Abstract
Nearly all cells express proteins that confer resistance to the mutagenic effects of oxidative DNA damage. The primary defense against the toxicity of oxidative nucleobase lesions in DNA is the base-excision repair (BER) pathway. Endonuclease III (EndoIII) is a [4Fe–4S] cluster-containing DNA glycosylase with repair activity specific for oxidized pyrimidine lesions in duplex DNA. We have determined the crystal structure of a trapped intermediate that represents EndoIII frozen in the act of repairing DNA. The structure of the protein–DNA complex provides insight into the ability of EndoIII to recognize and repair a diverse array of oxidatively damaged bases. This structure also suggests a rationale for the frequent occurrence in certain human cancers of a specific mutation in the related DNA repair protein MYH.
Keywords: crystal structure/DNA glycosylase/DNA repair/endonuclease III/[4Fe–4S] cluster
Introduction
Oxidative damage to cellular components is an unavoidable consequence of aerobic respiration. Reactive oxygen species produced as by-products of respiration cause mutations in the genome through oxidation of DNA bases (Lindahl, 1993). It is therefore not surprising that virtually all aerobic organisms have evolved the ability to repair oxidative DNA damage. The enzyme endonuclease III (EndoIII), encoded for by the nth gene, is a ubiquitous DNA repair enzyme that repairs oxidized pyrimidine base lesions in DNA. EndoIII is a bifunctional DNA glycosylase/β-lyase belonging to the helix–hairpin–helix (HhH)-GPD superfamily (Thayer et al., 1995; Nash et al., 1996) of DNA glycosylases. The bifunctional glycosylases are characterized by the dual nature of their biochemical activity, catalysing excision of a damaged nucleobase from DNA and subsequent strand nicking 3′ to the lesion (Dodson et al., 1994). EndoIII, like all DNA glycosylases, is responsible for initiating the base-excision repair (BER) pathway at sites of DNA base damage. After the lesion is processed by a DNA glycosylase, downstream enzymes in the BER cascade complete the repair process by completely removing the processed lesion nucleoside, then repolymerizing and resealing the DNA backbone (Lindahl and Wood, 1999). The burdens of lesion recognition and base removal thus fall entirely on the DNA glycosylases. In addition to the well documented role of EndoIII in the BER pathway, recent evidence from Arabidopsis thaliana has indicated that homologues of EndoIII in plants may play a direct role in chromosomal DNA demethlyation (Gong et al., 2002) or an indirect role in genomic imprinting (Choi et al., 2002).
EndoIII was first identified in Escherichia coli as a nuclease that generates single-stranded nicks in UV-irradiated double-stranded DNA (Radman, 1975, 1976; Gates and Linn, 1977). It was soon recognized to have both N-glycosylase (base excision) and AP endonuclease (strand nicking or lyase) activities (Warner et al., 1980; Katcher and Wallace, 1983), and to contain a [4Fe–4S] cluster (Cunningham et al., 1989). Further studies determined that EndoIII has a broad substrate specificity and is most active towards oxidized pyrimidine substrates in DNA such as thymine glycol (Demple and Linn, 1980; Katcher and Wallace, 1983; Breimer and Lindahl, 1984; Weiss and Cunningham, 1985; Higgins et al., 1987; Hatahet et al., 1994). We now know that the coupled activities of base excision and subsequent strand-nicking via a β-elimination mechanism (Bailly and Verly, 1987; Kow and Wallace, 1987; Mazumder et al., 1991) are performed by a single active site lysine in the bifunctional DNA glycosylase/β-lyase (Weiss and Grossman, 1987; Dodson et al., 1994). This lysine, residue 120 in the E.coli orthologue, was proposed to serve as the key catalytic moiety in EndoIII (Kuo et al., 1992); mutagenesis studies confirmed the requirement of this residue for glycosylase/β-lyase activity, and also demonstrated the importance of another conserved active site residue, Asp138 (Thayer et al., 1995). Cloning and overexpression of the E.coli protein (Asahara et al., 1989) led to the crystal structure of EndoIII (Kuo et al., 1992; Thayer et al., 1995)—the first determined for any DNA glycosylase.
A characteristic of the bifunctional DNA glycosylases is their ability to form an irreversible covalent attachment to a DNA substrate in the presence of borohydride ion through the reduction of the Schiff base intermediate (Dodson et al., 1994; Sun et al., 1995; Nash et al., 1996) (Figure 1). This ‘borohydride trapping’ procedure has been useful in the isolation (Piersen et al., 1995; Nash et al., 1996; Bruner et al., 1998) and characterization (Sun et al., 1995; Girard et al., 1997; Nash et al., 1997; Zharkov et al., 1997; Ikeda et al., 1998; Sidorkina and Laval, 2000) of numerous bifunctional DNA glycosylases; more recently, several crystal structures of borohydride-trapped intermediates have been reported (Fromme and Verdine, 2002; Gilboa et al., 2002; Zharkov et al., 2002; Fromme et al., 2003; Verdine and Norman, 2003). These covalently complexed structures are informative both in terms of DNA substrate recognition and catalytic mechanism.
Fig. 1. Reaction pathway catalysed by EndoIII. The Schiff base intermediate 2 can be intercepted in situ by borohydride to furnish the trapped complex 4, which is the subject of this study.
Here we report the crystal structure of the borohydride-trapped intermediate of Bacillus stearothermophilus EndoIII, representing the first structure of an EndoIII–DNA complex, and the first of any protein with an iron–sulfur cluster bound to DNA. We describe the present structure in relation to that of two related but distinct DNA complexes of HhH-GPD superfamily members, human 8-oxoguanine glycosylase (hOgg1) (Bruner et al., 2000), and E.coli 3-methyladenine glycosylase (AlkA) (Hollis et al., 2000).
Results and discussion
Overall structure of the complex and DNA binding mode
We determined two structures of the EndoIII–DNA trapped intermediate with different ‘estranged’ bases opposite the lesion, and one structure of a non-covalent EndoIII–DNA complex using MIRAS and difference-Fourier techniques (see Materials and methods). We refined the structures using native X-ray data to 1.7, 2.2 and 2.5 Å (estranged-G, estranged-A and THF-Iodine1, respectively) (Table I). The structure of the EndoIII–DNA complex is shown in Figure 2. EndoIII consists entirely of α-helices and connecting loops organized into two domains, the six-helix barrel domain and the [4Fe–4S] cluster domain, so named for their characteristic features (Kuo et al., 1992). The two domains lie astride the DNA duplex, with the lesion-containing strand sandwiched between them (Figure 2A and B). The DNA-binding surface of the enzyme bears few residues with formal charge (Figure 2D), unlike most DNA-binding proteins but similar to AlkA and hOgg1 (Bruner et al., 2000; Hollis et al., 2000). The overall structure of B.stearothermophilus EndoIII in complex with DNA is very similar to that of the unbound E.coli enzyme (Thayer et al., 1995) (RMSD = 2.7 Å for all main-chain atoms), with most of the differences being ascribable to the orthologous nature of the two proteins (43% sequence identity). This lack of significant structural reorganization upon DNA binding appears to be a common feature of DNA glycosylases, irrespective of the structural family to which they belong, judging from the present comparison of liganded versus unliganded Endo III, plus comparisons of hOgg1 (Bruner et al., 2000; Bjoras et al., 2002), AlkA (Labahn et al., 1996; Hollis et al., 2000), MutM (Fpg) (Sugahara et al., 2000; Fromme and Verdine, 2002; Gilboa et al., 2002; Zharkov et al., 2002) and Udg (Mol et al., 1995; Slupphaug et al., 1996).
Table I. Data collection, phasing and refinement statistics.
| Estranged-G | Estranged-A | Native1 | THF-Iodine1 | THF-Iodine2 | ||
|---|---|---|---|---|---|---|
| Data collection statisticsa | ||||||
| Radiation source | CHESS A1 | CHESS A1 | R-AxisIV++ | R-AxisIV++ | R-AxisIV++ | |
| Resolution (Å) | 30–1.7 | 30–2.2 | 50–2.2 | 50–2.5 | 50–3.0 | |
| Unique reflections | 33 490 | 15 614 | 15 279 | 17 421 | 11 213 | |
| Completeness (%)b | 98.6 (99.6) | 97.9 (99.2) | 98.4 (97.5) | 89.0 (83.1) | 96.9 (95.3) | |
| Redundancyb | 7.3 (4.4) | 5.1 (4.5) | 6.9 (6.7) | 3.3 (2.7) | 2.9 (2.6) | |
| Rmergeb,c | 0.057 (0.418) | 0.056 (0.341) | 0.055 (0.432) | 0.056 (0.271) | 0.105 (0.332) | |
| <I/σ>a |
23.8 (3.0) |
24.0 (4.4) |
|
30.6 (4.9) |
19.6 (3.7) |
10.4 (3.1) |
| Phasing statisticsd | ||||||
| Phasing powere (centric/acentric) | 0.60/0.63 | 0.45/0.43 | ||||
| Rcullisf (centric) | 0.70 | 0.74 | ||||
| MIR figure of meritg |
|
|
0.45 |
|
|
|
| Refinement and model statistics | ||||||
| PDB accession code | 1ORN | 1ORP | 1P59 | |||
| Resolution (Å) | 30–1.7 | 30–2.2 | 30–2.5 | |||
| Rworkb,h (%) | 23.5 (32.8) | 24.1 (34.8) | 21.4 (29.5) | |||
| Rfreeb,h (%) | 24.8 (32.1) | 26.2 (37.2) | 26.9 (34.1) | |||
| Mean B-value, all atoms (Å2) | 39.5 | 57.2 | 38.3 | |||
| R.m.s.d. from ideality | ||||||
| Bond lengths (Å) | 0.007 | 0.008 | 0.008 | |||
| Bond angles (°) | 1.1 | 1.1 | 1.2 | |||
| Dihedral angles (°) | 18.9 | 19.1 | 19.1 | |||
| Ramachandran ploti (%) | ||||||
| Most favoured | 94.8 | 91.7 | 91.6 | |||
| Additionally allowed | 5.2 | 8.3 | 8.4 | |||
| Generously allowed | 0.0 | 0.0 | 0.0 | |||
| Non-water atoms | 2083 | 2004 | 2083 | |||
| Water atoms | 105 | 71 | 31 | |||
aAll native data were processed so anomalous pairs were merged, while both derivatives were processed so as to keep anomalous pairs separate. Estranged-G, Estranged-A and Native1 are trapped-intermediate complexes, while the THF-Iodine complexes contain a non-covalent abasic site analogue inhibitor (tetrahydrofuran).
bValues in parentheses refer to the highest of 10 resolution bins: 1.76–1.70 Å for 1ORN, 2.28–2.20 Å for 1ORP and 2.59–2.50 Å for 1P59.
cRmerge = Σ|I – <I>|/Σ<I>, where I is the observed intensity.
dCalculated using Native1, Iodine1 and Iodine2 data to 3.0 Å by SOLVE (Terwilliger and Berendzen, 1999). ‘Better’ statistics were obtained with MLPHARE (CCP4), but maps produced with phases from MLPHARE were not interpretable.
ePhasing power = <|FH|>/<||FPH| – |FP + FH||>, where FP is the protein structure factor amplitude, FH is the heavy-atom structure factor amplitude and FPH is the structure factor amplitude of the heavy-atom derivative.
fRcullis = Σ||FPH| – |FP + FH||/Σ|FPH – FP|
gFigure of merit = <ΣP(α)eiα/ΣP(α)>, where α is the phase and P(α) is the phase probability distribution.
hRwork = Σ|Fo – Fc|/Σ|Fo|, where Fo and Fc are the observed and calculated structure factor amplitudes, respectively. Rfree was calculated based on 8% of the total data omitted throughout structure refinement.
iValues calculated using PROCHECK (Laskowski et al., 1993).
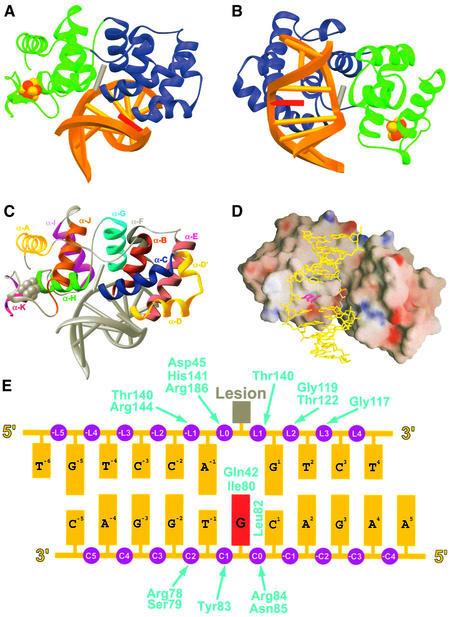
Fig. 2. Overall structure of the trapped complex. (A) Ribbon diagram, with the two domains of EndoIII in blue and green; the [4Fe–4S] cluster is shown in CPK format (Fe, rust; sulfur, yellow). The DNA duplex is shown in gold, with the lesion nucleoside modelled in dark grey (see text) and the estranged nucleoside in red. (B) Same as in (A), but from a different perspective. (C) Ribbon diagram with distinct colours for α-helices, from the same perspective as in (A). (D) Electrostatic surface representation (GRASP) (Nicholls et al., 1991) of the protein–DNA interface, from the same perspective as in (B). Red represents negatively charged surface, blue represents positive charge; only formal charges are represented. (E) DNA duplex used for crystallization, with phosphate groups in purple, the estranged base in magenta and the lesion in grey. Enzyme interactions are denoted by arrows. The numbering system corresponds to that used in the text; phosphates are numbered according to the strand on which they reside (L, lesion strand; C, complementary strand). A dihydrouridine deoxynucleoside was present at the location of the lesion during enzymatic processing, and this nucleoside sugar has become covalently and irreversibly linked to the protein via borohydride reduction.
The enzyme is bound primarily over the minor groove face of the DNA duplex, with the six-helix barrel domain invading the locally widened groove space (Figure 2A and B). The adjacent ends of helices α-B and α-C, plus the intervening loop segment, contact the estranged base and enforce extrusion of the lesion nucleoside from the DNA helix (Figures 2C, 3A and B). This invasion of the minor groove occurs at the site of a sharp bend (∼55°) in the DNA helical axis (Figure 2). Though the mode of DNA bending by EndoIII is quite similar to that seen in the AlkA and hOgg1 DNA complexes, the latter proteins introduce somewhat more drastic bends (∼70°). The overall mode of interaction of EndoIII with DNA is very similar to that seen with other members of the HhH-GPD family, thus establishing that these enzymes utilize a conserved DNA recognition motif, regardless of whether they contain a [4Fe–4S] cluster.
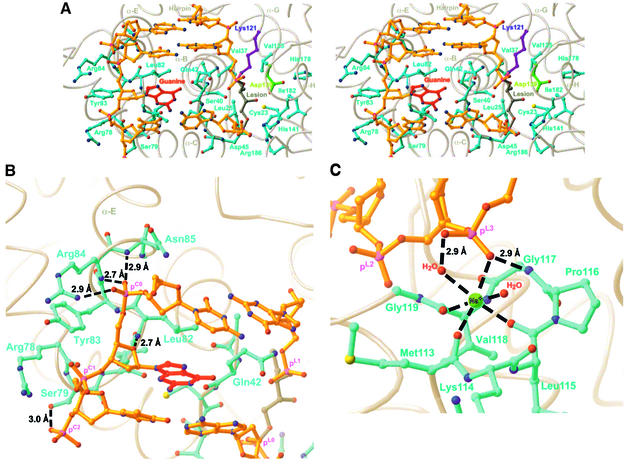
Fig. 3. (A) Global stereoview of the protein–DNA interface modelled as a ball-and-stick representation. Colouring of DNA as in Figure 2A, and side chains of the protein are coloured teal, except for Lys121 (purple) and Asp139 (green). (B) Interactions of the complementary strand of DNA with the protein. Hydrogen bonds are shown as dashed lines, and distances are as indicated. Note that phosphate group pC0 caps the N-terminus of helix α-E. (C) Role of the HhH-GPD hairpin element in DNA binding. The central metal ion is modelled as Na+, but is probably a magnesium ion under physiological conditions (both are known to exhibit octahedral geometry). Hydrogen bonds are shown as dashed lines with distances, and metal coordination is shown as dashed lines without distances (measured metal–ligand distances range from 2.3 to 2.6 Å).
There is one significant point of divergence between the EndoIII structure and that of the other structurally characterized HhH-GPD proteins. Whereas hOgg1 and AlkA contact the backbone of only the lesion-containing DNA strand, EndoIII contacts the backbone of both DNA strands. All of these proteins, including EndoIII, position the helix corresponding to α-E so that the cationic end of its helix dipole can make a favourable electrostatic interaction with the negatively charged backbone of the non-lesion-containing DNA strand (Bruner et al., 2000). However, neither AlkA nor hOgg1 use this helix to contact the DNA backbone directly. By contrast, in EndoIII, both α-E and the loop preceding it are in intimate contact with the non-lesion-containing strand. Specifically, the N-terminal end of α-E is engaged in a helix-capping interaction with the backbone phosphate pC0 (Figure 3B). Additional interactions are provided by Arg78, Ser79, Tyr83, Arg84 and Asn85.
All HhH-GPD proteins insert an amino acid side-chain from the N-terminus of α-E (or its counterpart) into the DNA duplex either at, or 5′ to, the estranged base. In EndoIII, this helix-intercalating residue is Leu81 (Figures 3B, 4A and B). HhH-GPD glycosylases also insert another residue from the aforementioned α-B/α-C loop into the DNA helix to fill the space vacated by the extrahelical nucleoside; in EndoIII, the lesion-intercalating role is served by Gln42 (Figures 3B, 4A and B), as predicted for the corresponding residue in MutY (Thayer et al., 1995). The side-chain amide of Gln41 π-stacks against the residue on the 3′-side of the lesion and hydrogen bonds with its sugar moiety. For comparison purposes, the role of Gln42 in EndoIII is performed by Leu125 in AlkA and Asn149 in hOgg1.
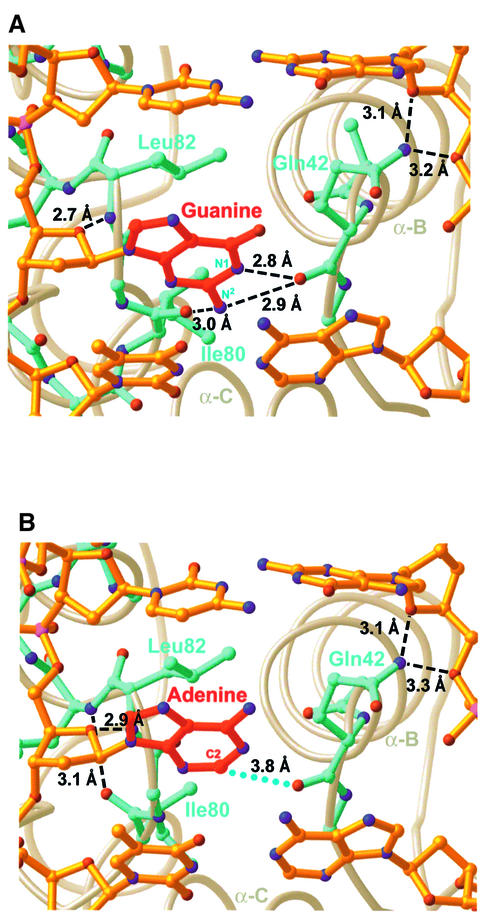
Fig. 4. Estranged base recognition. (A) Recognition of an estranged guanine base. (B) Recognition of an estranged adenine base. Dashed lines denote hydrogen bonds, with the indicated heavy atom distances; blue dots denote a van der Waals interaction. The hydrogen bond between the carbonyl of Ile80 and the amide of Tyr83 can be seen in this panel (and forms part of a type II turn), but is absent in (A).
The hallmark of the HhH-GPD DNA glycosylases is the HhH motif (Thayer et al., 1995), contained within the six-helix barrel domain (α-F through α-G in EndoIII). This motif plays an identical role in EndoIII as in the other structurally characterized members of its superfamily, namely binding to the lesion-containing DNA strand on the 3′-side of the lesion, and presenting the catalytic nucleophile (Lys121 in the B.stearothermophilus orthologue) to the substrate nucleoside inserted in the extrahelical active site (see below). The N-terminal end of helix α-G is capped by hydrogen bonding to the pL2 phosphate, and phosphate pL3 is coordinated to a metal ion held tightly in place through coordination to multiple atoms of the HhH hairpin element (Figure 3C). This metal ion, presumed to be Na+ in our structure, exhibits near-perfect octahedral geometry, being coordinated to three main-chain carbonyls, a non-bridging phosphate oxygen and two water molecules (Figure 3C). An essentially identical metal ion was observed in the AlkA co-crystal structure (Hollis et al., 2000), and is presumably present in hOgg1 as well, though it was originally thought to be a water molecule (Bruner et al., 2000). Thus, the hairpin itself presents a chelated metal ion to the DNA backbone for electrostatic association, a direct and most likely critical role in DNA binding.
Role of the iron–sulfur cluster
A subject of longstanding interest in EndoIII is its [4Fe–4S] iron–sulfur cluster (Cunningham et al., 1989). Early on, it was considered paradoxical that a DNA repair protein should deliberately deliver iron, a genotoxic oxygen-activating metal, to the surface of DNA. This issue was resolved by the finding that the iron–sulfur cluster of EndoIII appears to be redox-inert (Fu et al., 1992). Thus EndoIII became recognized as a rare example of a protein in which the iron–sulfur cluster serves to stabilize the protein fold, with no attendant redox function. Based on the crystal structure of EndoIII in the absence of DNA, plus accompanying mutagenesis data, one loop projecting from the cluster (residues C189KAQSPQC196 in B.stearothermophilus EndoIII) was proposed to interact with the DNA phosphate backbone (Thayer et al., 1995). A similar proposal was advanced in the case of the related [4Fe–4S]-containing glycosylase MutY, for which only a non-DNA-bound structure is available (Guan et al., 1998; Chepanoske et al., 2000). Inspection of the present EndoIII co-crystal structure, the first of a [4Fe–4S]-containing glycosylase bound to DNA, provides suggestive evidence in support of this hypothesis, as the cluster-bound loop presents its polar residues Lys190 and Gln192 in the general vicinity of the DNA backbone. Interestingly, most of the residues on this loop are not well ordered in the DNA-bound complex and furthermore are not strictly conserved among different EndoIII orthologues (Asahara et al., 1989; Sorokin et al., 1996; Aspinwall et al., 1997; Hilbert et al., 1997).
Recognition of the estranged base
Bacillus stearothermophilus EndoIII binds most tightly to lesion-containing DNA with a guanine opposite the lesion (estranged-G), somewhat less tightly to a duplex with an estranged adenine, and very weakly to DNA with either cytosine or thymine in the estranged position (data not shown), consistent with the primary role of EndoIII in repairing oxidized pyrimidines. We have determined two different trapped-intermediate complexes: one with an estranged guanine (estranged-G complex) and the other with an estranged adenine (estranged-A complex). Many DNA glycosylases exhibit at least some specificity for the estranged base, and indeed many co-crystal structures of glycosylase-DNA complexes exhibit specific intermolecular interactions between the estranged base and amino acid side-chains of the enzyme (Bruner et al., 2000; Fromme and Verdine, 2002; Zharkov et al., 2002). Instead of using side-chains to recognize the estranged base, EndoIII employs only backbone carbonyls (Figure 4A); a similar strategy is employed by the mismatch-specific uracil glycosylase MUG (Barrett et al., 1998). In the estranged-G structure, the nucleobase N1-H and N2-H atoms appear to donate hydrogen bonds to the backbone carbonyl of Gln42 (Figure 4A), and N2-H donates a hydrogen bond to the backbone carbonyl of Ile80. Most of these interactions are not available to an estranged adenine, due to its lone pair of electrons at N1 and lack of an exocyclic amine at position 2. Consequently, the estranged adenine is retracted from the protein backbone, and the protein conformation near Ile80 rearranges to form a type II turn (Figure 4B); the latter conformation is present in the structure of non-DNA-bound EndoIII (Thayer et al., 1995), enforced by a conserved glycine at position 81. These differences in interactions of the protein with the estranged base, and perhaps in DNA strain energy, are the likely source of the preference to bind G rather than A in the estranged position. Other factors, such as base-stacking energies of adenine versus guanine, may also play a role. These structures, however, provide no obvious explanation for purines being favoured over pyrimidines in the estranged position.
The active site
As is characteristic of all known DNA glycosylases acting on single-base lesions, the enzyme has extruded the lesion nucleoside from the DNA duplex and inserted it into an extrahelical active site on the enzyme. The covalent bond between the catalytic nucleophile, Lys121, and C1′ of the lesion is clearly evident from the electron density (Figure 5A and B). The other key catalytic residue, Asp139, is observed to serve a characteristic role as an N-terminal helix cap for helix α-H (Norman et al., 2003). Asp138 is also hydrogen-bonded to the O4′-hydroxyl group of the lesion sugar moiety, now available as a hydrogen-bond donor due to ring-opening in the step leading to Schiff base formation. Electron density corresponding to the dihydrouridine (DHU) base excised during the initiation step of repair is not visible in the active site, presumably because it was released following excision. Dihydrouracil free base did not load into the active site by either crystal soaking or co-crystallization, suggesting that this product base does not bind the active site tightly. This is in contrast to the active site of the borohydride-trapped hOgg1–DNA complex, which not only retains the expelled oxoG for hours but also binds purine analogues upon crystal soaking. We have reported that the product purine base assists β-lyase catalysis by hOgg1 (Fromme et al., 2003), explaining the need for persistent binding of the expelled nucleobase; however, we could find no biochemical evidence for product-assisted catalysis in the case of B.stearothermophilus EndoIII. Furthermore, in the absence of a product base, the rate of strand cleavage by EndoIII is several orders of magnitude faster than by hOgg1 (kobs > 0.5 min–1 and kobs = 0.001 min–1, respectively; see Materials and methods), indicating that EndoIII has no need to accelerate the β-lyase cascade by product assistance.
Fig. 5. (A) Stereoview of the final 1.7 Å, σA-weighted (Read, 1986) 2Fo – Fc electron density map contoured at 1.25 σ, superimposed on the active site region of the final estranged-G model. (B) Active site region of the enzyme. The C-N bond of the reduced Schiff base is shown in pink. Contacts between the 3′-phosphate and the amide protons of Gln42 and Thr140, and between the 5′-phosphate and the amide proton of Asp45 are not shown explicitly, for reasons of illustrative clarity. The positions of the C2′ protons, though not visible in experimental electron density maps, are inferred from the geometry of the heavy atom backbone. (C) Structural superimposition of the non-covalent complex (THF-Iodine1) onto the covalent estranged-G complex. The non-covalent complex is coloured similarly to (B) and the covalent complex (estranged-G) is dark grey.
The phosphate groups flanking the lesion nucleoside are contacted by several residues of the enzyme, thereby orienting the lesion nucleoside for insertion into the active site (Figure 5B). Namely, His141 hydrogen bonds to a non-bridging oxygen atom of the 5′-phosphate (pL0), an interaction strictly homologous to that made by His271 in hOgg1. The 5′-bridging oxygen of pL0 is also hydrogen bonded to Arg186. The amide protons of Asp45 and Thr140 are hydrogen bonded to the non-bridging oxygens of pL0 and pL1, respectively. Additional phosphate interactions are provided by an extensive network of ordered water molecules observed at the DNA–enzyme interface.
We also determined the structure of a non-covalent complex between EndoIII and a DNA duplex containing the tetrahydrofuran (THF) abasic site analogue. This structure does not represent any particular step in the EndoIII reaction pathway, but highlights any possible effects of covalent linkage between the enzyme and DNA. This structure is virtually identical to that of the covalent complex (r.m.s.d. of 0.4 Å for all atoms), except for minor differences in the active site region, some of which are due to the differences in covalent structure between the covalent and non-covalent complexes (Figure 5C). There are two other noticeable differences: (i) the side-chain conformation of Asp139 is rotated slightly but maintains its role as a helix cap for helix α-H; and (ii) Asp45 is situated closer to the lesion nucleotide in the non-covalent structures, due to a shift in the protein backbone at this position. The different Asp139 side-chain rotamer is possibly due to the slight difference in lesion nucleoside O4′ position between the covalent and non-covalent complex structures: in the former, the opened deoxyribose ring positions O4′ closer to Asp139, allowing interaction between the two. In the latter structure, the tetrahydrofuran ring constrains O4′ from close contact with Asp139. The acid group of Asp139 is directed towards Lys121, but does not make a close contact with this residue (4.6 Å between Nζ and Oδ2). Regarding the movement of Asp45, it is not clear what causes the change in position, but it is interesting in light of the presently proposed role of this residue in catalysis (see below).
The ring-opened sugar in our reduced intermediate complex is disposed such that one of the C-H bonds at C2′ (that to the pro-R proton) is trans-antiperiplanar to the C3′–O3′ bond, a favourable geometry for concerted β-elimination resulting in 3′-strand scission (Figure 4B). Using a published procedure (Fromme et al., 2003), we determined that B.stearothermophilus EndoIII does indeed stereospecifically remove the pro-R hydrogen at C2′ with 97% selectivity (±1.1% for a P-value of 0.05). Although the present structures reveal the geometry of a reduced species (Figure 1, ‘4’), and not of the actual Schiff base intermediate (Figure 1, ‘2’), it may be that the sugar adopts a similar conformation in both. Curiously, E.coli EndoIII has been reported to remove the pro-S hydrogen at C2′ (Mazumder et al., 1991).
What are the most likely candidates for an acid/base moiety to catalyse the elimination reaction via proton abstraction at C2′? The acid group of Asp139 is 4.5 Å away from C2′, but has been shown to be dispensable for β-lyase activity in other DNA glycosylases: Asp268 in hOgg1 (Norman et al., 2003) and Glu3 in EndoVIII (Zharkov et al., 2002). An ordered water molecule located only 3.8 Å away from C2′ is hydrogen bonded to the acid group of Asp45 (C2′-O distance 2.6 Å; Figure 5B). Asp45 is conserved in EndoIII across all species, from bacteria to humans, but is not conserved in the closely related MutY/MYH family of monofunctional DNA glycosylases, members of which are devoid of β-elimination activity. It is conceivable that the water molecule and Asp45 act as a proton shuttle to deprotonate C2′, a hypothesis made more appealing by the direct location of the water molecule above the pro-R proton at C2′.
Lesion–base recognition
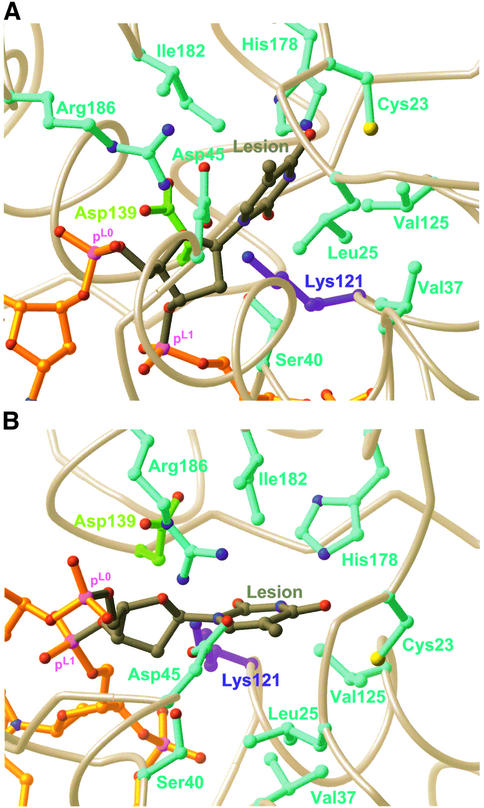
Though the present structures contain no lesion base, they do contain an active site pocket suitably disposed for interaction with a lesion base. To gain insight into the molecular origins of lesion-specific recognition, we modelled a thymine nucleoside into the active site, using as a reference structure the hOgg1-oxoG DNA recognition complex (Bruner et al., 2000) (EndoIII operates on multiple substrates, several of which are closely related in structure to thymine). We then mutated Lys121 to alanine, performed a CHARMm minimization (MacKerell et al., 1998), and then restored Lys121 in order to generate the model for Figure 6. The model is obviously not a suitable basis for speculation on specific details, but it does suggest some general comments regarding the nature of the lesion base recognition pocket. First, the pocket contains both polar residues (Cys23, Asp45, His178 and Arg176) and branched aliphatic hydrophobic residues (Leu25, Val37, Val125 and Ile182). The enzyme’s broad substrate specificity may be accounted for by the amphiphilic character of the recognition site. Enzymes like EndoIII and AlkA, which process a variety of structurally dissimilar lesions, may take advantage of the higher lability of the glycosidic bonds in their cognate lesions as a key aspect of their substrate recognition strategy (Berdal et al., 1998).
Fig. 6. (A) Structure of the active site, with a thymine base modelled into the base-recognition pocket. (B) Same as in (A), from a different perspective.
Implications for human cancer
A specific mutation in the BER gene encoding the human MutY homologue MYH is found in numerous unrelated patients displaying multiple colorectal adenomas and carcinoma (Al-Tassan et al., 2002; Jones et al., 2002). Though no structure of a MutY–DNA complex is available, the structure of the E.coli MutY catalytic core has been determined (Guan et al., 1998). It is therefore possible to map the primary human mutation (Y165C) onto the present structure of an EndoIII–DNA complex, using both sequence and structural alignments; the corresponding amino acid in B.stearothermophilus EndoIII is Leu82. As discussed above, the side chain of Leu82 wedges itself into the minor groove on the 5′-side of the estranged base, and is actually in van der Waals contact with this base (Figures 3A and B, 4A and B). In the case of MutY/MYH, the estranged base would be oxoG (its partner adenine residue would be inserted into the enzyme active site). It comes as no surprise that the residue corresponding to Leu82 should be critical for proper MutY function, because the EndoIII structure suggests that intercalation of this side chain stabilizes the highly deformed DNA duplex conformation to which the enzyme binds. A tyrosine residue is appropriate for this role because it can provide additional stabilization through π-stacking interactions; mutation of tyrosine to cysteine at this position would reduce both the steric and electronic ability of the residue to stabilize the DNA conformation necessary for efficient catalysis in MutY. It has been suggested that MYH might actually force extrusion of the estranged oxoG from the DNA helix (Bernards et al., 2002), thereby allowing it to be inserted into an estranged base recognition pocket on a separate domain found only in MutY orthologues. If so, a large residue such as Tyr could fill the void vacated by the estranged base, and a smaller residue such as Cys would be less effective in such a role.
Conclusions
The crystal structures presented here have elucidated the interaction of EndoIII with lesion-containing DNA. Comparison of the present structure to those of other family members has revealed some common themes of DNA recognition among the HhH-GPD superfamily of DNA glycosylases. Despite the substantial differences in the chemical nature of the lesions recognized, and in certain critical features of the protein folds, the overall architecture of the lesion-recognition complexes formed by these proteins is remarkably conserved. Finally, the present structure provides a rationale for understanding the association of the Y165C mutation in the human MYH gene with certain hereditary and spontaneous cancers.
Materials and methods
Cloning of endonuclease III and protein purification
The nth gene was identified in the B.stearothermophilus genome by a BLAST search (http://www.genome.ou.edu/bstearo.html). Colony PCR was performed on B.stearothermophilus cells using the primers 5′-GGCTAACATATGGTGCTGACAAAGCAACAAATCCGCTACTGT TTGG-3′ and 5′-TTATTATGCGGCCGCTCACTTCTGATTTGCCGC TTTTTCCTCCCG-3′, and the resulting product was inserted into the pET28 (Novagen) expression vector using NdeI and NotI (New England Biolabs). This resulted in an ORF with an N-terminal His tag and thrombin protease cleavage site followed by the full-length gene. The plasmid was transformed into C41(DE3) cells (Miroux and Walker, 1996), and expression was carried out at 37°C for 4 h following induction with 1 mM IPTG. Cells were harvested by centrifugation, resuspended in Lysis Buffer (50 mM sodium phosphate pH 8.0, 500 mM sodium chloride, 10 mM imidazole and 0.1% (v/v) βME) and frozen at –80°C for storage. All remaining steps were carried out at 4°C. The cell suspension was thawed in cold water and lysed by French press after the addition of 1 mM PMSF. DNase was added and the lysate was clarified by centrifugation at 20 000 g for 25 min. The clarified lysate was incubated with 2 ml of Ni2+-NTA resin (Qiagen) for 1 h. The resin was washed with lysis buffer supplemented with imidazole to 20 mM. His-tagged protein was eluted with a buffer containing 20 mM sodium phosphate pH 8.0, 40 mM Tris pH 8.0, 500 mM sodium chloride, 250 mM imidazole and 0.1% (v/v) βME. The elution was buffer-exchanged into a solution containing 20 mM Tris pH 7.4, 150 mM sodium chloride, 0.1% (v/v) βME, and loaded onto a MonoS column (Pharmacia) pre-equilibrated in the same buffer. A buffered sodium chloride gradient was applied to the column to elute His-tagged EndoIII. An overnight thrombin digest was performed to remove the His-tag and the protein was again purified using a MonoS column. Fractions containing EndoIII were pooled and buffer-exchanged into a solution containing 20 mM Tris pH 7.4, 500 mM sodium chloride and 0.1% (v/v) βME for storage (the protein was stable for ∼1 week at 4°C under these conditions). The purified protein was used in DNA cleavage assays as described previously (Norman et al., 2003), but using dihydrouridine-containing DNA as a substrate, in order to confirm that it possesses characteristic EndoIII enzymatic activity.
β-lyase rate measurement
32P-labelled abasic site-containing DNA was prepared as described previously (Fromme et al., 2003). Reactions between B.stearothermophilus EndoIII or hOgg1 and DNA were allowed to proceed under single turnover conditions at 8°C, and quenched with the addition of 1 vol. of 95% formamide, 1× TBE solution. We have previously demonstrated that hOgg1 Schiff base formation with an abasic site is fast and that subsequent β-lyase activity is rate limiting (Fromme et al., 2003). Reactions were resolved on 8 M urea, 20% PAGE gels in 1 × TBE, imaged using a phosphoimager, and reactant/product ratios were quantified using MacBas (Fuji). Rates were extracted using the equation: P(t) = P∞ – P∞ · e–kt, where P(t) is the amount of product formed at time t. The kobs for B.stearothermophilus EndoIII could only be determined as a lower limit, owing to the speed of the reaction.
DNA preparation, complex formation and crystallization
The DNA oligomers 5′-TGTCCAXGTCT-3′ [where X is either 5,6-dihydro-deoxyuridine (DHU) or a tetrahydrofuran (THF) abasic-site; phosphoramidites purchased from Glen Research] and 5′-AAG ACYTGGAC-3′ [where Y is either G (for estranged-G complex) or A (for estranged-A complex)] were synthesized on an ABI 392 DNA synthesizer using standard ‘ultra-mild’ reagents. The underlined positions in the sequences indicate where 5-iodo-deoxyuridine (phosphoramidite; purchased from Glen Research) replaced thymidine in DNA used for derivative crystals. All oligomers were purified by urea–PAGE gels, dissolved in 10 mM Tris pH 8.0, and annealed to each other at a concentration of 1 mM. The reductively trapped covalent intermediate complex was prepared by adding a solution containing 30 nmol of DHU-containing duplex DNA and 80 µmol of sodium borohydride to a solution containing 12 nmol of EndoIII, so that the final composition of the buffer solution was 5 mM Tris pH 7.4, 150 mM sodium chloride, 5% (v/v) glycerol, 0.03% (v/v) βME and 100 mM sodium borohydride. The reaction was allowed to proceed at 4°C until judged complete by SDS–PAGE analysis (<3 h). The covalent complex was separated from free DNA using a MonoQ column (Pharmacia), buffer exchanged into 20 mM Tris pH 7.4, 150 mM sodium chloride and 0.1% βME, and concentrated to 230 µM. For iodine derivative complexes, THF-containing iodinated duplex DNA was mixed in 1.5 molar excess with EndoIII to form a non-covalent complex in final buffer solution which was identical in composition to that of the covalent complex. All complexes were crystallized by the hanging drop vapour diffusion method at room temperature using a solution of 100 mM Tris pH 8.5, 20–25% (w/v) PEG 8000 and 5 mM βME. Crystals appeared after several days and grew to final dimensions of approximately 200 × 200 × 300 µm over several weeks. Crystals were serially transferred through solutions with increasing amounts of glycerol into a final cryoprotectant solution of 100 mM Tris pH 8.5, 25% (w/v) PEG 8000, 5 mM βME and 20% glycerol, and then frozen in liquid nitrogen for X-ray data collection.
Data collection and structure solution
Data were collected on a Rigaku R-Axis IV++ detector at Enanta Pharmaceuticals, and at the A1 beamline of the Cornell High Energy Synchrotron Source (CHESS). Data was processed and reduced using HKL2000 or HKL 1.97, respectively (Otwinowski and Minor, 1997). The crystals belong to the orthorhombic space group P21212 with unit cell dimensions of a = 68 Å, b = 107 Å and c = 42 Å. Data collection statistics are summarized in Table I. We were unable to solve the structure by molecular replacement methods [using the structure of 43% sequence-identical E.coli EndoIII (Thayer et al., 1995), Protein Data Bank (PDB) accession code 2ABK, and many variations thereof as a probe], despite attempting the search with several different software packages [CNS 1.0 (Brunger et al., 1998), and the programs Molrep (Vagin and Teplyakov, 1997), AMORE (Navaza, 1994) and BEAST (Read, 2001) bundled with CCP4 (CCP4, 1994)]. Single-wavelength anomalous data were collected on three different estranged-G crystals to yield one native dataset (‘Native1’; Table I) and two derivative datasets. The derivatives were crystals in which either two (‘THF-Iodine2’; Table I) or three (‘THF-Iodine1’; Table I) of the thymine bases were replaced with 5-iodo-uracil, and an abasic site analogue (THF) was present instead of a covalent enzyme–DNA bond. Iodine positions were unambiguously determined using anomalous data in SOLVE (Terwilliger et al., 1987; Terwilliger and Berendzen, 1999). Heavy-atom refinement (ignoring anomalous differences) and MIRAS phasing (using anomalous differences) were then carried out using data to 3.0 Å in SOLVE. Phases were improved using DM (Cowtan, 1994) in CCP4. Electron density maps calculated using these phases were poor, but density for the DNA was clearly observed. The lesion-containing DNA from the structure of a covalent hOgg1–DNA complex (Fromme et al., 2003) (PDB accession code 1HU0) was manually fit as a rigid body into the density using the iodine positions as a guide. Model phases were calculated to 2.2 Å from this DNA model and combined with MIRAS phases using SIGMAA (Read, 1986). Combined phases were improved with DM, and new electron density maps revealed clear protein backbone traces for ∼70% of the protein. Corresponding features of 2ABK were manually fit into the density and remodelled as appropriate. Iterative rounds of simulated-annealing and grouped B-factor refinement using CNS, and model building in Quanta98 (Molecular Simulations) were carried out until the Rfree (Brunger, 1993) dropped to 0.35. Individual B-factor and conjugant-gradient energy minimization refinement using CNS were then included in subsequent rounds. At this time, higher resolution data of the estranged-G complex and data of the estranged-A complex collected using synchrotron radiation became available. These structures were determined using difference-Fourier methods and the refinement was performed, beginning with simulated annealing in the same manner as detailed in the later stages above. In addition, the THF-Iodine1 data was used to refine a model of non-covalently bound enzyme–DNA complex. Water molecules were added to the models using both automated methods (in CNS) and manual inspection of difference maps. Amino acid side-chains of some surface residues were truncated at the α-, β-, γ- or δ-carbon position if electron density was not visible for the other atoms. The final models consist of amino acid residues 1–214 (estranged-G and THF-Iodine1) or 1–213 (estranged-A), and 21 nucleotides of DNA; phasing, refinement and model statistics are presented in Table I. Figures were prepared using Ribbons (Carson, 1997) and GRASP (Nicholls et al., 1991). Coordinates of the three complexes have been deposited in the PDB (1ORN for the estranged-G complex, 1ORP for the estranged-A complex, and 1P59 for the THF-Iodine1 complex).
Acknowledgments
Acknowledgements
We are indebted to Yakov Korkhin for assistance with data collection and invaluable advice. Bill Miller provided helpful assistance at the CHESS A1 beamline. We thank Aaron Handler for assistance with protein purification, gel shifts and crystal screens. We are grateful to Alexander Schiffer for advice on refinement of the iron–sulfur cluster in CNS. Wei Yang performed the energy minimization of the modelled substrate recognition complex. We thank Derek Norman and Steven Bruner for critical reading of the manuscript. We thank Enanta Pharmaceuticals for use of their X-ray generator and detector apparatus. J.C.F was funded by an National Science Foundation (NSF) pre-doctoral fellowship and a Merck-Wiley fellowship. This work is based upon research conducted at CHESS, which is supported by the NSF and the National Institutes of Health/National Institute of General Medical Sciences under award No. DMR 9713424.
References
- Al-Tassan N. et al. (2002) Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nat. Genet., 30, 227–232. [DOI] [PubMed] [Google Scholar]
- Asahara H., Wistort,P.M., Bank,J.F., Bakerian,R.H. and Cunningham,R.P. (1989) Purification and characterization of Escherichia coli endonuclease III from the cloned nth gene. Biochemistry, 28, 4444–4449. [DOI] [PubMed] [Google Scholar]
- Aspinwall R. et al. (1997) Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc. Natl Acad. Sci. USA, 94, 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly V. and Verly,W.G. (1987) Escherichia coli endonuclease III is not an endonuclease but a beta-elimination catalyst. Biochem. J., 242, 565–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T.E., Savva,R., Panayotou,G., Barlow,T., Brown,T., Jiricny,J. and Pearl,L.H. (1998) Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell, 92, 117–129. [DOI] [PubMed] [Google Scholar]
- Berdal K.G., Johansen,R.F. and Seeberg,E. (1998) Release of normal bases from intact DNA by a native DNA repair enzyme. EMBO J., 17, 363–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernards A.S., Miller,J.K., Bao,K.K. and Wong,I. (2002) Flipping duplex DNA inside out: a double base-flipping reaction mechanism by Escherichia coli MutY adenine glycosylase. J. Biol. Chem., 277, 20960–20964. [DOI] [PubMed] [Google Scholar]
- Bjoras M., Seeberg,E., Luna,L., Pearl,L.H. and Barrett,T.E. (2002) Reciprocal ‘flipping’ underlies substrate recognition and catalytic activation by the human 8-oxo-guanine DNA glycosylase. J. Mol. Biol., 317, 171–177. [DOI] [PubMed] [Google Scholar]
- Breimer L.H. and Lindahl,T. (1984) DNA glycosylase activities for thymine residues damaged by ring saturation, fragmentation, or ring contraction are functions of endonuclease III in Escherichia coli. J. Biol. Chem., 259, 5543–5548. [PubMed] [Google Scholar]
- Bruner S.D., Nash,H.M., Lane,W.S. and Verdine,G.L. (1998) Repair of oxidatively damaged guanine in Saccharomyces cerevisiae by an alternative pathway. Curr. Biol., 8, 393–403. [DOI] [PubMed] [Google Scholar]
- Bruner S.D., Norman,D.P. and Verdine,G.L. (2000) Structural basis for recognition and repair of the endogenous mutagen 8-oxoguanine in DNA. Nature, 403, 859–866. [DOI] [PubMed] [Google Scholar]
- Brunger A. (1993) Assessment of phase accuracy by cross validation: the free R value. Methods and applications. Acta Crystallogr. D, 49, 24–36. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Carson M. (1997) Ribbons. Methods Enzymol., 277, 493–505. [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 Suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chepanoske C.L., Golinelli,M.P., Williams,S.D. and David,S.S. (2000) Positively charged residues within the iron-sulfur cluster loop of E.coli MutY participate in damage recognition and removal. Arch. Biochem. Biophys., 380, 11–19. [DOI] [PubMed] [Google Scholar]
- Choi Y., Gehring,M., Johnson,L., Hannon,M., Harada,J.J., Goldberg,R.B., Jacobsen,S.E. and Fischer,R.L. (2002) DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell, 110, 33–42. [DOI] [PubMed] [Google Scholar]
- Cowtan K. (1994) ‘dm’: an automated procedure for phase improvement by density modification. Jt CCP4 ESF-EACBM Newslett. Protein Crystallogr., 31, 34–38. [Google Scholar]
- Cunningham R.P. et al. (1989) Endonuclease III is an iron-sulfur protein. Biochemistry, 28, 4450–4455. [DOI] [PubMed] [Google Scholar]
- Demple B. and Linn,S. (1980) DNA N-glycosylases and UV repair. Nature, 287, 203–208. [DOI] [PubMed] [Google Scholar]
- Dodson M.L., Michaels,M.L. and Lloyd,R.S. (1994) Unified catalytic mechanism for DNA glycosylases. J. Biol. Chem., 269, 32709–32712. [PubMed] [Google Scholar]
- Fromme J.C. and Verdine,G.L. (2002) Structural insights into lesion recognition and repair by the bacterial 8-oxoguanine DNA glycosylase MutM. Nat. Struct. Biol., 9, 544–552. [DOI] [PubMed] [Google Scholar]
- Fromme J.C., Bruner,S.D., Yang,W., Karplus,M. and Verdine,G.L. (2003) Product-assisted catalysis in base-excision DNA repair. Nat. Struct. Biol., 10, 204–211. [DOI] [PubMed] [Google Scholar]
- Fu W., O’Handley,S., Cunningham,R.P. and Johnson,M.K. (1992) The role of the iron-sulfur cluster in Escherichia coli endonuclease III. A resonance Raman study. J. Biol. Chem., 267, 16135–16137. [PubMed] [Google Scholar]
- Gates F.T. and Linn,S. (1977) Endonuclease from Escherichia coli that acts specifically upon duplex DNA damaged by ultraviolet light, osmium tetroxide, acid, or X-rays. J. Biol. Chem., 252, 2802–2807. [PubMed] [Google Scholar]
- Gilboa R., Zharkov,D.O., Golan,G., Fernandes,A.S., Gerchman,S.E., Matz,E., Kycia,J.H., Grollman,A.P. and Shoham,G. (2002) Structure of formamidopyrimidine-DNA glycosylase covalently complexed to DNA. J. Biol. Chem., 277, 19811–19816. [DOI] [PubMed] [Google Scholar]
- Girard P.M., Guibourt,N. and Boiteux,S. (1997) The Ogg1 protein of Saccharomyces cerevisiae: a 7,8-dihydro-8-oxoguanine DNA glycosylase/AP lyase whose lysine 241 is a critical residue for catalytic activity. Nucleic Acids Res., 25, 3204–3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z., Morales-Ruiz,T., Ariza,R.R., Roldan-Arjona,T., David,L. and Zhu,J.-K. (2002) ROS1, a Repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell, 111, 803–814. [DOI] [PubMed] [Google Scholar]
- Guan Y., Manuel,R.C., Arvai,A.S., Parikh,S.S., Mol,C.D., Miller,J.H., Lloyd,S. and Tainer,J.A. (1998) MutY catalytic core, mutant and bound adenine structures define specificity for DNA repair enzyme superfamily. Nat. Struct. Biol., 5, 1058–1064. [DOI] [PubMed] [Google Scholar]
- Hatahet Z., Kow,Y.W., Purmal,A.A., Cunningham,R.P. and Wallace,S.S. (1994) New substrates for old enzymes. 5-Hydroxy-2′-deoxycytidine and 5-hydroxy-2′-deoxyuridine are substrates for Escherichia coli endonuclease III and formamidopyrimidine DNA N-glycosylase, while 5-hydroxy-2′-deoxyuridine is a substrate for uracil DNA N-glycosylase. J. Biol. Chem., 269, 18814–18820. [PubMed] [Google Scholar]
- Higgins S.A., Frenkel,K., Cummings,A. and Teebor,G.W. (1987) Definitive characterization of human thymine glycol N-glycosylase activity. Biochemistry, 26, 1683–1688. [DOI] [PubMed] [Google Scholar]
- Hilbert T.P., Chaung,W., Boorstein,R.J., Cunningham,R.P. and Teebor,G.W. (1997) Cloning and expression of the cDNA encoding the human homologue of the DNA repair enzyme, Escherichia coli endonuclease III. J. Biol. Chem., 272, 6733–6740. [DOI] [PubMed] [Google Scholar]
- Hollis T., Ichikawa,Y. and Ellenberger,T. (2000) DNA bending and a flip-out mechanism for base excision by the helix–hairpin–helix DNA glycosylase, Escherichia coli AlkA. EMBO J., 19, 758–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S., Biswas,T., Roy,R., Izumi,T., Boldogh,I., Kurosky,A., Sarker,A.H., Seki,S. and Mitra,S. (1998) Purification and characterization of human NTH1, a homolog of Escherichia coli endonuclease III. Direct identification of Lys-212 as the active nucleophilic residue. J. Biol. Chem., 273, 21585–21593. [DOI] [PubMed] [Google Scholar]
- Jones S., Emmerson,P., Maynard,J., Best,J.M., Jordan,S., Williams,G.T., Sampson,J.R. and Cheadle,J.P. (2002) Biallelic germline mutations in MYH predispose to multiple colorectal adenoma and somatic G:C→T:A mutations. Hum. Mol. Genet., 11, 2961–2967. [DOI] [PubMed] [Google Scholar]
- Katcher H.L. and Wallace,S.S. (1983) Characterization of the Escherichia coli X-ray endonuclease, endonuclease III. Biochemistry, 22, 4071–4081. [DOI] [PubMed] [Google Scholar]
- Kow Y.W. and Wallace,S.S. (1987) Mechanism of action of Escherichia coli endonuclease III. Biochemistry, 26, 8200–8206. [DOI] [PubMed] [Google Scholar]
- Kuo C.F., McRee,D.E., Fisher,C.L., O’Handley,S.F., Cunningham,R.P. and Tainer,J.A. (1992) Atomic structure of the DNA repair [4Fe–4S] enzyme endonuclease III. Science, 258, 434–440. [DOI] [PubMed] [Google Scholar]
- Labahn J., Scharer,O.D., Long,A., Ezaz-Nikpay,K., Verdine,G.L. and Ellenberger,T.E. (1996) Structural basis for the excision repair of alkylation-damaged DNA. Cell, 86, 321–329. [DOI] [PubMed] [Google Scholar]
- Laskowski R.J., Macarthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–290. [Google Scholar]
- Lindahl T. (1993) Instability and decay of the primary structure of DNA. Nature, 362, 709–715. [DOI] [PubMed] [Google Scholar]
- Lindahl T. and Wood,R.D. (1999) Quality control by DNA repair. Science, 286, 1897–1905. [DOI] [PubMed] [Google Scholar]
- MacKerell A.D. Jr et al. (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B, 102, 3586–3616. [DOI] [PubMed] [Google Scholar]
- Mazumder A., Gerlt,J.A., Absalon,M.J., Stubbe,J., Cunningham,R.P., Withka,J. and Bolton,P.H. (1991) Stereochemical studies of the β-elimination reactions at aldehydic abasic sites in DNA: endonuclease III from Escherichia coli, sodium hydroxide and Lys-Trp-Lys. Biochemistry, 30, 1119–1126. [DOI] [PubMed] [Google Scholar]
- Miroux B. and Walker,J.E. (1996) Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol., 260, 289–298. [DOI] [PubMed] [Google Scholar]
- Mol C.D., Arvai,A.S., Slupphaug,G., Kavli,B., Alseth,I., Krokan,H.E. and Tainer,J.A. (1995) Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. Cell, 80, 869–878. [DOI] [PubMed] [Google Scholar]
- Nash H.M., Bruner,S.D., Scharer,O.D., Kawate,T., Addona,T.A., Spooner,E., Lane,W.S. and Verdine,G.L. (1996) Cloning of a yeast 8-oxoguanine DNA glycosylase reveals the existence of a base-excision DNA-repair protein superfamily. Curr. Biol., 6, 968–980. [DOI] [PubMed] [Google Scholar]
- Nash H.M., Lu,R., Lane,W.S. and Verdine,G.L. (1997) The critical active-site amine of the human 8-oxoguanine DNA glycosylase, hOgg1: direct identification, ablation and chemical reconstitution. Chem. Biol., 4, 693–702. [DOI] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Norman D.P.G., Chung,S.J. and Verdine,G.L. (2003) Structural and biochemical exploration of a critical amino acid in human 8-oxoguanine glycosylase. Biochemistry, 42, 1564–1572. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Piersen C.E., Prince,M.A., Augustine,M.L., Dodson,M.L. and Lloyd,R.S. (1995) Purification and cloning of Micrococcus luteus ultraviolet endonuclease, an N-glycosylase/abasic lyase that proceeds via an imino enzyme–DNA intermediate. J. Biol. Chem., 270, 23475–23484. [DOI] [PubMed] [Google Scholar]
- Radman M. (1975) Endonuclease III: an endonuclease from Escherichia coli that introduces single polynucleotide chain scissions in ultraviolet-irradiated DNA. Basic Life Sci., 5A, 197–200. [DOI] [PubMed] [Google Scholar]
- Radman M. (1976) An endonuclease from Escherichia coli that introduces single polynucleotide chain scissions in ultraviolet-irradiated DNA. J. Biol. Chem., 251, 1438–1445. [PubMed] [Google Scholar]
- Read R.J. (1986) Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. A, 42, 140–149. [Google Scholar]
- Read R.J. (2001) Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D, 57, 1373–1382. [DOI] [PubMed] [Google Scholar]
- Sidorkina O.M. and Laval,J. (2000) Role of the N-terminal proline residue in the catalytic activities of the Escherichia coli Fpg protein. J. Biol. Chem., 275, 9924–9929. [DOI] [PubMed] [Google Scholar]
- Slupphaug G., Mol,C.D., Kavli,B., Arvai,A.S., Krokan,H.E. and Tainer,J.A. (1996) A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature, 384, 87–92. [DOI] [PubMed] [Google Scholar]
- Sorokin A., Azevedo,V., Zumstein,E., Galleron,N., Ehrlich,S.D. and Serror,P. (1996) Sequence analysis of the Bacillus subtilis chromosome region between the serA and kdg loci cloned in a yeast artificial chromosome. Microbiology, 142, 2005–2016. [DOI] [PubMed] [Google Scholar]
- Sugahara M., Mikawa,T., Kumasaka,T., Yamamoto,M., Kato,R., Fukuyama,K., Inoue,Y. and Kuramitsu,S. (2000) Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. EMBO J., 19, 3857–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun B., Latham,K.A., Dodson,M.L. and Lloyd,R.S. (1995) Studies on the catalytic mechanism of five DNA glycosylases. Probing for enzyme-DNA imino intermediates. J. Biol. Chem., 270, 19501–19508. [DOI] [PubMed] [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T.C., Kim,S.-H. and Eisenberg,D. (1987) Generalized method of determining heavy-atom positions using the difference Patterson function. Acta Crystallogr. A, 43, 1–5. [Google Scholar]
- Thayer M.M., Ahern,H., Xing,D., Cunningham,R.P. and Tainer,J.A. (1995) Novel DNA binding motifs in the DNA repair enzyme endonuclease III crystal structure. EMBO J., 14, 4108–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A. and Teplyakov,A. (1997) MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr., 30, 1022–1025. [Google Scholar]
- Verdine G.L. and Norman,D.P.G. (2003) Covalent trapping of protein–DNA complexes. Annu. Rev. Biochem., 72, 337–336. [DOI] [PubMed] [Google Scholar]
- Warner H.R., Demple,B.F., Deutsch,W.A., Kane,C.M. and Linn,S. (1980) Apurinic/apyrimidinic endonucleases in repair of pyrimidine dimers and other lesions in DNA. Proc. Natl Acad. Sci. USA, 77, 4602–4606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B. and Cunningham,R.P. (1985) Genetic mapping of nth, a gene affecting endonuclease III (thymine glycol-DNA glycosylase) in Escherichia coli K-12. J. Bacteriol., 162, 607–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss B. and Grossman,L. (1987) Phosphodiesterases involved in DNA repair. Adv. Enzymol., 60, 1–34. [DOI] [PubMed] [Google Scholar]
- Zharkov D.O., Rieger,R.A., Iden,C.R. and Grollman,A.P. (1997) NH2-terminal proline acts as a nucleophile in the glycosylase/AP-lyase reaction catalyzed by Escherichia coli formamidopyrimidine-DNA glycosylase (Fpg) protein. J. Biol. Chem., 272, 5335–5341. [DOI] [PubMed] [Google Scholar]
- Zharkov D.O., Golan,G., Gilboa,R., Fernandes,A.S., Gerchman,S.E., Kycia,J.H., Rieger,R.A., Grollman,A.P. and Shoham,G. (2002) Structural analysis of an Escherichia coli endonuclease VIII covalent reaction intermediate. EMBO J., 21, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]