Stumm,J., Fulga,T.A., Sinning,I. and Dobberstein,B. (2002) Distinct modes of signal recognition particle interaction with the ribosome. Science, 297, 1345–1348. [DOI] [PubMed] [Google Scholar]
- Powers T. and Walter,P. (1995) Reciprocal stimulation of GTP hydrolysis by two directly interacting GTPases. Science, 269, 1422–1424. [DOI] [PubMed] [Google Scholar]
- Rapiejko P.J. and Gilmore,R. (1997) Empty site forms of the SRP54 and SR alpha GTPases mediate targeting of ribosome-nascent chain complexes to the endoplasmic reticulum. Cell, 89, 703–713. [DOI] [PubMed] [Google Scholar]
- Römisch K., Webb,J., Lingelbach,K., Gausepohl,H. and Dobberstein,B. (1990) The 54-kD protein of signal recognition particle contains a methionine-rich RNA binding domain. J. Cell Biol., 111, 1793–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rösch K., Naeher,D., Laird,V., Goder,V. and Spiess,M. (2000) The topogenic contribution of uncharged amino acids on signal sequence orientation in the endoplasmic reticulum. J. Biol. Chem., 275, 14916–14922. [DOI] [PubMed] [Google Scholar]
- Sakaguchi M., Tomiyoshi,R., Kuroiwa,T., Mihara,K. and Omura,T. (1992) Functions of signal and signal–anchor sequences are determined by the balance between the hydrophobic segment and the N-terminal charge. Proc. Natl Acad. Sci. USA, 89, 16–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A.S., Rottier,P.J.M. and Rose,J.K. (1988) Evidence for the loop model of signal-sequence insertion into the endoplasmic reticulum. Proc. Natl Acad. Sci. USA, 85, 7592–7596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Raden,D., Mandon,E.C. and Gilmore,R. (2000) Role of Sec61alpha in the regulated transfer of the ribosome-nascent chain complex from the signal recognition particle to the translocation channel. Cell, 100, 333–343. [DOI] [PubMed] [Google Scholar]
- Spiess M. (1995) Heads or tails—what determines the orientation of proteins in the membrane. FEBS Lett., 369, 76–79. [DOI] [PubMed] [Google Scholar]
- van Klompenburg W., Nilsson,I., von Heijne,G. and de Kruijff,B. (1997) Anionic phospholipids are determinants of membrane protein topology. EMBO J., 16, 4261–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. (1986) The distribution of positively charged residues in bacterial inner membrane proteins correlates with the transmembrane topology. EMBO J., 5, 3021–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlberg J.M. and Spiess,M. (1997) Multiple determinants direct the orientation of signal–anchor proteins: The topogenic role of the hydrophobic signal domain. J. Cell Biol., 137, 555–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P. and Johnson,A.E. (1994) Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell. Biol., 10, 87–119. [DOI] [PubMed] [Google Scholar]
- Wang L. and Dobberstein,B. (1999) Oligomeric complexes involved in translocation of proteins across the membrane of the endoplasmic reticulum. FEBS Lett., 457, 316–322. [DOI] [PubMed] [Google Scholar]
- Wessels H.P., Beltzer,J.P. and Spiess,M. (1991) Analysis of protein topology in the endoplasmic reticulum. Methods Cell Biol., 34, 287–302. [DOI] [PubMed] [Google Scholar]
- Zopf D., Bernstein,H.D., Johnson,A.E. and Walter,P. (1990) The methionine-rich domain of the 54 kd protein subunit of the signal recognition particle contains an RNA binding site and can be crosslinked to a signal sequence. EMBO J., 9, 4511–4517. [DOI] [PMC free article] [PubMed] [Google Scholar]