Abstract
The transcription factor Clock (Clk) plays a critical role in animal circadian rhythms. Genetic studies defining its function have relied on two dominant negative alleles, one in Drosophila and one in mice. Here we describe a novel recessive allele of Drosophila Clock, Clkar. Homozygous Clkar flies are viable and behaviorally arrhythmic. The Clkar phenotype is caused by a splice site mutation that severely disrupts splicing and reduces Clk activity. Despite the behavioral arrhythmicity, molecular oscillations are still detectable in Clkar flies. Transcription analysis indicates potent effects of Clkar on levels and amplitude of transcriptional oscillations. Taken together with other data, we propose that Clk makes a major contribution to the strength and amplitude of circadian rhythms.
Keywords: amplitude/circadian rhythms/Clock/mutant allele/transcription
Introduction
The persistence of daily rhythmic behaviors without external temporal cues is a manifestation of circadian clocks. Although only approximating 24 h periodicity under constant conditions, these clocks achieve precise 24 h cycles by synchronizing to solar light cycles. Circadian pacemakers facilitate anticipation of these and other environmental events, and rhythmicity present throughout the animal and plant kingdoms punctuates its value to species fitness.
The investigation of fruit fly (Drosophila melanogaster) clocks has revealed many evolutionarily conserved pacemaker components (Allada et al., 2001). Studies on the biochemistry of the timekeeping mechanism have focused on transcriptional regulation. It is believed that the basic helix–loop–helix (bHLH) transcription factors CLOCK (CLK) and CYCLE (CYC) directly bind to upstream E boxes (CACGTG) and activate transcription of the period (per) and timeless (tim) genes (Hao et al., 1997; Allada et al., 1998; Darlington et al., 1998; Rutila et al., 1998; Wang et al., 2001). This view is based on strong biochemical evidence and on the phenotype of one semi-dominant allele of Clk. PER and TIM proteins subsequently feed back and inhibit transcriptional activation by CLOCK and CYCLE (Darlington et al., 1998; Lee et al., 1998, 1999). A similar feedback loop exists in mammals, including humans. Of note, circadian transcription studies in vivo have relied heavily on two dominant negative (antimorphic) alleles of Clock, one in Drosophila (ClkJrk) and one in mouse (King et al., 1997a,b; Allada et al., 1998).
Further studies have implicated a second feedback loop in circadian timing. Like per and tim, Clk and cry RNA levels also oscillate with respect to time of day (Bae et al., 1998; Darlington et al., 1998; Emery et al., 1998). However, these oscillations are antiphase to those of per and tim, suggesting that they are indirect targets of the Clk–cyc system. This is consistent with the levels of the Clk and cry RNAs in ClkJrk and cyc0 mutants; they are high, whereas the levels of per and tim RNAs are low (Emery et al., 1998; Glossop et al., 1999). It has been proposed that these genes, per and tim on the one hand and Clk on the other, define two interdependent transcriptional feedback loops. Two Clk-dependent transcription factors VRILLE and PDP1 are thought to mediate rhythmic Clk transcription (Cyran et al., 2003; Glossop et al., 2003). Transcriptional oscillations are thought to emerge from the dynamic interplay of these feedback loops, leading to behavioral and physiological rhythms.
Several aspects of circadian gene expression are also subject to post-transcriptional control, including RNA and protein stability, as well as protein phosphorylation (Dembinska et al., 1997; So and Rosbash, 1997; Kim et al., 2002). Protein levels and phosphorylation states of PER and TIM oscillate with time of day (reviewed in Allada et al., 2001). Doubletime (Dbt), a casein kinase I epsilon homolog; shaggy, a glycogen synthase kinase-3 homolog; and casein kinase 2(CK2), appear to phosphorylate PER and TIM (Kloss et al., 1998, 2001; Price et al., 1998; Martinek et al., 2001; Lin et al., 2002). These additional layers of feedback make it difficult to untangle the roles of different mechanisms in determining rhythm period, phase and amplitude.
Here we describe a second mutant allele of Drosophila Clk, Clkar, that is fully viable and recessive. Although behaviorally arrhythmic, molecular oscillations persist with reduced levels and amplitude, consistent with the reported role of Clk in transcriptional regulation. However, the phase of these oscillations is not substantially altered. These data are consistent with previous reports of ClkJrk heterozygotes, which exhibit modest changes in period in combination with reductions in rhythmicity as well as a reduced amplitude of gene expression (Allada et al., 1998). We propose that PER/TIM and its associated kinases primarily regulate period and phase, whereas CLK/CYC primarily regulates rhythmic amplitude.
Results
Identification of a novel recessive Clock allele
As part of a search for novel genes involved in circadian rhythmicity, we screened ethyl methane sulfonate (EMS) mutagenized perL flies for alterations in circadian locomotor activity (Rutila et al., 1996). One line homozygous for a mutagenized third chromosome, first called 1(7), was arrhythmic. Although initially described in a perL background, 1(7) is similarly arrhythmic in a wild-type background (Table I; see Clkar). This phenotype maps to the third chromosome and is recessive, i.e., homozygotes do not exhibit robust rhythms, whereas heterozygotes are virtually indistinguishable from wild type (Table I; see Clkar).
Table I. Circadian locomotor rhythms in Clk mutants.
| Genotype | %Ra | Periodb | nc |
|---|---|---|---|
| +/+ | 97 | 23.98 ± 0.05 | 34 |
| Clkar/+ | 94 | 24.19 ± 0.06 | 51 |
| Clkar/Clkar | 1d | ARe | 161 |
| D1/+ | 65 | 25.62 ± 0.11 | 23 |
| Clkar/D1 | 0 | AR | 26 |
| ClkJrk/+ | 75 | 24.67 ± 0.13 | 16 |
| Clkar/ClkJrk | 0 | AR | 80 |
| crygal4-Clkar/UASClock-Clkar | 60 | 22.58 ± 0.07 | 25 |
| ClkJrk/ClkJrk | 0 | AR | 23 |
| crygal4-ClkJrkUASClock-ClkJrk | 18 | 22.33 ± 0.13 | 37 |
a% rhythmic.
b± indicates SEM.
cNumber of flies analyzed.
dSingle weakly rhythmic fly.
eAR = arrhythmic.
Meiotic recombination mapping placed the 1(7) phenotype near the circadian rhythm gene Clock (Clk; data not shown). We were able to separate a female recessive sterile found in the original 1(7) stock from the circadian rhythm phenotype by meiotic recombination. However, we were unable to identify wild-type recombinants between 1(7) and ClkJrk (0/112 recombinants). To determine if 1(7) is an allele of Clk, we performed complementation analysis with a deletion (D1), that removes the Clk locus and ClkJrk. Neither D1 nor ClkJrk complemented the arrhythmicity of 1(7), consistent with the notion that 1(7) is an allele of Clk. As a result, we renamed 1(7) Clkar (Table I). The observation that the heterozygous D1 period phenotype (∼25.5 h) is more severe than that of heterozygous Clkar further suggests that Clkar is a hypomorphic allele, although alternative interpretations must also be considered (see Discussion).
Rescue of Clkar using GAL4/UAS
To confirm this assignment we rescued the arrhythmic phenotype of Clkar using the GAL4/UAS system. We generated a single transgenic Clk line driven by the upstream activating sequence (UAS) for GAL4 (UASClk) and combined it with various rhythm-relevant gal4 drivers. Overexpression of Clk driven by period promoter-GAL4 (pergal4) or timeless promoter-GAL4 (timgal4) resulted in early larval lethality (Emery et al., 1998; Kaneko and Hall, 2000). Heterozygous lines expressing Clk driven by pigment dispersing factor promoter-GAL4 (pdfgal4) or cryptochrome promoter-GAL4 (crygal4) were adult viable and therefore were assessed for behavioral rescue of Clkar (Emery et al., 2000; Park et al., 2000). pdfgal4 is expressed in the central brain in pacemaker neurons (small and large ventral lateral neurons; sLNv, lLNv), whereas crygal4 is expressed in a slightly broader distribution, extending to a dorsal group of lateral neurons (LNd; Emery et al., 2000; Park et al., 2000).
Expression of Clk by pdfgal4 in a Clkar background did not result in significant rescue of rhythmicity (data not shown). On the other hand, crygal4-driven expression of Clk resulted in rescue in the rhythmicity of a majority of these flies (Table I). The rescued flies exhibited a slightly short period, similar to periods in flies with crygal4-driven expression of Clk in a wild-type background (data not shown). The period shortening with increased Clk expression is consistent with the long periods of flies with only a single dose of Clk (Table I; see D1/+). BAC transgenic mice containing extra copies of Clock also exhibit short periods (Antoch et al., 1997). We obtained similar results in a ClkJrk background: crygal4-driven Clk expression was able to rescue the rhythmicity of ClkJrk (18% rhythmic), although more weakly than Clkar (60% rhythmic), consistent with the antimorphic effects of ClkJrk.
Altered Clock sequence, transcripts and function in Clkar
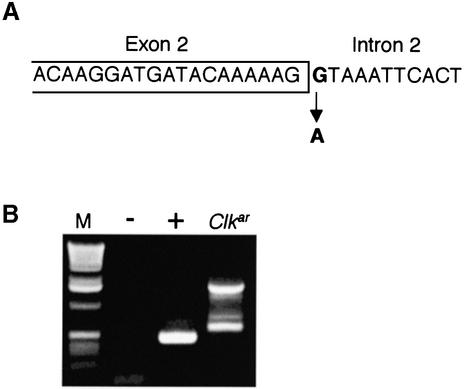
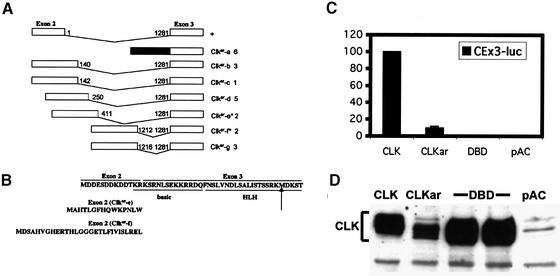
Given the strong evidence that Clkar is an allele of Clk, we searched coding exons and exon–intron boundaries for EMS-induced base changes, comparing Clkar with sibs. We identified a single mutation at the 5′ splice site of the second intron, destroying the GT dinucleotide required for efficient splicing (Figure 1A). The mutation is a G to A transition classically found in EMS-induced alleles (Bentley et al., 2000). We examined Clk spliceforms across the second intron in the Clkar mutant using reverse transcriptase–polymerase chain reaction (RT–PCR). RT–PCR across this intron identified a single band of the appropriate size in wild-type flies (Figure 1B). In Clkar, multiple bands are observed, none of which correspond by electrophoretic migration to that seen in wild type, consistent with the observed splice site mutation. Splice junctions between other coding exons were not grossly perturbed as assayed by RT–PCR (data not shown). Exon 2 encodes for the N-terminal 13 amino acids, including the first two amino acids of the basic region (Figure 2B; KR). The exons beyond exon 2 encode the remainder of the CLK protein, including most of the basic region, the PAS dimerization motif and the glutamine-rich activation domain (Allada et al., 1998). To determine if these altered Clkar transcripts can produce functional CLK protein, we sequenced Clkar cDNAs. To produce essentially full-length CLK, altered transcripts must splice properly into exon 3. We therefore sequenced Clkar cDNAs to determine if there were any intact open reading frames upstream of exon 3. In 18/22 cases, there is a stop codon in-frame and upstream of the remaining Clk gene in the Clkar cDNA (Figure 2A). Assuming initiation from the first downstream methionine within exon 3, translation would result in a CLK protein lacking the basic DNA binding domain (DBD) and a portion of the helix–loop–helix dimerization motif. In 4/22 clones, an upstream methionine codon is in-frame with the remainder of the Clk gene (Figure 2A and B; see Clkar-e and f). Assuming initiation from this methionine, translation of these transcripts would result in a CLK protein with novel N-termini: two of 15 amino acids and two of 28 amino acids (Figure 2B). In all four cases, only the first two amino acids of the basic DBD are altered.
Fig. 1. Alterations in Clk DNA and RNA in Clkar flies. (A) Sequence change present at the 5′ splice site of intron 2. (B) Altered splicing of intron 2 in Clkar as shown by RT–PCR across intron 2. – indicates negative control (no RNA). + indicates single band in wild type corresponding to RNA without intron 2 (spliced).
Fig. 2. Analysis of splice junctions in Clkar transcripts. (A) Schematic of splicing of exons 2 and 3 flanking intron 2. Black box indicates the presence of intron and absence of any detectable splicing of intron 2. Altered transcripts present in Clkar alphabetically are indicated as well as the number of clones of that type identified out of 22 clones analyzed. Nucleotide numbers of intron (1–1281 in wild type) indicate sites of splice junctions. Asterisks indicate clones with potential in-frame methionines from the altered upstream exon 2. (B) Conceptual translation of altered transcripts. Top line indicates translation of wild-type transcript from first methionine located in exon 2. Basic DBD (basic) and helix–loop–helix DNA binding and dimerization motif is indicated (HLH) in the translated product. Potential new N-termini from altered exon 2 in Clkar-e and -f are indicated from Clkar transcripts. (C) Activation of a triplicated per circadian enhancer by CLKar in S2 cells. CLK, CLOCK; DBD, CLOCK without a DBD; pAC, empty pActin vector; CEx3-luc is a triplicated circadian enhancer derived from the per promoter; y-axis indicates luminescence relative to CLK where CLK = 100%. Measurements displayed are averages of two independent experiments. Error bars indicate SEM. Some error bars were not visible due to low variability. (D) Expression of CLOCKar in transfected cells.
To determine the functional consequence of the splice site mutation, the Clkar gene, including the mutant second intron, was subcloned into an S2 tissue culture expression vector under the control of the Actin promoter (pAc). Consistent with this mutation, transfection of the Clkar clone resulted in a substantial reduction in transcriptional activation relative to wild-type Clk. Nonetheless, this Clkar clone did activate transcription well above that achieved with pAc alone (Figure 2C). We observed comparable levels of activation by Clkar with a full-length per promoter construct (BG-luc) and an enhancer derived from the tim promoter (timenh-LUC; see below; data not shown). Given that a large fraction of Clkar transcripts should produce Clk without its DBD, we sought to determine if this form can activate transcription. However, cloning of this particular isoform into the pAc vector gave rise to little activation relative to Clkar (Figure 2C). Because differences in activation may derive from differences in protein levels, we compared expression of each of these forms by western blot analysis (Figure 2D). The wild-type and DBD forms of CLK are comparably expressed, whereas the CLKar clone has reduced protein levels. Inefficient splicing to the AG of exon 3 might hinder expression of CLKar, which probably contributes to the reduced activity of CLKar relative to wild-type CLK (∼10%; Figure 2C). Nonetheless, CLKar has low but significant activity, and the data suggest that this activity derives from a minor, essentially full-length form of the protein, such as those from Clkar-e and f or perhaps other unidentified forms (Figure 2B). We conclude that Clkar is a hypomorphic allele with residual activity.
No evident behavioral rhythmicity in Clkar
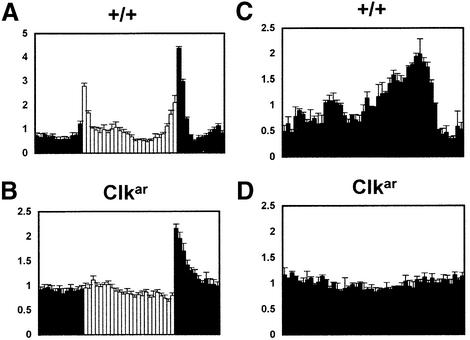
Consistent with the hypomorph interpretation was a more detailed analysis of the homozygous Clkar behavioral rhythms (Figure 3). Under light: dark conditions (Figure 3A and B), these mutant flies manifested a highly abnormal activity pattern: no lights-on startle response but a robust lights-off startle response. This feature was indistinguishable from that of ClkJrk flies and substantiates the connection between Clk and responses to light–dark (LD) transitions (Allada et al., 1998). In contrast, wild-type flies demonstrate anticipation of lights-on and lights-off peaks, evidence of an underlying endogenous clock, as well as ‘startle’ responses to both lights-on and lights-off events. There was also no measurable rhythmic activity of Clkar in constant darkness (DD), even during the first two days of DD (Figure 3C and D). This contrasts markedly with the rhythmic activity of pdf01 flies during the first two days of DD (Renn et al., 1999). In sum, the homozygous Clkar behavioral data showed no evidence of rhythmicity and were essentially identical to those of homozygous ClkJrk flies, consistent with a strong hypomorphic etiology.
Fig. 3. Abnormal diurnal and circadian locomotor behavior in Clkar flies. Normalized plots of locomotor activity in LD (A and B) and first two days of constant darkness (C and D; see Materials and methods) for wild type (+; A and C; n = 16) and Clkar flies (B and D; n = 16). Light and dark bars correspond to times of lights-on and -off. Error bars indicate SEM.
Persistent molecular oscillations in Clkar
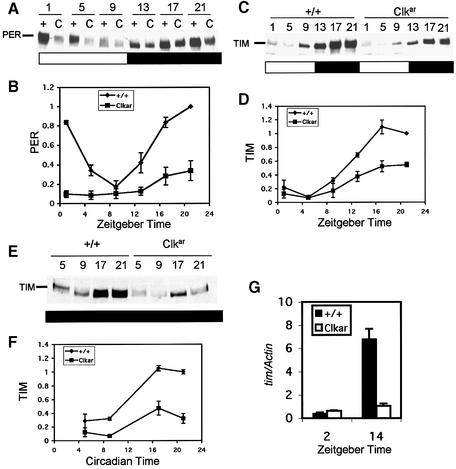
To determine the effect of Clkar on molecular oscillations, we examined timeless and period protein by western blotting under LD conditions (Figure 4). In Clkar, PER and TIM cycle robustly under LD conditions, despite a decrease in protein levels relative to wild-type flies (Figure 4A–D). As light has previously been shown to suppress TIM levels and contribute to protein oscillations, we also assayed TIM under DD conditions (first full day of DD). To our surprise, TIM cycling persisted (Figure 4E and F), indicating the presence of a circadian clock not evident at the behavioral level (Figure 3D). We made similar observations of reduced but oscillating PER levels in DD in the Clkar mutant (data not shown). The reduction in peak levels of PER and TIM was consistent with a reduction in Clk transcriptional activation. As confirmation, we measured tim transcript levels using real-time quantitative PCR. Consistent with our western results, we found a strong reduction in peak levels of tim (Figure 4G; Zeitgeber time 14).
Fig. 4. Molecular oscillations in Clkar. Western blots of PERIOD (PER; A and B) and TIM (C–F) under 12 h light:12 h dark (A–D) and constant darkness (E and F) conditions. Zeitgeber Time (ZT; B and D) 0 is lights-on and 12 is lights-off. Circadian time (F) indicates time in constant darkness where Circadian Time 0 is 12 h after lights-off. PER quantification (B) is from four independent experiments (three for ZT 1 and 13). TIM quantification in LD (D) is from three independent experiments (two for ZT 1 and 13) and in DD (F) from two independent experiments. Error bars indicate SEM tim transcript oscillations as measured by real-time quantitative PCR (see Materials and methods). (G) Levels of tim transcript measured with respect to Actin control (tim/Actin).
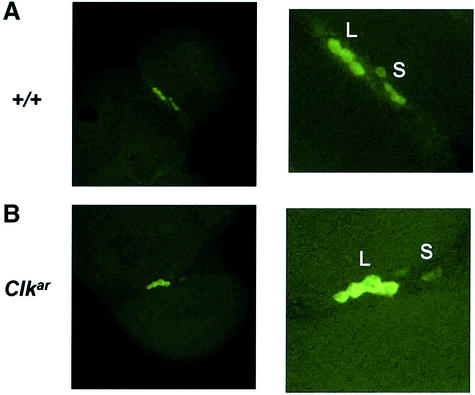
The absence of behavioral rhythms in the context of intact molecular rhythms has been previously observed in mutants that lack PDF-expressing lateral neurons (Hardin et al., 1992). We therefore performed fluorescent in situ hybridization for pdf transcripts to assess the presence of large and small pacemaker lateral neurons. Consistent with previous reports in ClkJrk mutants, we found reductions in pdf expression specifically in the small ventral lateral neurons (Figure 5). Nonetheless, we were able to visualize both large and small groups of pdf neurons. Thus, the absence of behavioral rhythms is not due to a complete failure of pacemaker lateral neurons to develop.
Fig. 5. pdf expression in Clkar. Fluorescent in situ hybridization for pdf transcript in wild type (A) and Clkar (B) reveals labeling of large (L) and small (S) ventral lateral neurons (LNs) under low (left panel) and high (right panel) magnifications. These two groups of neurons can be detected in both wild type and Clkar, although levels are reduced in the small neurons in Clkar. High magnification image of Clkar is enhanced to ease visualization of the small LNs.
Reduction in cycling amplitude and levels of per and tim enhancer activity
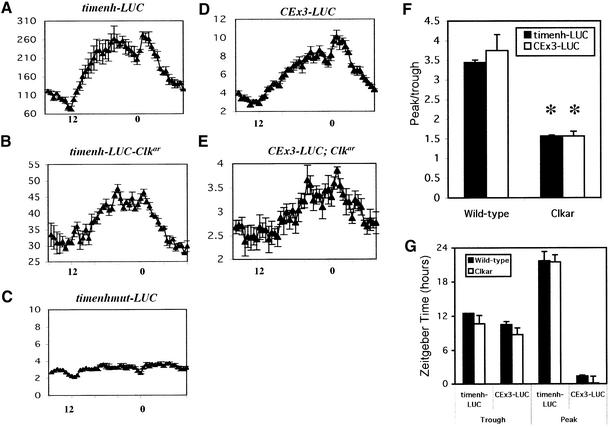
To more directly address CLK activity, we examined the cycling of tim and per enhancers in Clkar mutants, using luciferase-mediated bioluminescence as a reporter (Brandes et al., 1996). The tim promoter contains an E-box target site for CLK/CYC that is required for full rescue and high level activity (McDonald et al., 2001; Wang et al., 2001). We cloned a small fragment (–756 to –604) encompassing this E-box upstream of DNA encoding the firefly luciferase gene (timenh-LUC). We observed robust cycling, demonstrating the presence of a cycling enhancer (Figure 6A). Consistent with the notion that the E-box is a functional target of CLK, we found that the bioluminescence levels are dramatically reduced in Clkar (Figure 6B). However, they are not as low as a version of this enhancer in which the E-box has been mutated (CACGTG to CTCGAG; timenhmut-LUC) consistent with the notion that Clkar is not a null allele (Figure 6C).
Fig. 6. Reduction in cycling levels and amplitude of per and tim enhancers in Clkar. timenh-LUC indicates transgenic flies containing a tim enhancer fused to luciferase. timenhmut-LUC is the same tim enhancer with the E-box site (CACGTG) mutated. CEx3-LUC indicates a 69 bp circadian enhancer from the per promoter triplicated and fused to luciferase. Luciferase activity is expressed in thousands of counts per minute for the tim enhancers (timenh-LUC, timenhmut-LUC) and millions of counts per minute for the per enhancer (CEx3-LUC). Error bars indicate SEM. Zeitgeber time is indicated on the x-axis with ZT0 as time of lights-on and ZT12 as time of lights-off. For timenh-LUC (A), n = 13, for timenh-LUC-Clkar (B), n = 16, for timenhmut-LUC (C), n = 17, for CEx3-LUC (D), n = 7 and for CEx3-LUC; Clkar (E), n = 5. (F) Amplitude assessment in wild type and Clkar. Error bars indicate the SEM across days. An asterisk indicates statistically significant result (P = 0.0001 for timenh; P = 0.002 for CEx3). (G) Phase assessment in wild type and Clkar. Times of average peak and trough values are indicated. SEM over several days indicated. No statistically significant differences detected between wild type and Clkar.
Despite the large reduction in expression levels, cycling bioluminescence was still apparent in timenh-LUC, Clkar flies. There was a large reduction in cycling amplitude relative to wild-type flies as measured by peak-to-trough ratios (Figure 6F), consistent with the large effect on cycling amplitude (Figure 5G). However, the phase was not dramatically affected as measured by times of peak and trough luminescence each day (Figure 6G). We also examined the activity of a triplicated circadian enhancer from the per promoter (CEx3-LUC; Hao et al., 1997; So et al., 2000). This enhancer contains a functional E-box and confers robust cycling on the luciferase reporter (Figure 6D). When assayed in a Clkar background, we observed luciferase cycling, also with reduced levels and amplitude (Figure 6E and F). Again, phase is not strongly affected (Figure 6G). These data are similar to those shown above for PER and TIM levels, suggesting that Clkar rhythms have reduced amplitudes and expression levels but only modest effects on phase.
Discussion
Here we describe the first recessive allele of Clock. The only previously reported allele, ClkJrk, is semi-dominant and behaves as a dominant negative (Allada et al., 1998). As a result, it is possible that ClkJrk may exert its effects through a gain-of-function effect. The strongest genetic evidence for a Clk requirement for normal rhythms is the long period of flies heterozygous for a deletion of the Clk locus (Allada et al., 1998). However, homozygous D1 flies do not live to adulthood, and behavioral rhythms therefore cannot be assessed in Clk-null animals. Even if they were arrhythmic, an essential contribution of Clk to rhythms would remain uncertain because the D1 deletion removes several genes. These most likely include Pdp1 and Hn, which are candidate circadian rhythm genes and are located within 40 kb (right) and 10 kb (left) of Clk, respectively (Neckameyer and White, 1992; Florez et al., 1996; McDonald and Rosbash, 2001). The D1 deletion was initially identified by its failure to complement the eye pigment phenotype of Hn, which lies just to the left of Clk (Grasso, 1996). Furthermore, D1 fails to complement mutant loci to the right of Pdp1 and is therefore likely to remove this gene as well (Allada et al., 1998). The heterozygous phenotype of the D1 deletion may therefore be due to the absence of one or more of these genes. A similar ambiguity exists in mammals, as there is only a single semi-dominant allele and no hypomorphic or null alleles of the mouse Clock locus. All of these considerations make this new recessive arrhythmic Clk allele important and indicate that Clk is indeed required for behavioral rhythmicity.
Part of the proof that Clkar is truly a Clk mutant comes from the rescue with heterozygous combinations of crygal4 and UASClk. We were, however, unable to effect successful rescue with other rhythm-relevant gal4 drivers: timgal4 and pergal4 were lethal in combination with UASClk, and pdfgal4 had no effect on the arrhythmic Clkar phenotype. The pdfgal4 driver was also unable to rescue the arrhythmic ClkJrk background. We also tested the rescuing ability of pdfgal4 in flies trans-heterozygous for the Clk mutants and the Clk deletion, D1: in these genetic backgrounds, pdfgal4 also failed to rescue either ClkJrk or Clkar (data not shown). The most obvious difference between crygal4 and pdfgal4 is a more widespread expression pattern in the case of crygal4, which extends to another more dorsal group of neurons, the LNds (Renn et al., 1999; Emery et al., 2000). Locomotor activity rhythms may therefore require wild-type Clk expression in more than just the restricted pdf locations.
Another possible difference between the cry and pdf drivers is RNA cycling: cry mRNA cycles with a roughly similar amplitude and phase to Clk, whereas the pdf gene undergoes no substantial transcriptional fluctuations (Park and Hall, 1998). Although Clk and cry mRNA cycling may contribute to circadian function, we do not favor this explanation. The stability of GAL4 should prevent significant oscillations in protein levels, even if mRNA levels undergo robust cycling. This suggests that rhythmic transcription of Clk is unnecessary for behavioral rhythmicity and is consistent with other experiments indicating that the timing and levels of Clk oscillation can be substantially altered without strong effects on circadian behavior (Kim et al., 2002). This conclusion is also consistent with the notion that the Clk transcriptional loop is less important than the original per–tim loop for behavioral rhythms.
We note, however, that GAL4-mediated rescue of Clk is only partial, with 60% rhythmicity and shortened periods. This is comparable to the GAL4-mediated rescue of per0; tim0 double mutants in which rhythmicity is 40–50% and periods range from 24–27 h (Yang and Sehgal, 2001). The similarly poor % rhythmicity values may reflect comparable contributions of mRNA cycling, per and tim in one case and Clk in the other. Alternatively, mRNA cycling may be largely irrelevant to penetrance; the poor rescue may result from inappropriate GAL4 levels (too high or too low) or from some other inadequate feature of GAL4-mediated expression.
The molecular assays in Clkar indicate bona fide rhythms with a predominant effect on circadian rhythm amplitude and no more than a modest effect on phase or period. With circadian per and tim enhancers, we observed reduced enhancer activity and a reduced cycling amplitude in a Clkar background, consistent with the role of Clk in regulating these enhancers (Figure 6). Nonetheless, the phase of oscillating bioluminescence is similar to that of wild-type flies. The presence of molecular rhythms contrasts with the absence of detectable behavioral rhythms. We favor the notion that this reflects a level or amplitude reduction below a critical threshold for behavioral rhythmicity. The absence of anticipation of LD transitions makes it very unlikely that an effect restricted to the lateral neurons, the absence of the neuropeptide PDF for example, is primarily responsible for the phenotype. This is also because LD behavioral rhythms are largely normal in flies devoid of PDF or the pacemaker lateral neurons (Renn et al., 1999). Moreover, we demonstrate both large and small PDF-expressing lateral pacemaker neurons are present in Clkar (Figure 5). However, we did observe a reduction in pdf expression in the small lateral neurons that may contribute to Clkar arrhythmicity in constant darkness.
Previous results with ClkJrk also support a role for Clk in defining rhythmic amplitude. ClkJrk heterozygotes reveal a dominant reduction in the amplitude of molecular rhythms with little apparent change in phase (Allada et al., 1998). These heterozygotes also exhibit reductions in rhythmic behavior with only slightly long periods. Indeed, Clk overexpression results in a selective increase in the amplitude of per RNA oscillations (Kim et al., 2002). This modest effect on period or phase of varying Clk activity is similar to the phenotype of transgenic strains missing the per promoter or expressing per and tim from constitutive promoters (Frisch et al., 1994; Yang and Sehgal, 2001). These strains also have reasonable periods (22–26 h) with poor rhythm amplitudes, as evidenced by the poor penetrance of rhythmicity. One argument for a role for Clk in period control is the phenotype of the D1 heterozygote (∼25.5 h period). However, even this altered period is within the limited range of altering per or tim transcription. Taken together, these data suggest that substantial changes in Clock gene transcription have limited effects on circadian period. Separate control of circadian rhythm amplitude on the one hand and period (or phase) on the other is also consistent with anatomical experiments in both the fly and mammalian system (Liu et al., 1991; Low-Zeddies and Takahashi, 2001).
We propose that the post-transcriptional phosphorylation turnover feedback loop involving several Clock components (e.g. per, tim, the protein kinase Dbt) is predominantly responsible for period determination. Excluding null alleles that are either arrhythmic or lethal, Flybase lists mutant alleles of per, tim and Dbt which exhibit period alterations ranging from 16–30 h for per (8 mutant alleles), 21–33 h for tim (8 mutant alleles) and 18–29 h for Dbt (5 mutant alleles; Flybase Consortium, 2002). Indeed, the only Dbt allele that fails to exhibit rhythmicity as a homozygote, displays a potent period-altering phenotype as a heterozygote (Rothenfluh et al., 2000). More recent additions to this list are the protein kinases shaggy (Martinek et al., 2001) and CK2. Indeed, one mutant allele of CK2α, CK2αTik, exhibits one of the strongest dominant period effects of any rhythm mutant (Lin et al., 2002). These large period effects contrast with the transcription factor mutants (Clk and cyc). Their phenotypes indicate that near-normal periods are maintained despite large protein level changes.
Materials and methods
Fly stocks, mutagenesis, mapping and behavioral analysis
perL;ry506 flies were used for mutagenesis (Rutila et al., 1996). pdfGAL4, crypiGAL4, timGAL4 have been previously described (Emery et al., 2000; Kaneko and Hall, 2000; Park et al., 2000). Flies were entrained for 2–5 days of 12 h light: 12 h dark before monitoring of activity in constant darkness for at least 5 days (Hamblen et al., 1986). Circadian periods were determined by χ-square periodograms analysis. For high-resolution analysis of diurnal and circadian behavior, data for individual flies is normalized to total activity and then the normalized activity of several flies and days is averaged together to produce single-day activity plots.
Construction of transgenic flies
Clock was first tagged with hemagglutinin (HA) epitope by PCR cloning. Briefly, a C-terminal fragment of Clock was PCR-amplifed using pSK(–) Clock cDNA and an oligonucleotide with HA epitope and XhoI site and an internal oligonucleotide. The amplified fragment was digested with ClaI/XhoI and ligated to ClaI/XhoI-digested pSKClock vector to generated pSK(–) ClockHA. ClockHA was subsequently ligated into pUAST (EagI/XhoI) to generate pUAS-ClockHA.
tim enhancer luciferase flies were constructed by PCR and subcloning into luciferase vector. A NotI/EcoRI fragment containing the E-box region of the timeless promoter (position –756 to –604; McDonald et al., 2001) and a EcoRI/SalI fragment containing the heat-shock minimal promoter (hs43) were obtained by PCR and enzymatic digestion, and cloned in front of the luciferase cDNA in pBluescript (Stanewsky et al., 1997). From this intermediate construct, a NotI/KpnI fragment was excised and cloned in pCasperR4. The resulting construct is called pCasp-timenh-luc. The pCasp-timenhmut-luc construct is identical to pCasp-timenh-luc, expect that the E-box (CACGTG) is mutated to an XhoI site (CTCGAG). These two luciferase constructs were sequenced prior to injection to ensure their integrity. y w; Ki pp (ry+ delta 2–3)/+ embryos were injected with DNAs. A single line (UASClk) was obtained as a third chromosome insert. HA epitope is not immunologically detectable using western blotting or immunoprecipitation with anti-HA antibodies.
Genomic DNA sequencing and transcript analysis
Genomic DNA was prepared from Clkar flies and wild-type sibs (Allada et al., 1998). Clk coding region exons and intron–exon boundaries were amplified by PCR and subjected to DNA sequencing. Total RNA was prepared from adult wild-type and Clkar heads (Rutila et al., 1996). Superscript II RNase H– and primers flanking intron 2 of Clk were used to generate cDNA by RT–PCR. PCR fragments were subcloned by TA cloning into pCR (Invitrogen). Individual clones were prepared and subjected to DNA sequencing to analyze intron–exon structure.
Transfection assays
Transfection assays performed as in McDonald et al. (2001). PAcClock (250 ng), pAc-Bgal (10–100 ng) and CEx3-LUC (100–500 ng) were used for transfections.
Western analysis
Western analysis was performed using rabbit anti-PER (1:10 000) and rat anti-TIM (1:5–10 000) antibodies (Allada et al., 1998). Rabbit anti-CLK antibody was obtained commercially (Alpha Diagnostic, San Antonio, TX). Cells transfected with CLK were pelleted and boiled in sample buffer. Samples were run on SDS–PAGE transferred and membranes were blocked in 5% milk/TBST. Blots were probed at 1 µg/1 ml in 1% milk/TBST overnight at 4°C and subsequently probed with 1:500 anti-rabbit secondary antibodies for 2 h at room temperature.
Real-time quantitative RT–PCR for tim transcript levels
Total RNA was prepared from adult wild-type and Clkar heads (Rutila et al., 1996). cDNA derived from this RNA was used as template for a quantitative real-time PCR assay performed on the Corbett Research Rotor-Gene 2000 real-time cycler. The PCR mixture contained Platinum Taq polymerase (Life Technologies), optimized concentrations of Sybrgreen and the following primers: tim primers, 5′-CCTTTTCGTACACAGATGCC-3′ and 5′-GGTCCGTCTGGTGATCCCAG-3′; actin primers, 5′-TGCAGCGGATAACTAGAAACTACTC-3′ and 5′-CAAAGGAGCCTCAGTAAGCAAG-3′. Cycling parameters were 95°C for 3 min, followed by 40 cycles of 95°C for 30 s, 55°C for 45 s and 72°C for 45 s. Fluorescence intensities were plotted against the number of cycles by using an algorithm provided by the manufacturer. tim and actin mRNA levels were quantitated using a calibration curve based upon dilution of concentrated cDNA from wild-type flies at ZT14. tim values/levels were normalized with the actin signal. For each condition three (in the case of wild-type ZT14, Clkar ZT2 and ZT14) or two (for wild-type ZT2) independent RNA samples were analyzed. For each RNA sample, four independent RT–PCR experiments were performed, in each real-time PCR experiment the samples were evaluated in triplicate. The mean of the four measurements for each RNA sample was determined and the tim/actin ratio was calculated. The tim/actin ratios for each of the three independent experiments (or two in the case of wild type at ZT2) were also averaged to give the data shown in Figure 4G.
pdf in situ mRNA hybridization on adult brain whole mounts
Adult fly brains were dissected in phosphate-buffered saline and fixed in 4% paraformaldehyde for 30 min at room temperature. After pre-hybridization for minimum of 2 h in Hybrix (50% formamide, 5× SSD, 100 µg/ml tRNA, 100 µg/ml ssDNA, 0.1% Tween-20) at 55°C, the brains were incubated with probes overnight at 55°C. The pdf probe used corresponds to nucleotides 282–570. Antisense RNA probes were synthesized and labeled using digoxigenin (pdf) RNA labeling kit from Boehringer Mannheim. The probes were hydrolyzed in sodium bicarbonate buffer and stored in Hybrix at –20°C until use. The hybridized RNA signals were detected using fluorescent tyramides (NEN LifeScience). Brains were mounted in glycerol with 4% n-propyl gallate and examined by confocal microscopy.
Luciferase reporting and analysis
Single fly luciferase activity reporting was performed under 12 h light: 12 h dark conditions (Brandes et al., 1996). Data from several flies were averaged together. Due to the decrement in bioluminescence activity observed over several days, we then performed detrending analysis as follows. We fit the data to a linear curve and obtained a slope and intercept. We then calculate the location of the fitted curve at the end of the experiment [yend = slope*number of measurements (n) + y-intercept (yint; luminescence)]. Where the fitted line starts (the yint) and where the line ends (yend) are used to calculate the % increase over the course of the experiment required to correct for the decrement [(yint /yend –1)*100%]. The % correction is then divided by the total number of measurements in the experiment (n). Each value is then corrected by a percentage, depending on the time from the beginning of the experiment. Thus based on a linear change in the % correction, early time points are increased by a small percentage and measurements late in the experiment are increased by a relatively large percentage. As a result, the detrended data would have a slope of approximately zero, i.e., no decrement. The averaged and detrended data from three to four days were then averaged together to produce a single day of data to facilitate phase comparisons. Standard error of the mean (SEM) is that of these 3–4 days averaged.
Assessments of amplitude and amplitude are performed on the data averaged from several flies without detrending. For each day, the peak and trough values are noted and peak/trough ratios are calculated for each day and averaged over 3–4 days of the experiment. This ratio reflects the amplitude of the oscillation. The times at which these peak and trough values are reached are also averaged over 3–4 days of the experiment to get an assessment of the phase of the oscillation. The SEM is that of these days averaged. Statistical comparisons were performed using Student’s t-test.
Acknowledgments
Acknowledgements
We thank Patrick Emery for the timenh-LUC and timenhmut-LUC lines and Kevin Keegan for assistance with data analysis. We thank P.Emery and Jeff Hall for comments on the manuscript. R.A. was supported by a Howard Hughes Medical Institute Postdoctoral Fellowship for Physicians and Burroughs Wellcome Career Award in the Biomedical Sciences.
References
- Allada R., White,N.E., So,W.V., Hall,J.C. and Rosbash,M. (1998) A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell, 93, 791–804. [DOI] [PubMed] [Google Scholar]
- Allada R., Emery,P., Takahashi,J.S. and Rosbash,M. (2001) Stopping time: the genetics of fly and mouse circadian clocks. Annu. Rev. Neurosci., 24, 1091–1119. [DOI] [PubMed] [Google Scholar]
- Antoch M.P. et al. (1997) Functional identification of the mouse circadian Clock gene by transgenic BAC rescue. Cell, 89, 655–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K., Lee,C., Sidote,D., Chuang,K.Y. and Edery,I. (1998) Circadian regulation of a Drosophila homolog of the mammalian Clock gene: PER and TIM function as positive regulators. Mol. Cell. Biol., 18, 6142–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley A., MacLennan,B., Calvo,J. and Dearolf,C.R. (2000) Targeted recovery of mutations in Drosophila. Genetics, 156, 1169–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes C., Plautz,J.D., Stanewsky,R., Jamison,C.F., Straume,M., Wood,K.V., Kay,S.A. and Hall,J.C. (1996) Novel features of Drosophila period Transcription revealed by real-time luciferase reporting. Neuron, 16, 687–692. [DOI] [PubMed] [Google Scholar]
- Cyran S.A., Buchsbaum,A.M., Reddy,K.L., Lin,M.C., Glossop,N.R., Hardin,P.E., Young,M.W., Storti,R.V. and Blau,J. (2003) vrille, Pdp1 and dClock form a second feedback loop in the Drosophila circadian clock. Cell, 112, 329–341. [DOI] [PubMed] [Google Scholar]
- Darlington T.K., Wager-Smith,K., Ceriani,M.F., Staknis,D., Gekakis,N., Steeves,T.D.L., Weitz,C.J., Takahashi,J.S. and Kay,S.A. (1998) Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science, 280, 1599–1603. [DOI] [PubMed] [Google Scholar]
- Dembinska M.E., Stanewsky,R., Hall,J.C. and Rosbash,M. (1997) Circadian cycling of a PERIOD–beta-galactosidase fusion protein in Drosophila: evidence for cyclical degradation. J. Biol. Rhythms, 12, 157–172. [DOI] [PubMed] [Google Scholar]
- Emery P., So,W.V., Kaneko,M., Hall,J.C. and Rosbash,M. (1998) CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell, 95, 669–679. [DOI] [PubMed] [Google Scholar]
- Emery P., Stanewsky,R., Helfrich-Forster,C., Emery-Le,M., Hall,J.C. and Rosbash,M. (2000) Drosophila CRY is a deep brain circadian photoreceptor. Neuron, 26, 493–504. [DOI] [PubMed] [Google Scholar]
- Florez J.C., Seidenman,K.J., Barrett,R.K., Sangoram,A.M. and Takahashi,J.S. (1996) Molecular cloning of chick pineal tryptophan hydroxylase and circadian oscillation of its mRNA levels. Brain Res. Mol. Brain Res., 42, 25–30. [DOI] [PubMed] [Google Scholar]
- Flybase Consortium (2002) The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res., 30, 106–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch B., Hardin,P.E., Hamblen-Coyle,M.J., Rosbash,M. and Hall,J.C. (1994) A promoterless period gene mediates behavioral rhythmicity and cyclical per expression in a restricted subset of the Drosophila nervous system. Neuron, 12, 555–570. [DOI] [PubMed] [Google Scholar]
- Glossop N.R., Lyons,L.C. and Hardin,P.E. (1999) Interlocked feedback loops within the Drosophila circadian oscillator. Science, 286, 766–768. [DOI] [PubMed] [Google Scholar]
- Glossop N.R., Houl,J.H., Zheng,H., Ng,F.S., Dudek,S.M. and Hardin,P.E. (2003) VRILLE feeds back to control circadian transcription of Clock in the Drosophila circadian oscillator. Neuron, 37, 249–261. [DOI] [PubMed] [Google Scholar]
- Grasso G., Bordne,D. and White,K. (1996) Drosophila Info. Serv., 77, 94–96. [Google Scholar]
- Hamblen M. et al. (1986) Germ-line transformation involving DNA from the period locus in Drosophila melanogaster: overlapping genomic fragments that restore circadian and ultradian rhythmicity to per0 and per– mutants. J. Neurogenet., 3, 249–291. [DOI] [PubMed] [Google Scholar]
- Hao H., Allen,D.L. and Hardin,P.E. (1997) A circadian enhancer mediates PER-dependent mRNA cycling in Drosophila melanogaster. Mol. Cell. Biol., 17, 3687–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardin P.E., Hall,J.C. and Rosbash,M. (1992) Behavioral and molecular analyses suggest that circadian output is disrupted by disconnected mutants in D.melanogaster. EMBO J., 11, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko M. and Hall,J.C. (2000) Neuroanatomy of cells expressing clock genes in Drosophila: transgenic manipulation of the period and timeless genes to mark the perikarya of circadian pacemaker neurons and their projections. J. Comp. Neurol., 422, 66–94. [DOI] [PubMed] [Google Scholar]
- Kim E.Y., Bae,K., Ng,F.S., Glossop,N.R., Hardin,P.E. and Edery,I. (2002) Drosophila CLOCK protein is under posttranscriptional control and influences light-induced activity. Neuron, 34, 69–81. [DOI] [PubMed] [Google Scholar]
- King D.P., Vitaterna,M.H., Chang,A.M., Dove,W.F., Pinto,L.H., Turek,F.W. and Takahashi,J.S. (1997a) The mouse Clock mutation behaves as an antimorph and maps within the W19H deletion, distal of Kit. Genetics, 146, 1049–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King D.P. et al. (1997b) Positional cloning of the mouse circadian clock gene. Cell, 89, 641–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloss B., Price,J.L., Saez,L., Blau,J., Rothenfluh,A., Wesley,C.S. and Young,M.W. (1998) The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell, 94, 97–107. [DOI] [PubMed] [Google Scholar]
- Kloss B., Rothenfluh,A., Young,M.W. and Saez,L. (2001) Phosphoryl ation of period is influenced by cycling physical associations of double-time, period and timeless in the Drosophila clock. Neuron, 30, 699–706. [DOI] [PubMed] [Google Scholar]
- Lee C., Bae,K. and Edery,I. (1998) The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation and interactions with the PER–TIM complex. Neuron, 21, 857–867. [DOI] [PubMed] [Google Scholar]
- Lee C., Bae,K. and Edery,I. (1999) PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK–CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol. Cell. Biol., 19, 5316–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J.M., Kilman,V.L., Keegan,K., Paddock,B., Emery-Le,M., Rosbash,M. and Allada,R. (2002) A role for casein kinase 2alpha in the Drosophila circadian clock. Nature, 420, 816–820. [DOI] [PubMed] [Google Scholar]
- Liu X., Yu,Q.A., Huang,Z.S., Zwiebel,L.J., Hall,J.C. and Rosbash,M. (1991) The strength and periodicity of D.melanogaster circadian rhythms are differentially affected by alterations in period gene expression. Neuron, 6, 753–766. [DOI] [PubMed] [Google Scholar]
- Low-Zeddies S.S. and Takahashi,J.S. (2001) Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell, 105, 25–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinek S., Inonog,S., Manoukian,A.S. and Young,M.W. (2001) A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell, 105, 769–779. [DOI] [PubMed] [Google Scholar]
- McDonald M.J. and Rosbash,M. (2001) Microarray analysis and organization of circadian gene expression in Drosophila. Cell, 107, 567–578. [DOI] [PubMed] [Google Scholar]
- McDonald M.J., Rosbash,M. and Emery,P. (2001) Wild-type circadian rhythmicity is dependent on closely spaced E boxes in the Drosophila timeless promoter. Mol. Cell. Biol., 21, 1207–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckameyer W.S. and White,K. (1992) A single locus encodes both phenylalanine hydroxylase and tryptophan hydroxylase activities in Drosophila. J. Biol. Chem., 267, 4199–4206. [PubMed] [Google Scholar]
- Park J.H. and Hall,J.C. (1998) Isolation and chronobiological analysis of a neuropeptide pigment-dispersing factor gene in Drosophila melanogaster. J. Biol. Rhythms, 13, 219–228. [DOI] [PubMed] [Google Scholar]
- Park J.H., Helfrich-Forster,C., Lee,G., Liu,L., Rosbash,M. and Hall,J.C. (2000) Differential regulation of circadian pacemaker output by separate clock genes in Drosophila. Proc. Natl Acad. Sci. USA, 97, 3608–3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J.L., Blau,J., Rothenfluh,A., Abodeely,M., Kloss,B. and Young,M.W. (1998) double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell, 94, 83–95. [DOI] [PubMed] [Google Scholar]
- Renn S.C., Park,J.H., Rosbash,M., Hall,J.C. and Taghert,P.H. (1999) A pdf neuropeptide gene mutation and ablation of PDF neurons each cause severe abnormalities of behavioral circadian rhythms in Drosophila. Cell, 99, 791–802. [DOI] [PubMed] [Google Scholar]
- Rothenfluh A., Abodeely,M. and Young,M.W. (2000) Short-period mutations of per affect a double-time-dependent step in the Drosophila circadian clock. Curr. Biol., 10, 1399–1402. [DOI] [PubMed] [Google Scholar]
- Rutila J.E., Zeng,H., Le,M., Curtin,K.D., Hall,J.C. and Rosbash,M. (1996) The timSL mutant of the Drosophila rhythm gene timeless manifests allele-specific interactions with period gene mutants. Neuron, 17, 921–929. [DOI] [PubMed] [Google Scholar]
- Rutila J.E., Suri,V., Le,M., So,W.V., Rosbash,M. and Hall,J.C. (1998) CYCLE is a second bHLH–PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell, 93, 805–814. [DOI] [PubMed] [Google Scholar]
- So W.V. and Rosbash,M. (1997) Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J., 16, 7146–7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So W.V., Sarov-Blat,L., Kotarski,C.K., McDonald,M.J., Allada,R. and Rosbash,M. (2000) takeout, a novel Drosophila gene under circadian clock transcriptional regulation. Mol. Cell. Biol., 20, 6935–6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanewsky R., Jamison,C.F., Plautz,J.D., Kay,S.A. and Hall,J.C. (1997) Multiple circadian-regulated elements contribute to cycling period gene expression in Drosophila. EMBO J., 16, 5006–5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G.K., Ousley,A., Darlington,T.K., Chen,D., Chen,Y., Fu,W., Hickman,L.J., Kay,S.A. and Sehgal,A. (2001) Regulation of the cycling of timeless (tim) RNA. J. Neurobiol., 47, 161–175. [DOI] [PubMed] [Google Scholar]
- Yang Z. and Sehgal,A. (2001) Role of molecular oscillations in generating behavioral rhythms in Drosophila. Neuron, 29, 453–467. [DOI] [PubMed] [Google Scholar]