Abstract
Oligonucleosomal fragmentation of chromosomes in dying cells is a hallmark of apoptosis. Little is known about how it is executed or what cellular components are involved. We show that crn-1, a Caenorhabditis elegans homologue of human flap endonuclease-1 (FEN-1) that is normally involved in DNA replication and repair, is also important for apoptosis. Reduction of crn-1 activity by RNA interference resulted in cell death phenotypes similar to those displayed by a mutant lacking the mitochondrial endonuclease CPS-6/endonuclease G. CRN-1 localizes to nuclei and can associate and cooperate with CPS-6 to promote stepwise DNA fragmentation, utilizing the endonuclease activity of CPS-6 and both the 5′–3′ exonuclease activity and a previously uncharacterized gap-dependent endonuclease activity of CRN-1. Our results suggest that CRN-1/FEN-1 may play a critical role in switching the state of cells from DNA replication/repair to DNA degradation during apoptosis.
Keywords: apoptosis/Caenorhabditis elegans/DNA degradation/EndoG/Fen-1
Introduction
Programmed cell death (apoptosis) is an evolutionarily conserved process that is important for development and homeostasis in metazoans. One hallmark of apoptosis is the fragmentation of chromosomal DNA in dying cells into a ‘ladder’ of oligonucleosomal-length fragments (Wyllie, 1980; Zhang and Xu, 2002). Two nucleases, DFF40 (40 kDa DNA fragmentation factor)/CAD (caspase-activated deoxyribonuclease) (Liu et al., 1997, 1998; Enari et al., 1998) and mitochondrial endonuclease G (EndoG) (Li et al., 2001), have been implicated in mediating apoptotic DNA degradation in mammals. DFF40/CAD is activated during apoptosis following caspase cleavage of its cognate inhibitor DFF45/ICAD (inhibitor of CAD) (Liu et al., 1997; Enari et al., 1998) and then associates with nuclear proteins such as histone H1 and HMG proteins to promote cleavage of internucleosomal DNA (Liu et al., 1998; Zhang and Xu, 2002). EndoG is released from mitochondria during apoptosis and translocates to nuclei to mediate DNA fragmentation through a caspase- and DFF40-independent pathway (Li et al., 2001). At physiological ionic strengths EndoG is a poor endonuclease (Widlak et al., 2001), thus it is likely that EndoG cooperates with other nuclear factors to efficiently mediate apoptotic DNA degradation.
Molecular genetic analyses in Caenorhabditis elegans have led to identification of over a dozen genes involved in specification of cell death (ces-1 and ces-2), activation of the cell death machinery (egl-1, ced-9, ced-4 and ced-3) and engulfment of cell corpses (ced-1, -2, -5, -6, -7, -10 and -12) (Horvitz, 1999). In addition, two nucleases, CPS-6 and NUC-1, an EndoG homologue and a type II DNase, respectively, have been implicated in mediating apoptotic DNA degradation in C.elegans, based on the observations that TUNEL (TdT-mediated dUTP nick end labeling)-positive nuclei accumulate in cps-6 or nuc-1 mutants (Wu et al., 2000; Parrish et al., 2001). These observations also indicate that CPS-6 and NUC-1 probably play an important role in resolving TUNEL-reactive DNA breaks generated by some upstream nucleases. In mammals, DFF40/CAD has been implicated in generating TUNEL-reactive DNA breaks (McIlroy et al., 2000); however, despite extensive searching, no DFF40/CAD homologue has been identified in C.elegans (The C.elegans sequencing consortium, 1998; Wu et al., 2000; our unpublished observations). In addition to the TUNEL phenotype, reducing cps-6 activity delays appearance of embryonic cell corpses during development and enhances the cell killing defect of other cell death mutants, suggesting that cps-6 is important for normal progression of apoptosis and can promote cell killing (Parrish et al., 2001). It is not clear how CPS-6 activity is regulated and how it is involved in apoptotic DNA degradation.
In contrast to apoptotic DNA degradation, which leads to fragmentation and destruction of nuclear DNA, DNA repair and replication maintain genome stability and fidelity. Several genes have been identified that play important roles in DNA damage repair and/or replication, including FEN-1 (flap endonuclease-1), which functions in both DNA replication and repair (Harrington and Lieber, 1994; Lieber, 1997). Mutations that disrupt the activity of Saccharomyces cerevisiae Rad27, a homologue of human FEN-1, result in conditional lethality, S-phase arrest and sensitivity to genotoxic stress (Reagan et al., 1995; Vallen and Cross, 1995; Tishkoff et al., 1997). Additionally, the mutant yeast displays a mutator phenotype, presumably due to defective DNA repair (Reagan et al., 1995; Tishkoff et al., 1997). Human FEN-1 possesses both a structure-specific endonuclease activity that specifically cleaves DNA flaps, bifurcated structures composed of double-stranded DNA and a displaced single-strand, and a 5′–3′ exonuclease activity that is specific for double-stranded DNA and may be important for processing Okazaki fragments during DNA replication (Harrington and Lieber, 1994; Bambara et al., 1997; Lieber, 1997).
Since EndoG/CPS-6 defines a conserved DNA degradation pathway (Li et al., 2001; Parrish et al., 2001), we further investigated the mechanisms by which CPS-6/EndoG affects apoptosis. Here we report the identification and characterization of a CPS-6/EndoG cofactor, crn-1, which encodes a C.elegans FEN-1 homologue. We show that CRN-1 plays an important but unexpected role in mediating apoptotic DNA fragmentation and is important for normal progression of apoptosis. We demonstrate that CRN-1 can physically associate with CPS-6 and CPS-6/CRN-1 protein interaction can stimulate both proteins’ nuclease activities in vitro. These findings establish that CRN-1, a protein implicated in DNA replication and repair, can function as a co-factor for the apoptotic nuclease CPS-6 and may provide an important link between DNA repair/replication and apoptotic DNA degradation.
Results
Identification of CRN-1
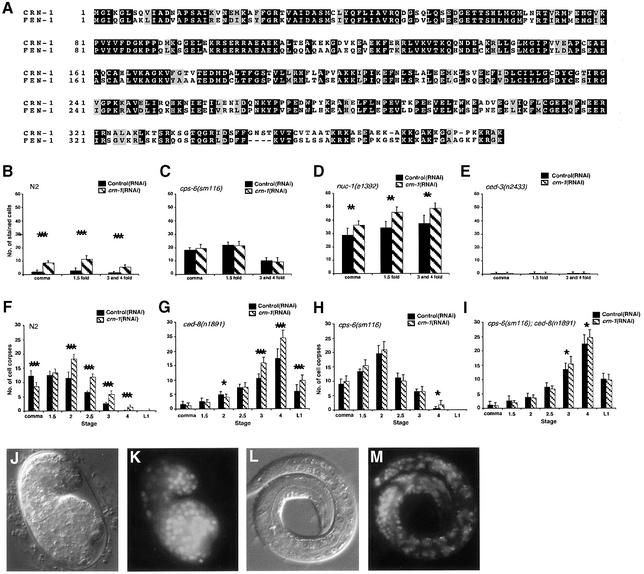
We have conducted a genome-wide screen using RNA interference (RNAi) to identify nucleases involved in apoptotic DNA degradation in C.elegans. From a screen of 77 candidate genes, nine apoptotic nucleases were identified, including two previously known nucleases, CPS-6 and NUC-1 (Parrish and Xue, 2003). One of them, crn-1 (cell death related nuclease 1), is a worm homologue of FEN-1 (Figure 1A), which normally functions in DNA replication and damage repair (Harrington and Lieber, 1994; Bambara et al., 1997; Lieber, 1997). crn-1 is essential for nematode development; crn-1(RNAi)-treated L1 larvae developed normally but laid predominantly dead eggs (95% penetrance). Animals treated at later larval stages (L2 and L3), thus with reduced exposure to crn-1(RNAi), had many surviving progeny (data not shown). These viable, crn-1(RNAi)-treated embryos accumulated TUNEL-positive nuclei throughout embryonic development (Figure 1B) and this TUNEL phenotype can be suppressed by the ced-3(n2433) mutation (Figure 1E), which blocks almost all cell deaths in C.elegans, indicating that the TUNEL-positive nuclei observed in crn-1(RNAi) embryos correspond to apoptotic cells and that crn-1 is involved in apoptotic DNA degradation. Interestingly, crn-1(RNAi) did not enhance the TUNEL phenotype of the cps-6(sm116) mutant but did so in nuc-1(e1392) mutants (Figure 1C and D), suggesting that crn-1 may function in the same DNA degradation pathway as cps-6, which is different from that of nuc-1 (Wu et al., 2000; Parrish et al., 2001). This observation also indicates that TUNEL-positive nuclei observed in crn-1(RNAi)-treated embryos are unlikely to be cells that undergo abnormal cell death due to defects in DNA replication or repair, as the latter scenario should result in more TUNEL-positive nuclei in cps-6(sm116); crn-1(RNAi) animals. The finding that crn-1(RNAi) results in a TUNEL phenotype that is qualitatively similar to that displayed by the cps-6(sm116) mutant suggests that, like cps-6, crn-1 is also involved in resolving the TUNEL-reactive DNA breaks generated during apoptosis.
Fig. 1. crn-1 encodes a FEN-1-like nuclear protein important for C.elegans apoptosis. (A) Alignment of CRN-1 and human FEN-1. Black shaded residues are identical and gray shaded residues are similar in two proteins. (B–E) TUNEL assays. N2 (B), cps-6(sm116) (C), nuc-1(e1392) (D) and ced-3(n2433) (E) animals were treated with control(RNAi) (filled bars) or crn-1(RNAi) (hatched bars) and their progeny were stained with TUNEL. The stages of embryos examined were comma, 1.5-fold and 3- and 4-fold. The y axis represents the mean number of TUNEL-positive cells present in the embryos (at least 12 embryos were scored at each stage). (F–I) Time course analysis of embryonic cell corpses. N2 (F), ced-8(n1891) (G), cps-6(sm116) (H), cps-6(sm116); ced-8(n1891) (I) animals were treated with control(RNAi) or crn-1(RNAi) and their progeny were scored for cell corpses in comma, 1.5-, 2-, 2.5-, 3-, 4-fold stage embryos and early L1 larvae. At least 15 animals were scored for each stage. (B to I) Data derived from control and crn-1(RNAi) treatment at the same stage were compared using unpaired t-test. *P < 0.05; **P < 0.002; ***P < 0.0001. All other points had P values > 0.05. Error bars indicate one standard deviation (SD). (J–M) Nuclear localization of CRN-1. Nomarski (J and L) and GFP fluorescent (K and M) images of a 1.5-fold stage transgenic embryo and a L1 transgenic larva are shown.
CRN-1 affects progression of apoptosis and can promote cell killing in C.elegans
A time-course analysis of embryonic cell corpses indicated that crn-1 affected the normal timing of apoptosis like cps-6 (Parrish et al., 2001). Specifically, crn-1(RNAi) delayed appearance of embryonic cell corpses during development, shifting the peak of cell corpses from the comma embryonic stage in wild-type animals to the 2-fold embryonic stage in crn-1(RNAi) animals (Figure 1F). To rule out the possibility that the delay of cell corpse appearance observed in crn-1(RNAi) animals was due to a delay in embryonic development, we monitored the appearance of embryonic cell corpses in N2 animals treated with RNAi of several genes (F31E8.6, Y47D3A.29, Y57A10A.13) that results in embryonic lethality similar to that caused by crn-1(RNAi) or RNAi that targets other predicted XPG-family DNA repair proteins (F45G2.3, F57B10.6) (Parrish and Xue, 2003). In all cases, we observed a wild-type profile of embryonic cell corpses in viable RNAi-treated embryos (data not shown). Thus, the shifted peak of cell corpses following crn-1(RNAi) is not likely to be a result of delayed embryonic development or defects in DNA repair machinery.
In addition, we found that crn-1(RNAi) enhanced the delay-of-cell-death phenotype of the ced-8(n1891) mutant (Stanfield and Horvitz, 2000), further increasing the numbers of late-appearing cell corpses in ced-8(n1891) embryos (Figure 1G). However, crn-1(RNAi) treatment did not enhance the delayed corpse appearance phenotype of cps-6(sm116) or cps-6(sm116); ced-8(n1891) mutants (Figure 1H and I), further suggesting that crn-1 and cps-6 may function in the same pathway to promote apoptosis.
We also examined whether crn-1(RNAi) could prevent cell death and generate extra ‘undead’ cells in the anterior pharynx of C.elegans (Parrish et al., 2001). On its own, crn-1(RNAi) did not block apoptosis, since few extra cells were seen in crn-1(RNAi) animals (Table I). Importantly, we did not observe any cell loss in crn-1(RNAi) animals, suggesting that crn-1(RNAi) does not cause ectopic cell deaths (data not shown). However, crn-1(RNAi) could enhance the cell killing defect of other cell death mutants, including mutants partially or strongly defective in two essential cell-killing genes, ced-3 and ced-4 (Table I). For example, a mean of only 1.6 extra cells was seen in the anterior pharynx of weak ced-3(n2447) mutants, compared with a mean of 2.7 extra cells seen in ced-3(n2447); crn-1(RNAi) animals (Table I). crn-1(RNAi) similarly enhanced cell survival in several other mutants including weak ced-4(n2273) mutant and strong ced-3(n2433) and ced-4(n1162) mutants (Table I). However, crn-1(RNAi) did not increase the number of extra cells observed in cps-6(sm116), cps-6(sm116); ced-3(n2447) or cps-6(sm116); ced-4(n2273) mutants (Table I). These results provide further evidence that crn-1 and cps-6 may promote programmed cell death through the same pathway.
Table I. crn-1 promotes cell killing in C.elegans.
| Strain | No. scored | Extra cells | |
|---|---|---|---|
| |
|
Mean ± SEM |
Range |
| N2; control(RNAi) | 18 | 0 | 0 |
| N2; crn-1(RNAi) | 22 | 0.09 ± 0.06 | 0–1 |
| ced-8(n1891); control(RNAi) | 16 | 0.87 ± 0.16 | 0–2 |
| ced-8(n1891); crn-1(RNAi)b | 15 | 1.50 ± 0.25 | 0–3 |
| ced-3(n2447); control(RNAi) | 16 | 1.56 ± 0.25 | 0–3 |
| ced-3(n2447); crn-1(RNAi)c | 16 | 2.69 ± 0.28 | 1–4 |
| ced-3(n2433); control(RNAi) | 15 | 13.3 ± 0.42 | 11–16 |
| ced-3(n2433); crn-1(RNAi) | 15 | 14.2 ± 0.36 | 12–16 |
| ced-4(n2273); control(RNAi) | 15 | 3.04 ± 0.37 | 1–6 |
| ced-4(n2273); crn-1(RNAi)b | 16 | 4.50 ± 0.38 | 2–7 |
| ced-4(n1162); control(RNAi) | 17 | 12.7 ± 0.43 | 10–15 |
| ced-4(n1162); crn-1(RNAi)b | 15 | 13.7 ± 0.39 | 11–15 |
| cps-6(sm116); control(RNAi) | 18 | 0.06 ± 0.06 | 0–1 |
| cps-6(sm116); crn-1(RNAi) | 15 | 0.07 ± 0.07 | 0–1 |
| cps-6(sm116); ced-8(n1891); control(RNAi) | 17 | 1.35 ± 0.18 | 0–3 |
| cps-6(sm116); ced-8(n1891); crn-1(RNAi) | 16 | 1.31 ± 0.18 | 0–2 |
| cps-6(sm116); ced-3(n2447); control(RNAi)a | 16 | 2.63 ± 0.28 | 1–5 |
| cps-6(sm116); ced-3(n2447); crn-1(RNAi)a | 15 | 2.74 ± 0.28 | 1–5 |
| cps-6(sm116); ced-4(n2273); control(RNAi)a | 15 | 3.80 ± 0.29 | 2–6 |
| cps-6(sm116); ced-4(n2273); crn-1(RNAi)a | 15 | 4.00 ± 0.29 | 2–6 |
aThese strains also contain dpy-5(e61).
bNumbers of extra cells from animals treated with control(RNAi) and crn-1(RNAi) were compared using unpaired t-test, P < 0.01.
cP < 0.002, unpaired t-test.
CRN-1 localizes to nuclei in C.elegans
FEN-1 is important for DNA replication, participating in Okazaki fragment processing (Li et al., 1995; Bambara et al., 1997; Lieber, 1997) and DNA damage repair, including base excision repair (Lieber, 1997; Kim et al., 1998; Shibata and Nakamura, 2002). Loss-of-function mutations in Rad-27, the S.cerevisiae FEN-1 homologue, cause conditional lethality, a mutator phenotype and sensitivity to genotoxic stress, underscoring its importance in genome maintenance (Reagan et al., 1995; Vallen and Cross, 1995; Tishkoff et al., 1997). Likewise, C.elegans crn-1 is essential for viability, suggesting a possible developmental role in DNA replication and repair. We have analyzed crn-1 expression using a fusion protein composed of CRN-1 and green fluorescent protein (CRN-1::GFP) under the control of its own promoter (Pcrn-1) and found that CRN-1 was ubiquitously expressed in C.elegans, beginning early in embryogenesis and lasting through late larval stages (data not shown). Importantly, CRN-1::GFP was found exclusively in nuclei (Figure 1J–M), consistent with its role in mediating chromosome fragmentation during apoptosis and a possible role in DNA replication and repair.
CRN-1 interacts with CPS-6 in vitro
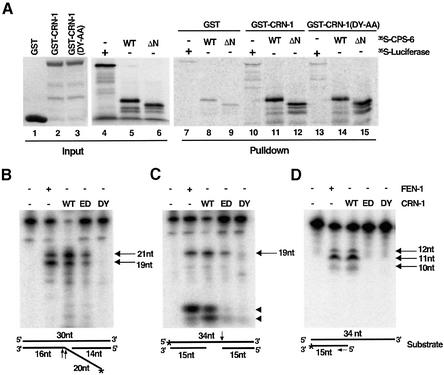
Since cps-6 and crn-1 appear to act in the same cell death pathway (Figure 1; Table I), we investigated whether CPS-6 and CRN-1 directly interact in vitro. As shown in Figure 2A, a CRN-1 glutathione S-transferase (GST) fusion protein [GST–CRN-1(21–382)] bound full-length, [35S]methionine-labeled CPS-6 in a GST fusion protein pulldown assay. This CRN-1/CPS-6 interaction is specific, since GST alone did not bind CPS-6 and GST–CRN-1(21–382) did not pull down an unrelated protein, luciferase (Figure 2A). Furthermore, CRN-1 interacted well with CPS-6(21–308), a truncated version of CPS-6 that lacks the mitochondria targeting sequence and localizes to nuclei instead of mitochondria [the mitochondria targeting sequence of EndoG, and probably CPS-6, is cleaved off following its import into mitochondria (Li et al., 2001; Parrish et al., 2001)], suggesting that the mature form of CPS-6 can interact with CRN-1. These data further suggest that CPS-6 and CRN-1 may function together at the same step of apoptotic DNA degradation.
Fig. 2. Nuclease activities of CRN-1 and its interaction with CPS-6. (A) CRN-1 binds CPS-6. Purified GST or GST-fusion proteins (5 µg each) were used to precipitate [35S]methionine-labeled proteins as indicated. WT indicates the wild-type CPS-6 protein. ΔN denotes CPS-6(21–308). 30% of input [35S]methionine-labeled proteins is shown. (B) CRN-1 has flap endonuclease activity. FEN-1 or CRN-1 proteins (wild type or mutant) synthesized in the reticulocyte lysate were incubated with the labeled flap substrate, which is schematized below the image (lengths of oligonucleotides are indicated and * indicates the position of 32P-labeling). Cleavage products (19 and 21 nt) and their respective cleavage sites on the substrate (1 nucleotide 5′ or 3′ of the branch point) are indicated by arrows. WT, wild-type CRN-1; ED, CRN-1(E160D); DY, CRN-1(DY-AA). (C) CRN-1 possesses a previously uncharacterized gap-dependent endonuclease activity. A different labeled substrate was incubated with FEN-1 or CRN-1 proteins. The 19 nt endonucleolytic cleavage product and its corresponding cleavage site on the substrate are indicated with an arrow. The low-molecular-weight bands (indicated with arrowheads) observed at the bottom of the gel are products resulting from CRN-1 5′–3′ exonuclease digestion of the labeled 5′ blunt end. Neither CRN-1(E160D) nor CRN-1(DY-AA) is capable of generating these products since both lack the 5′–3′ exonuclease activity. (D) CRN-1 has 5′–3′ exonuclease activity. A 3′-end-labeled substrate was incubated with FEN-1 or CRN-1 proteins. The sizes of multiple cleavage products (indicated by arrows) are consistent with successive removal of 1 nt from the 5′ end of the labeled strand (indicated by an arrow) by the exonuclease. Reactions from panels B–D were resolved on 12% polyacrylamide/7 M urea gels and visualized using phosphorimager.
CRN-1 is a flap endonuclease and a 5′–3′ exonuclease like FEN-1 and possesses a new gap-dependent endonuclease activity
FEN-1 is a structure-specific endonuclease that processes DNA flaps (bifurcated structures composed of double-stranded DNA and a displaced single strand) and a 5′–3′ exonuclease (Harrington and Lieber, 1994). Both nuclease activities are important for the function of FEN-1 in DNA damage repair and replication (Harrington and Lieber, 1994; Nolan et al., 1996; Shen et al., 1996; Lieber, 1997). Like FEN-1, CRN-1 cleaved a synthetic flap substrate, generating two characteristic cleavage products of 19 and 21 nucleotides (Figure 2B) (Harrington and Lieber, 1994). Furthermore, mutations in CRN-1 (D233A and Y234A; DY-AA) that alter conserved residues important for FEN-1 nuclease activities (Hosfield et al., 1998) also abolished the flap endonuclease activity of CRN-1 (Figure 2B), confirming that CRN-1 has flap endonuclease activity like FEN-1.
Interestingly, we found that both CRN-1 and FEN-1 possessed a second substrate-specific endonuclease activity that was not reported previously. Both proteins could endonucleotically cleave a double-stranded DNA substrate with a 4 nt single-stranded gap at the 3′ end of the gap (Figure 2C). This new gap-dependent endonuclease activity was also observed with a substrate that has 32 bp double-stranded DNA regions flanking a similar 4 nt gap (data not shown) and was lost in the CRN-1(DY-AA) mutant protein (Figure 2C).
We next tested if CRN-1 has 5′–3′ exonuclease activity like FEN-1 (Harrington and Lieber, 1994). We found that both FEN-1 and CRN-1 could process a labeled 5′ blunt end of a double-stranded oligonucleotide substrate to generate low molecular weight labeled nucleotides (bottom bands indicated by arrowheads in Figure 2C), indicative of 5′–3′ exonuclease activity. Additionally, both FEN-1 and CRN-1 cleaved a double-stranded substrate containing a 5′ recessed end and a labeled 3′ blunt end to generate a ladder of labeled products resulting from 5′–3′ exonuclease digestion (Figure 2D). In both assays, CRN-1(DY-AA) lacked 5′–3′ exonuclease activity (Figure 2C and D). Interestingly, a mutation (E160D) in CRN-1 specifically abolished its 5′–3′ exonuclease activity but only partially reduced its flap or gap-dependent endonuclease activity (Figure 2B–D). The similarities of CRN-1 and FEN-1 in their nuclease activities suggest that CRN-1 is a functional homologue of FEN-1.
CRN-1 enhances CPS-6 nuclease activity in vitro
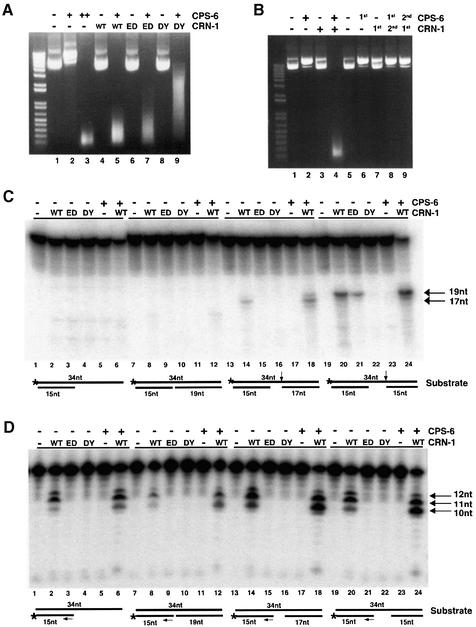
Since CRN-1 and CPS-6 interacted in vitro, we tested whether they affect each other’s activity using a plasmid cleavage assay (Li et al., 2001). At a low concentration (50 ng), CPS-6 caused single-stranded nicking of plasmid DNA, generating products with slower mobility (Figure 3A, lane 2). At a 5-fold higher concentration (250 ng), CPS-6 further fragmented plasmid DNA, generating a smear of smaller products (lane 3). CRN-1 alone had no detectable plasmid nicking or cleaving activity, even at very high concentrations (lane 4; data not shown). However, adding CRN-1 to a reaction where CPS-6 alone only induced plasmid nicking resulted in complete plasmid degradation and thus an ∼5-fold increase in nuclease activity (lane 5), suggesting that CRN-1 could potentiate CPS-6 nuclease activity. Interestingly, WAH-1, the C.elegans homologue of AIF (apoptosis-inducing factor), also enhances CPS-6 nuclease activity in vitro (Wang et al., 2002). However, WAH-1 did not interact with CRN-1 or affect CRN-1 activity and could not further stimulate CPS-6 activity in the presence of CRN-1 (data not shown), suggesting that CRN-1 and WAH-1 may use a similar mechanism to stimulate CPS-6 nuclease activity.
Fig. 3. CRN-1 and CPS-6 cooperate to promote DNA degradation. (A) Plasmid cleavage assay. CPS-6 (‘+’ denotes 50 ng, ‘++’ denotes 250 ng) or CRN-1 proteins (250 ng each) was incubated either alone or together as indicated with plasmid DNA (1 µg). WT, wild-type CRN-1; ED, CRN-1(E160D); DY, CRN-1(DY-AA). (B) Simultaneous presence of CRN-1 and CPS-6 is important for DNA degradation. In lanes 2–4, plasmid DNA was mock-treated with buffer and passed over Ni2+ NTA resin to simulate the depletion step. His6CPS-6 or CRN-1-His6 was subsequently incubated either alone or together with plasmid DNA for 30 min. In lanes 6–9, His6CPS-6 (lanes 6 and 8) or CRN-1-His6 (lanes 7 and 9) was first incubated with plasmid DNA for 30 min, depleted using Ni2+ NTA resin (1st), and plasmid DNA was subsequently incubated with CRN-1-His6 (2nd; lane 8) or His6CPS-6 (2nd; lane 9), or mock treated (buffer alone; ‘–’) for another 30 min. His6CPS-6 (50 ng) and 250 ng of CRN-1-His6 were used in all reactions. (C) CPS-6 enhances CRN-1 gap-dependent endonuclease activity. CRN-1 proteins (WT or mutants) or CPS-6 were incubated either alone or together, as indicated, with different substrates (schematized below reactions in which they were used). The endonucleolytic product sizes (indicated with arrows) increase with the increasing lengths of the single-stranded gaps in the substrates. (D) CPS-6 enhances CRN-1 5′–3′ exonuclease activity. Reactions were carried out as in (C) except that substrates were 3′-end labeled to monitor 5′–3′ exonuclease activity. Twelve, 11 and 10 nt exonucleolytic products were most prominent (indicated with arrows). In the presence of both CRN-1 and CPS-6, additional, smaller products were also visible. Reactions from panels C and D were resolved on 15% polyacrylamide/7 M urea gels and visualized using phosphorimager.
We next examined whether CRN-1 nuclease activities are important for enhancing CPS-6 activity by including the exonuclease-defective CRN-1(E160D) protein or nuclease-defective CRN-1(DY-AA) protein in plasmid cleavage reactions with CPS-6. We found that loss of CRN-1 exonuclease activity attenuated the ability of CRN-1 to stimulate CPS-6 activity, as shown by the presence of larger DNA fragments (Figure 3A, lane 7), and the loss of both endonuclease and exonuclease activities of CRN-1 further reduced the ability of CRN-1 to activate CPS-6 (lane 9), suggesting that both the exonuclease and the endonuclease activities of CRN-1 contribute to the stimulation of CPS-6 activity. However, the nuclease-defective CRN-1(DY-AA) protein was still capable of enhancing CPS-6 activity, albeit less potently than wild-type CRN-1. Since CRN-1(DY-AA) bound CPS-6 as well as wild-type CRN-1 (Figure 2A), association of CRN-1(DY-AA) with CPS-6 may be sufficient to stimulate CPS-6 nuclease activity, possibly by changing the conformation of the CPS-6 protein. Taken together, these observations indicate that CRN-1 can enhance CPS-6 nuclease activity through both nuclease-dependent and -independent mechanisms.
We also tested if CPS-6 and CRN-1 act in a sequential manner to cleave DNA by pre-incubating plasmid DNA with CPS-6, depleting CPS-6 from the reaction before adding CRN-1, or reversing the incubation order of the two proteins. In both cases, we were unable to observe any enhancement of the CPS-6 activity by CRN-1 (Figure 3B), indicating that CRN-1 and CPS-6 need to be simultaneously present to synergistically promote DNA degradation.
CPS-6 enhances CRN-1 gap-dependent endonuclease and 5′–3′ exonuclease activities
We next examined whether CPS-6 could enhance CRN-1 nuclease activities. Interestingly, CPS-6 had no effect on CRN-1 flap endonuclease activity (data not shown), but could enhance the gap-dependent endonuclease activity of CRN-1 (Figure 3C). We found that ungapped single-stranded DNA substrates (lane 2) or double-stranded DNA substrates with a nick (no gap; lane 8) or a 1 nt gap (data not shown) were not endonucleotically cleaved by CRN-1. The gap-dependent endonuclease activity was detectable when the gap size was increased to 2 nt, and was stronger when the gap size was increased to 4 nt (lanes 14 and 20). Although CPS-6 was unable to process any of these substrates on its own, it enhanced the gap-dependent endonuclease activity of CRN-1 by >2-fold, as quantified from the intensities of the cleavage products using phosphorimager analysis (lanes 18 and 24). In addition, a nuclease-defective CPS-6 mutant protein [CPS-6 (D134A, Y135A); data not shown] failed to enhance or reduce CRN-1 gap-dependent endonuclease activity (data not shown), suggesting that the nuclease activity of CPS-6 most likely contributes to the stimulation of CRN-1 gap-dependent endonuclease activity.
Interestingly, CRN-1 had somewhat similar substrate preferences for its 5′–3′ exonuclease activity. Although it could process 5′ recessed ends (Figure 3D, lane 2) or 5′ ends near a nick (lane 8) or 1 nt gap (data not shown) in double-stranded substrates, CRN-1 had stronger 5′–3′ exonuclease activity when the single-stranded gap was 2 or 4 nt long (lanes 14 and 20). In these two cases, addition of CPS-6 significantly enhanced CRN-1 exonuclease activity, generating smaller cleavage products (lanes 18 and 24). Again, CPS-6 alone had no activity in processing any of the substrates (Figure 3D).
We also tested the possibility that the CRN-1 activities are modulated by the CED-3 caspase, a likely activator of many cell death execution events including apopototic DNA degradation, but failed to find any significant CED-3-mediated cleavage of CRN-1 or modulation of CRN-1 nuclease activities (data not shown). Taken together, these observations suggest that CPS-6 can enhance both the new endonuclease and 5′–3′ exonuclease activities of CRN-1 in a gap-dependent manner and offer important insights into the type of substrates or cleavage intermediates that CRN-1/CPS-6 might process or generate during apoptosis (see Discussion).
CRN-1 can induce ectopic cell deaths in a CPS-6-dependent manner
We next examined whether CRN-1 nuclease activities and its interaction with CPS-6 are important for its activity in promoting programmed cell death. We found that expression of crn-1 in six non-essential touch receptor neurons under the control of the mec-7 gene promoter (Pmec-7crn-1) induced ectopic neuronal death (Savage et al., 1989), killing ∼16–20% (when expressed from low copy transgenes) or ∼60–77% (when expressed from high copy transgenes) of the posterior lateral microtubule (PLM) touch receptor neurons (Table II). Ectopic expression of CRN-1(E160D) or CRN-1(DY-AA) in touch cells only weakly induced cell killing (Table II), suggesting that both the exonuclease and the endonuclease activities of CRN-1 are important for its killing activity. Importantly, crn-1-induced ectopic neuronal deaths were significantly inhibited by the cps-6(sm116) or the ced-3(n2433) mutation (Table II), indicating that CRN-1 induced cell death through existing apoptotic programs mediated by cps-6 and ced-3. Furthermore, the nuclease-defective CPS-6 protein [CPS-6(D134A, Y135A)], which by itself has very weak cell killing activity (Table II), significantly inhibited CRN-1 killing activity when co-expressed with CRN-1 in touch cells, suggesting that the mutant CPS-6 protein may dominant-negatively inhibit crn-1 cell killing (Table II). These results provide further evidence that CPS-6 and CRN-1 function together to promote apoptosis.
Table II. Overexpression of crn-1 induces ectopic cell killing.
| Straina | High concentrationb |
Low concentrationb |
||
| |
Array |
% PLM survivalc |
Array |
% PLM survivalc |
| N2 | 100 | |||
| N2; control(RNAi) | 100 | |||
| N2; crn-1(RNAi) | 97 | |||
| ced-3(n2433) | 100 | |||
| cps-6(sm116) | 100 | |||
| N2; Pmec-7CRN-1 | 1 | 38 | 1 | 82 |
| 2 | 40 | 2 | 84 | |
| 3 | 23 | 3 | 80 | |
| N2; Pmec-7 CRN-1(E160D) | 1 | 80 | 1 | 93 |
| 2 | 90 | 2 | 90 | |
| 3 | 87 | 3 | 90 | |
| N2; Pmec-7 CRN-1(DY-AA) | 1 | 83 | 1 | 100 |
| 2 | 87 | 2 | 100 | |
| 3 | 90 | 3 | 97 | |
| ced-3(n2433); Pmec-7CRN-1 | 1 | 75 | 1 | 97 |
| 2 | 70 | 2 | 97 | |
| 3 | 69 | 3 | 100 | |
| cps-6(sm116); Pmec-7CRN-1 | 1 | 66 | 1 | 93 |
| 2 | 72 | 2 | 93 | |
| 3 | 60 | 3 | 97 | |
| N2; Pmec-7 CPS-6(DY-AA) | 1 | 93 | 1 | 100 |
| 2 | 93 | 2 | 97 | |
| 3 | 87 | 3 | 100 | |
| N2; Pmec-7CRN-1/Pmec-7 | 1 | 69 | 1 | 97 |
| CPS-6(DY-AA) | 2 | 75 | 2 | 93 |
| 3 | 67 | 3 | 100 | |
aAll strains contain an integrated transgene, bzIs8, which directs GFP expression in six mechanosensory neurons under the control of the mec-4 gene promoter and is used to help identify the PLML/R neurons. N2 is the wild-type strain. CPS-6(DY-AA) denotes CPS-6(D134A, Y135A). CRN-1(DY-AA) denotes CRN-1(D233A, Y234A). crn-1(RNAi) did not result in any ectopic neuronal deaths.
bTransgenes were injected at a concentration of 50 µg/ml (high concentration) or 5 µg/ml (low concentration) with pRF4 (at 50 µg/ml), a dominant co-injection marker (Mello et al., 1992).
cPLM survival was scored in transgenic L4 larvae based on the number of fluorescent PLMR/L neurons detected using a fluorescent Nomarski microscope. At least 15 animals were scored for each transgenic line.
Discussion
CRN-1 is a CPS-6 co-factor
We have shown here that CRN-1, a new apoptotic nuclease identified from a functional genomic screen, associates and cooperates with another apoptotic nuclease CPS-6 to promote apoptotic DNA degradation. This conclusion is supported by the in vitro observations that CRN-1 bound CPS-6 and these two proteins mutually stimulated each other’s nuclease activities. It is reinforced by the in vivo observations that crn-1(RNAi) caused cell death defects very similar to those caused by the cps-6(sm116) mutation, including an apoptotic DNA degradation defect (the TUNEL phenotype), that crn-1(RNAi) did not enhance the cell death defects observed in cps-6(sm116) mutants, and that crn-1 induced ectopic cell death in a cps-6-dependent manner. These findings suggest that CRN-1, a nuclear protein, may serve as a nuclear co-factor for CPS-6 to promote apoptotic DNA degradation.
Interestingly, reduction of activity of any of the three apoptotic nucleases discussed in this study, CPS-6, CRN-1 and NUC-1, results in very similar DNA degradation phenotype (accumulation of TUNEL-positive nuclei in affected embryos), indicating that they all play a role in resolving TUNEL-reactive DNA degradation intermediates. However, several lines of evidence indicate that cps-6/crn-1 and nuc-1 may function in different DNA degradation pathways and at different stages of apoptosis. Firstly, both cps-6(sm116); nuc-1(e1392) double mutant embryos and nuc-1(e1392); crn-1(RNAi) embryos have higher numbers of TUNEL-positive cells than those of single mutant embryos or crn-1(RNAi) embryos alone (Figure 1D; Parrish et al., 2001), suggesting that cps-6/crn-1 act in a different DNA degradation pathway from nuc-1. Secondly, nuc-1 is dispensable for the killing process and the timing of apoptosis, whereas both cps-6 and crn-1 are important for normal progression of apoptosis and can contribute to cell killing (Wu et al., 2000; Parrish et al., 2001). Thus cps-6/crn-1 may act at an earlier stage of apoptosis than nuc-1. Thirdly, NUC-1 appears to function in multiple DNA degradation processes, including digestion of DNA from ingested bacteria in intestines, a non-apoptotic event (Wu et al., 2000). In contrast, CPS-6 appears to be specialized for apoptotic DNA degradation (Parrish et al., 2001; data not shown). Finally, using various cytological markers, we have found that CPS-6 functions earlier than NUC-1 relative to other apoptotic events (our unpublished observations), consistent with the hypothesis that cps-6/crn-1 acts in an earlier stage of apoptosis.
CRN-1 and CPS-6 cooperate to mediate stepwise DNA degradation
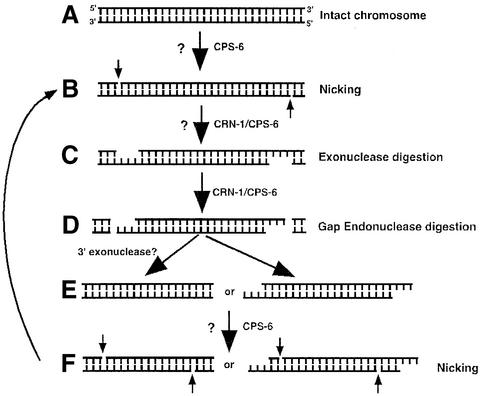
CRN-1 is a structure-specific nuclease that cleaves DNA flaps and also possesses a 5′–3′ exonuclease activity. In this study, we uncovered an additional structure-specific endonuclease activity associated with both CRN-1 and FEN-1: the gap-dependent endonuclease activity. In contrast, CPS-6 is an endonuclease that generates single-stranded nicks in double-stranded DNA substrates and has low enzymatic activity at physiological ionic strengths (Widlak et al., 2001). So how do these two drastically different nucleases work together to promote apoptotic DNA degradation in C.elegans? On the one hand, we found that CRN-1 can potently enhance CPS-6 nuclease activity through both CRN-1 nuclease-independent and -dependent mechanisms. Firstly, nuclease-defective CRN-1(DY-AA) can still bind CPS-6 and partially stimulate CPS-6 activity, suggesting that either association of CRN-1 with CPS-6 or interactions among CRN-1, CPS-6 and their substrate may account for such a stimulatory activity. In this case, it could be similar to WAH-1-mediated enhancement of CPS-6 because WAH-1 binds CPS-6 and is not a nuclease itself (Wang et al., 2002). Indeed, WAH-1 cannot further increase the activity of CPS-6 in the presence of CRN-1 (data not shown). Additionally, both the exonuclease and the endonuclease activities of CRN-1 contribute to the stimulation of CPS-6 activity, since loss of either exonuclease or endonuclease activity attenuates the ability of CRN to stimulate CPS-6. In this regard, these findings are consistent with the observations that the activity of EndoG can be enhanced by an endonuclease, DNase I and an exonuclease, ExoIII (Widlak et al., 2001). On the other hand, CPS-6 can also specifically enhance the 5′–3′ exonuclease activity and the gap-dependent endonuclease activity of CRN-1 but not its flap endonuclease activity, which appears to be specialized for DNA replication/repair (Bambara et al., 1997; Lieber, 1997). Thus through such a unique combination, CPS-6 and CRN-1 can act cooperatively to initiate and facilitate DNA fragmentation (Figure 4), starting with the nicking of double-strand DNA by CPS-6 (and most likely other nicking nucleases, Figure 4A), followed by the expansion of single-stranded DNA nicks into single-stranded DNA gaps by CRN-1 5′–3′ exonuclease activity (with the help of other exonucleases; Figure 4B) and the subsequent double-stranded DNA breaks by CRN-1 gap-dependent endonuclease activity (Figure 4C).
Fig. 4. Molecular model for CRN-1/CPS-6-mediated chromosome fragmentation during apoptosis. (A) Intact chromosomal DNA is likely nicked by CPS-6 (aided by CRN-1) and/or another nuclease (denoted by ?). (B) Following nicking, the 5′–3′ exonuclease activities of CRN-1 (aided by CPS-6), and possibly, other exonucleases (?) turn the nicks into gaps. (C) The resulting gapped substrates are cleaved by CRN-1 gap-dependent endonuclease activity (aided by CPS-6), resulting in fragmented substrates (D) which either are further processed by a 3′–5′ exonuclease(s) (?) (E) or can be directly processed (F) through similar steps (A–D) to generate smaller DNA fragments.
However, CPS-6 and CRN-1 are unlikely to act alone in vivo and almost certainly need help at various steps of DNA fragmentation. For example, an additional nuclease(s) may participate in initiating apoptotic DNA degradation by nicking the double-stranded chromosomal DNA (Figure 4A). An additional exonuclease(s) would open the nicks created by the initiator nucleases (Figure 4B), which allows CRN-1, a 5′–3′ exonuclease that works best on 2 to 4 nt gap substrates (Figure 3D), to further expand the single-stranded DNA gaps (Figure 4B). Nevertheless, our detailed mechanistic study of how CRN-1 and CPS-6 work together to mediate DNA degradation provides the first glimpse on how various components of the apoptotic DNA degradation machinery may act cooperatively to promote step-by-step apoptotic DNA degradation.
CRN-1/FEN-1 may define a critical switch from DNA repair/replication to apoptotic DNA degradation
CRN-1 encodes a homologue of human FEN-1 and yeast Rad27p, which are nucleases important for DNA replication and damage repair (Bambara et al., 1997; Lieber, 1997). The requirement of crn-1 for C.elegans embryonic development and the finding that CRN-1 possesses key biochemical properties characteristic of FEN-1 suggest that crn-1 plays an important role in DNA replication/repair in C.elegans, in addition to its newly discovered role in apoptosis. Therefore, crn-1 may serve as a critical switch between DNA replication/repair and degradation, two opposing biological events. Normally, CRN-1/FEN-1 assists in nuclear DNA replication and repair to maintain genome stability and fidelity. Pro-apoptotic stimuli could trigger translocation of CPS-6/EndoG from mitochondria to nuclei, facilitating association of CPS-6/EndoG with CRN-1/FEN-1 in nuclei, which then transforms CRN-1/FEN-1 from a genome stabilizer to a genome destroyer. Thus, CRN-1/FEN-1 could act as a ‘double agent’ like cytochrome c in regulating both cell survival and cell death.
Materials and methods
Caenorhabditis elegans strains and genetics
Caenorhabditis elegans animals were cultured and maintained using standard procedures (Brenner, 1974). All alleles used in this study have been described previously (Parrish et al., 2001). Germline transformation experiments were carried out as described (Mello et al., 1991). Constructs were injected into host strains (at 5–50 µg/ml) with pRF4 (at 50 µg/ml), which is used as a co-injection marker (Mello et al., 1991).
RNAi experiments
RNAi experiments were carried out using a bacterial feeding method (Parrish et al., 2001). Briefly, L2 hermaphrodite larvae were transferred to plates seeded with bacteria expressing either control or crn-1 dsRNA and the progeny of treated animals were scored for RNAi phenotypes.
Quantification of cell corpses, extra cells and PLM neurons
Cell corpses in the head region of embryos or larvae and extra cells in anterior pharynges of L3 hermaphrodites were scored using Nomarski optics (Parrish et al., 2001). PLM survival was scored in the presence of an integrated array, bzIs8, using a Nomarski microscope equipped with epifluorescence. bzIs8 contains the Pmec-4GFP construct, which directs GFP expression in six mechanosensory neurons under the control of the mec-4 gene promoter (M.Driscoll, personal communication). CRN-1 proteins (wild-type or mutant) or the CPS-6 protein were expressed under the control of the mec-7 promoter (Savage et al., 1989) to assay for their killing activities.
Molecular biology
Standard procedures (Sambrook et al., 1989) were used for plasmid construction and a Quick-change mutagenesis kit (Stratagene) was used to generate mutations. To generate the Pcrn-1 crn-1::gfp construct, the entire crn-1 coding region with 3491 bp of 5′ upstream region was amplified through PCR using an expand long template PCR kit (Roche) and then subcloned into the pPD95.77 vector via its SphI and SalI sites. Other detailed procedures, including sequences of oligonucleotide primers used for PCR and mutagenesis, are available upon request.
Protein purification
Recombinant His6CPS-6, CRN-1-His6, or GST–CRN-1 proteins were expressed in Escherichia coli BL21(DE3)pLysS and purified using similar procedures as described previously (Parrish et al., 2000).
Endonuclease and exonuclease assays
For flap endonuclease assays, three oligonucleotides were annealed to generate a flap substrate (Harrington and Lieber, 1994): FLAP (5′- GATGTCAAGCAGTCCTAACTTTGAGGCAGAGTCC-3′), FLAP-Br (5′- GGACTCTGCCTCAAGACGGTAGTCAACGTG-3′) and FLAP-Adj (5′-CACGTTGACTACCGTC-3′). For 5′–3′ exonuclease and gap-dependent endonuclease assays, FLAP-Br Long (5′-GGACTCTGCCTCAAGACGGTAGTCAACGTGGTGTG-3′) and FLAP-Blunt (5′-CTTGAGGCAGAGTCC-3′) were annealed to generate a double-strand substrate with a 5′ recessive end. FLAP 3′ AS (5′-CACACCACGTTGACTACCGT-3′) or its derivatives with 1, 2 or 4 fewer nucleotides at the 3′ end were annealed with FLAP-Br Long and FLAP-Blunt to generate double-stranded substrates with a nick or various single-stranded gaps. Prior to annealing, one of the oligonucleotides was either 5′-end labeled with [γ-32P]ATP using T4 polynucleotide kinase or 3′-end labeled with [α-32P]cordycepin-5′-triphosphate using terminal deoxynucleotide transferase and subsequently purified on 7 M urea–polyacrylamide gels. For each substrate, 2 pmol of unlabeled oligonucleotides were mixed with 50 nmol of labeled oligonucleotides in annealing buffer (10 mM Tris pH 8.0, 50 mM KCl and 1 mM EDTA), heated to 80°C, and slowly cooled down to 25°C to facilitate annealing. Annealing efficiency was monitored on 5% non-denaturing polyacrylamide gels. Nuclease assays were carried out in 50 mM Tris pH 8.0, 5 mM MgCl2, 0.5 mM β-mercaptoethanol and 0.1 µg/ml bovine serum albumin. Labeled substrate (100 fmol) was incubated with 0.2 µl of the indicated protein (synthesized using Promega TNT-coupled reticulocyte lysate system). Reactions were incubated at 30°C for 45 min, resolved on 7 M urea/15% polyacrylamide gels, and analyzed using a phosphorimager (Molecular Dynamics). Imagequant software was used to quantify the apparent amounts of substrates and cleavage products observed on a given gel. Relative changes in a specific nuclease activity were calculated based on the amount of substrate cleaved (measured by substrate disappearance) and the amount of cleavage products generated for given lanes within a single gel and the obtained values were confirmed by multiple independent experiments. Using this quantification method, CPS-6 was found to consistently enhance CRN-1 gap-dependent endonuclease activity and 5′–3′ exonuclease activity by ∼2- to 3-fold.
Plasmid cleavage assay
Recombinant proteins were incubated with 1 µg of plasmid DNA in 20 mM HEPES pH 7.5, 10 mM KCl, 3 mM MgCl2 and 0.5 mM dithiothreitol for 0.5–1.5 h at 37°C and reactions were resolved on 1% agarose gels. To assay the enhancement of CPS-6 plasmid cleavage activity mediated by CRN-1, the amount of CPS-6 required to fully degrade plasmid DNA in 30 min was defined as 1 unit (250 ng). In the presence of 250 ng of CRN-1, only 0.2 units (50 ng) of CPS-6 were required to achieve complete plasmid digestion, thus CPS-6 had 5-fold higher activity in this assay in the presence of excess CRN-1.
GST pulldown experiments
GST fusion protein pulldown assays were performed as described (Parrish et al., 2000). Briefly, purified GST or GST fusion proteins (5 µg each) immobilized on glutathione–Sepharose beads (Amersham) were incubated with [35S]methionine-labeled proteins at 4°C for 2 h. The beads were washed extensively and the bound proteins were resolved on 12% SDS–PAGE and visualized by autoradiography.
Acknowledgments
Acknowledgements
We thank Y.Kohara, A.Fire and M.Driscoll for reagents and strains, M.Winey and Y.C.Wu for critical reading of the manuscript. This study was supported by a NSF Graduate Research Fellowship (to J.P.), an NCI grant CA 85344 (to B.S.), an NIH grant GM59083, a DOD grant DAMD17-01-1-0214, a Burroughs Wellcome Fund Career Award and a Searle Scholar Award (to D.X.).
References
- Bambara R.A., Murante,R.S. and Henricksen,L.A. (1997) Enzymes and reactions at the eukaryotic DNA replication fork. J. Biol. Chem., 272, 4647–4650. [DOI] [PubMed] [Google Scholar]
- Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics, 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enari M., Sakahira,H., Yokoyama,H., Okawa,K., Iwamatsu,A. and Nagata,S. (1998) A caspase-activated DNase that degrades DNA during apoptosis and its inhibitor ICAD. Nature, 391, 43–50. [DOI] [PubMed] [Google Scholar]
- Harrington J.J. and Lieber,M.R. (1994) The characterization of a mammalian DNA structure-specific endonuclease. EMBO J., 13, 1235–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvitz H.R. (1999) Genetic control of programmed cell death in the nematode Caenorhabditis elegans. Cancer Res., 59, 1701s–1706s. [PubMed] [Google Scholar]
- Hosfield D.J., Mol,C.D., Shen,B. and Tainer,J.A. (1998) Structure of the DNA repair and replication endonuclease and exonuclease FEN-1: coupling DNA and PCNA binding to FEN-1 activity. Cell, 95, 135–146. [DOI] [PubMed] [Google Scholar]
- Kim K., Biade,S. and Matsumoto,Y. (1998) Involvement of flap endonuclease 1 in base excision DNA repair. J. Biol. Chem., 273, 8842–8848. [DOI] [PubMed] [Google Scholar]
- Li L.Y., Luo,X. and Wang,X. (2001) Endonuclease G is an apoptotic DNase when released from mitochondria. Nature, 412, 95–99. [DOI] [PubMed] [Google Scholar]
- Li X., Li,J., Harrington,J., Lieber,M.R. and Burgers,P.M. (1995) Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. J. Biol. Chem., 270, 22109–22112. [DOI] [PubMed] [Google Scholar]
- Lieber M.R. (1997) The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. BioEssays, 19, 233–240. [DOI] [PubMed] [Google Scholar]
- Liu X., Zou,H., Slaughter,C. and Wang,X. (1997) DFF, a heterodimeric protein that functions downstream of caspase-3 to trigger DNA fragmentation during apoptosis. Cell, 89, 175–184. [DOI] [PubMed] [Google Scholar]
- Liu X., Li,P., Widlak,P., Zou,H., Luo,X., Garrard,W.T. and Wang,X. (1998) The 40-kDa subunit of DNA fragmentation factor induces DNA fragmentation and chromatin condensation during apoptosis. Proc. Natl Acad. Sci. USA, 95, 8461–8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlroy D., Tanaka,M., Sakahira,H., Fukuyama,H., Suzuki,M., Yamamura,K., Ohsawa,Y., Uchiyama,Y. and Nagata,S. (2000). An auxiliary mode of apoptotic DNA fragmentation provided by phagocytes. Genes Dev., 14, 549–558. [PMC free article] [PubMed] [Google Scholar]
- Mello C.C., Krame,J.M., Stinchcomb,D. and Ambros,V. (1991) Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J., 10, 3959–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan J.P., Shen,B., Park,M.S. and Sklar,L.A. (1996) Kinetic analysis of human flap endonuclease-1 by flow cytometry. Biochemistry, 35, 11668–11676. [DOI] [PubMed] [Google Scholar]
- Parrish J.Z. and Xue,D. (2003) Functional genomic analysis of apoptotic DNA degradation in C.elegans. Mol. Cell, 11, 987–996. [DOI] [PubMed] [Google Scholar]
- Parrish J., Metters,H., Chen,L. and Xue,D. (2000) Demonstration of the in vivo interaction of key cell death regulators by structure-based design of second-site suppressors. Proc. Natl Acad. Sci. USA, 97, 11916–11921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish J., Li,L., Klotz,K., Ledwich,D., Wang,X. and Xue,D. (2001) Mitochondrial endonuclease G is important for apoptosis in C.elegans. Nature, 412, 90–94. [DOI] [PubMed] [Google Scholar]
- Reagan M.S., Pittenger,C., Siede,W. and Friedberg,E.C. (1995) Characterization of a mutant strain of Saccharomyces cerevisiae with a deletion of the RAD27 gene, a structural homolog of the RAD2 nucleotide excision repair gene. J. Bacteriol., 177, 364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Savage C., Hamelin,M., Culotti,J.G., Coulson,A., Albertson,D.G. and Chalfie,M. (1989) mec-7 is a beta-tubulin gene required for the production of 15-protofilament microtubules in Caenorhabditis elegans. Genes Dev., 3, 870–881. [DOI] [PubMed] [Google Scholar]
- Shen B., Nolan,J.P., Sklar,L.A. and Park,M.S. (1996) Essential amino acids for substrate binding and catalysis of human flap endonuclease 1. J. Biol. Chem., 271, 9173–9176. [DOI] [PubMed] [Google Scholar]
- Shibata Y. and Nakamura,T. (2002) Defective flap endonuclease 1 activity in mammalian cells is associated with impaired DNA repair and prolonged S phase delay. J. Biol. Chem., 277, 746–754. [DOI] [PubMed] [Google Scholar]
- Stanfield G.M. and Horvitz,H.R. (2000) The ced-8 gene controls the timing of programmed cell deaths in C.elegans. Mol. Cell, 5, 423–433. [DOI] [PubMed] [Google Scholar]
- The C.elegans Sequencing Consortium (1998) Genome sequence of the nematode C.elegans: a platform for investigating biology. Science, 282, 2012–2018. [DOI] [PubMed] [Google Scholar]
- Tishkoff D.X., Filosi,N., Gaida,G.M. and Kolodner,R.D. (1997) A novel mutation avoidance mechanism dependent on S.cerevisiae RAD27 is distinct from DNA mismatch repair. Cell, 88, 253–263. [DOI] [PubMed] [Google Scholar]
- Vallen E.A. and Cross,F.R. (1995) Mutations in RAD27 define a potential link between G1 cyclins and DNA replication. Mol. Cell. Biol., 15, 4291–4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Yang,C., Chai,J., Shi,Y. and Xue,D. (2002) Mechanisms of AIF-mediated apoptotic DNA degradation in Caenorhabditis elegans. Science, 298, 1587–1592. [DOI] [PubMed] [Google Scholar]
- Widlak P., Li,L.Y., Wang,X. and Garrard,W.T. (2001) Action of recombinant human apoptotic endonuclease G on naked DNA and chromatin substrates: cooperation with exonuclease and DNase I. J. Biol. Chem., 276, 48404–48409. [DOI] [PubMed] [Google Scholar]
- Wu Y.C., Stanfield,G.M. and Horvitz,H.R. (2000) NUC-1, a Caenorhabditis elegans DNase II homolog, functions in an intermediate step of DNA degradation during apoptosis. Genes Dev., 14, 536–548. [PMC free article] [PubMed] [Google Scholar]
- Wyllie A.H. (1980) Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature, 284, 555–556. [DOI] [PubMed] [Google Scholar]
- Zhang J. and Xu,M. (2002) Apoptotic DNA fragmentation and tissue homeostasis. Trends Cell Biol., 12, 84–89. [DOI] [PubMed] [Google Scholar]