Abstract
The myelin-derived proteins Nogo, MAG and OMgp limit axonal regeneration after injury of the spinal cord and brain. These cell-surface proteins signal through multi-subunit neuronal receptors that contain a common ligand-binding glycosylphosphatidylinositol-anchored subunit termed the Nogo-66 receptor (NgR). By deletion analysis, we show that the binding of soluble fragments of Nogo, MAG and NgR to cell-surface NgR requires the entire leucine-rich repeat (LRR) region of NgR, but not other portions of the protein. Despite sharing extensive sequence similarity with NgR, two related proteins, NgR2 and NgR3, which we have identified, do not bind Nogo, MAG, OMgp or NgR. To investigate NgR specificity and multi-ligand binding, we determined the crystal structure of the biologically active ligand-binding soluble ectodomain of NgR. The molecule is banana shaped with elongation and curvature arising from eight LRRs flanked by an N-terminal cap and a small C-terminal subdomain. The NgR structure analysis, as well as a comparison of NgR surface residues not conserved in NgR2 and NgR3, identifies potential protein interaction sites important in the assembly of a functional signaling complex.
Keywords: axon outgrowth/leucine-rich repeats/ligand binding/Nogo-66 receptor
Introduction
Many spinal cord and brain injuries damage axons without causing prominent neuronal loss. The inability of axons to re-extend in the injured CNS produces profound and persistent functional deficits in numerous clinical cases. Nevertheless, the axons of the adult CNS are capable of regeneration when provided with a suitable environment, such as a growth-permissive peripheral nerve grafts (Richardson et al., 1980; David and Aguayo, 1981). Multiple factors contribute to the observed lack of spontaneous regeneration. Both astroglial scar tissue (Snow et al., 1990; Dou and Levine, 1994; Davies et al., 1997) and CNS myelin (Schwab and Caroni, 1988; Savio and Schwab, 1989; Bandtlow et al., 1990) have recognized roles in blocking CNS axon growth.
Protein fractionation studies identified three cell-surface molecules in CNS myelin with axon outgrowth inhibitory activity, Nogo-A (GrandPre et al., 2000; Huber and Schwab, 2000; Prinjha et al., 2000), MAG (McKerracher et al., 1994; Mukhopadhyay et al., 1994), and OMgp (Wang et al., 2002a). Each of these proteins has been shown to directly collapse axonal growth cones and to inhibit axonal extension. Nogo-A is the longest isoform of Nogo, a large transmembrane protein member of the reticulon family. The molecule is divided into three regions separated by two hydrophobic segments (McGee and Strittmatter, 2003). Its large N-terminal domain (N-Nogo) resides in the cytoplasmic space, as does its small C-terminal domain (C-Nogo), whereas a short stretch of 66 amino acids (aa), termed Nogo-66, forms an extracellular loop detectable on the oligodendrocyte surface (GrandPre et al., 2000; McGee and Strittmatter, 2003). Independently, this Nogo-66 domain and the N-Nogo domain can inhibit axons in vitro (Fournier et al., 2001, 2003). Several lines of evidence demonstrate the relevance of Nogo to axon regeneration in vivo. Antibodies directed against Nogo-A can promote axon growth and plasticity in the adult brain (Schnell and Schwab, 1990; Thallmair et al., 1998; Raineteau et al., 1999). A Nogo-66 peptide antagonist increases axonal sprouting and functional recovery after spinal cord injury (GrandPre et al., 2002; Li and Strittmatter, 2003). Expression of Nogo in peripheral myelinating Schwann cells slows the otherwise robust pace of axon regeneration (Pot et al., 2002; Kim et al., 2003a). Some strains of mice lacking Nogo-A exhibit CNS regenerative axonal growth after spinal cord section (Kim et al., 2003b).
MAG is a member of the immunglobulin superfamily expressed in myelinating cells (McKerracher et al., 1994; Mukhopadhyay et al., 1994). The protein inhibits in vitro axonal growth in a fashion similar to Nogo-A, but does not share sequence similarity to Nogo-A. Mice lacking MAG exhibit enhanced axonal regeneration under specialized circumstances (Bartsch et al., 1995; Schafer et al., 1996). OMgp is the third myelin-derived axon outgrowth inhibitor (Wang et al., 2002a). It is a glycosylphos phatidylinositol (GPI)-anchored leucine-rich repeat (LRR) protein without detectable sequence similarity to Nogo-A or MAG. Its role has not been studied in vivo.
We identified Nogo-66 receptor (NgR) as a high-affinity Nogo-66 binding site required for inhibition of axon extension (Fournier et al., 2001). Surprisingly, this receptor has also been shown to mediate the inhibitory activities of two other myelin-derived proteins, MAG and OMgp (Domeniconi et al., 2002; Liu et al., 2002; Wang et al., 2002a). This is particularly striking, since there is no sequence similarity between these NgR ligands. NgR is a GPI-anchored molecule that belongs to the family of LRR proteins (Fournier et al., 2001; McGee and Strittmatter, 2003). It contains eight central LRRs flanked by a cysteine-rich C-terminal subdomain (LRRCT) and by a smaller leucine-rich N-terminal subdomain (LRRNT). This 311 aa NgR region is necessary and sufficient for Nogo-66 binding (Fournier et al., 2002). The most C-terminal 100 NgR residues are most likely unstructured on their own, but may participate in the interactions of Nogo with its co-receptors (Fournier et al., 2002; Wang et al., 2002b). NgR is concentrated in lipid rafts, and soluble recombinant NgR has affinity for cell-surface NgR (Fournier et al., 2002; Liu et al., 2002), but the exact valence of the protein in vivo is not clear. Since NgR lacks a cytoplasmic domain, it was predicted early on that it signals through the action of co-receptors (Fournier et al., 2001), one of which was recently identified as the neurotrophin receptor p75NTR (Wang et al., 2002b; Wong et al., 2002). p75NTR was first described as an NGF-binding protein and was subsequently shown to interact with multiple ligands and to modulate the activity of several Trk family tyrosine kinases (Dechant and Barde, 2002; Roux and Barker, 2002). It associates with NgR both alone and in complex with some of the NgR ligands (Wang et al., 2002b; Wong et al., 2002). This association depends on both the LRR domains and more carboxyl regions of the NgR (Wang et al., 2002b).
Since NgR mediates the signaling of all characterized myelin-derived inhibitory factors, an understanding of the molecular details of its receptor/ligand and receptor/co-receptor interactions is an essential step on the road to developing clinically effective therapeutics to promote recovery after adult CNS injury. Therefore, we identified and examined two proteins closely related to NgR for myelin inhibitor-binding activity. We also determined the crystal structure of the N-terminal region of NgR comprising the LRRCT, LRR and LRRNT subdomains of the protein. A comparison of structural features and non-conserved residues in the closely related but inactive NgR-like proteins provides insight into the basis of NgR molecular interactions.
Results and discussion
Identification of the MAG-binding domain in NgR
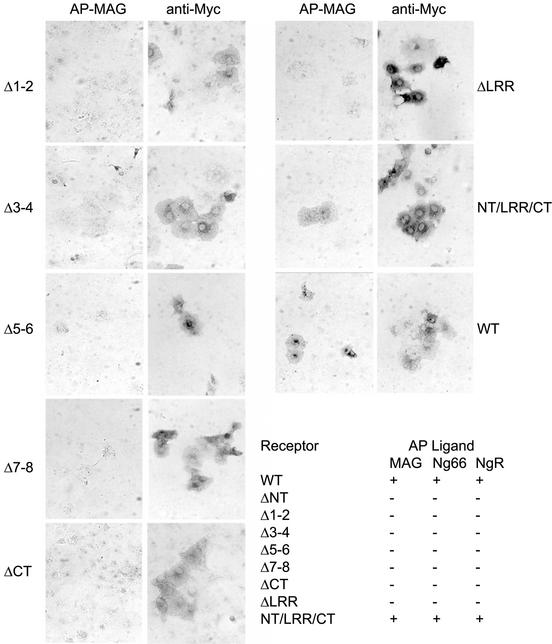
A series of NgR deletion mutants was previously generated and screened to identify regions required for Nogo-66 and NgR binding (Fournier et al., 2002). These deletions remove specific modular domains in toto. Such studies demonstrated that deletion of the LRRNT subdomain, any two LRR subdomains or the LRRCT subdomain abrogates binding activity. To investigate the relationship between the MAG, Nogo-66 and NgR (NgR self-association) binding sites, we examined the ability of soluble AP-MAG ligand to bind to each of the deletion mutants of NgR. Previous results indicate that MAG interacts with the NgR LRR region (Liu et al., 2002), but the interactions with the individual subdomains had not been studied. The data presented in Figure 1 shows that MAG binding is lost by deletion of the same regions as for Nogo-66 or NgR. Thus, the LRRNT/LRR/LRRCT domain as a whole is required for binding Nogo-66, MAG or soluble NgR. Despite this detailed similarity, the Nogo-66 and MAG binding sites are probably not identical, since NEP1-40 blocks Nogo-66 but not MAG activity (Liu et al., 2002). The localization of distinct binding sites within the same LRRNT/LRR/LRRCT domain emphasizes the need for further and more detailed analysis to better understand the molecular basis of NgR function.
Fig. 1. MAG-binding region in NgR. COS-7 cells were transfected with wild-type (WT) NgR or NgR deletion mutant plasmids and tested for AP-MAG binding. WT NgR and LRRNT/LRR/LRRCT-expressing COS-7 cells bind AP-MAG, whereas other NgR deletion mutants do not. Table I summarizes the ligand-binding attributes of the different NgR mutants from the present work and from Fournier et al. (2002). Deletions were as follows: NgR-ΔNT, residues 27–57; NgR-Δ1–2, residues 58–105; NgR-Δ3–4, residues 106–154; NgR-Δ5–6, residues 155–202; NgR-Δ7–8, residues 203–250; NgR-ΔCT, residues 260–310; NgR-ΔLRR, residues 27–310; and NT/LRR/CT, residues 311–445.
Identification of NgR2 and NgR3
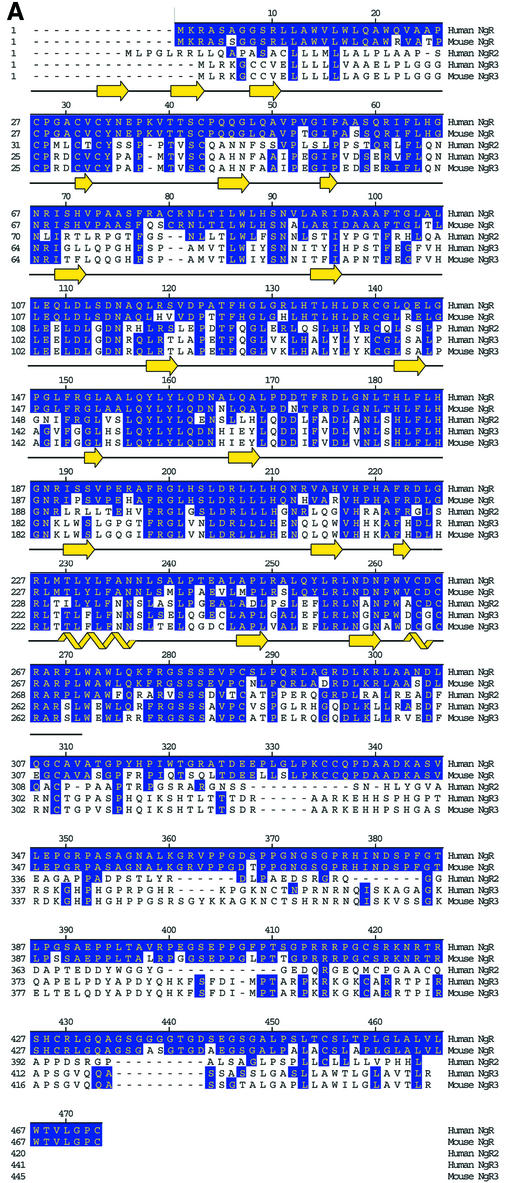
Since there are numerous ligands for NgR, we considered the possibility that NgR might be part of a family of proteins. Initial scans of Genbank cDNA and completed genomic sequences at the time of NgR identification did not reveal any sequences predicted to encode proteins with greater than ∼35% aa identity within the LRR region. This moderate degree of similarity is unlikely to reflect functional homology, since all recognized LRR domains (>200 proteins) share 25–35% aa identity. Therefore, we scanned unfinished genomic sequence databases and found two possible NgR-related sequences (Figure 2). Parts of these sequences are found in cDNA databases, consistent with expression of the proteins in vivo. One human sequence predicts a protein (NgR-related protein-2, NgR2) with 55% LRR identity with the NgR, and a mouse sequence predicts another protein (NgR-related protein-3, NgR3) with 55% LRR identity to both NgR and human NgR2. Later genomic searches with mouse NgR3 allowed identification of the human NgR3 sequence. NgR2 and NgR3 sequences have identical overall architectures as NgR. They all encode a signal sequence, followed by an LRRNT, eight LRRs, an LRRCT, an ∼100 aa unique linker region and a predicted GPI anchorage site. We obtained full-length cDNA for both human NgR2 and human and mouse NgR3 (Figure 2A). These two NgR-related proteins were recently identified independently by another group (Pignot et al., 2003), who has named them NgRH1 and NgRH2 (see Note added in proof).
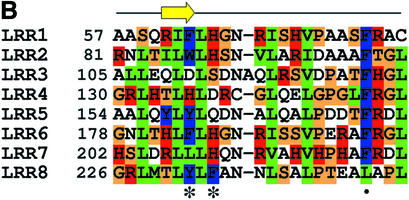
Fig. 2. Comparison of NgR, NgR2 and NgR3 protein sequence. (A) Alignment of the amino acid sequence of the human and mouse NgR, human NgR2 and human and mouse NgR3. Regions of sequence identity are indicated. Secondary structure present in the NgR (27–311) crystal structure is indicated. The black line indicates the extent of the crystallographic model. (B) Sequence alignment of the individual NgR repeats that constitute the LRR domain (Fournier et al., 2001). Conserved structurally important residues are in green, and the residues with exposed aromatic and polar side chains are in blue and red, respectively. Asterisks indicate the positions of the aromatic and histidine exposed residues highlighted in Figure 6. The dot denotes the position of the conserved phenylalanine present in ‘typical’ LRRs.
NgR2 and NgR3 do not bind known NgR ligands
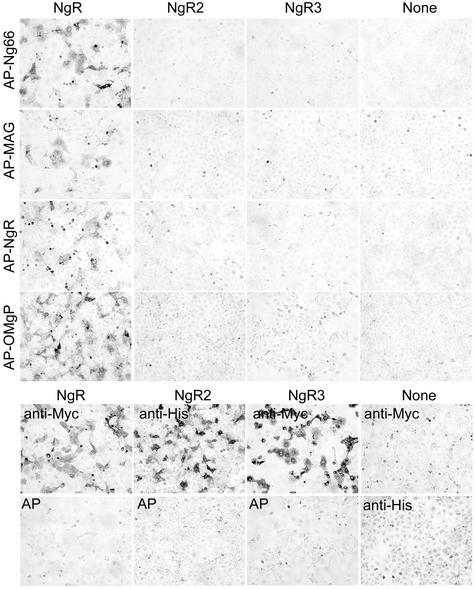
To determine whether the sequence and the expected structural homology between NgR, NgR2 and NgR3 extends to their ligand-binding specificities, each of these proteins was expressed in COS-7 cells and examined for binding to known NgR ligands (Figure 3). Cells transfected with His-NgR2 or Myc-NgR3 clearly express the epitope-tagged protein on their surface, but do not bind AP-Nogo-66, AP-MAG or AP-OMgp under conditions that produce strong binding to NgR. The absence of AP-NgR binding to NgR2 and NgR3 indicates that they are unlikely to exist in a complex with NgR that would allow for indirect participation in myelin inhibitor signaling. Although these three proteins form a sequence-related family, their functions appear to have diverged during evolution. We therefore conclude that a proportion of the residues that differ between NgR and NgR2 plus NgR3 must contribute to the NgR-selective binding of Nogo-66, MAG and OMgp.
Fig. 3. NgR2 and NgR3 do not bind NgR ligands. COS-7 cells were transfected with vectors for Myc-NgR, His-NgR2, Myc-NgR3 or None and then stained with AP-tagged soluble ligands as indicated (upper half). The AP-ligand concentration was 10 nM AP-Nogo-66, 20 nM AP-MAG, 20 nM AP-NgR and 20 nM AP-OMgp. Note the expression of NgR2 and NgR3 without detectable Nogo-66, MAG, NgR or OMgp binding. In the lower half, the expression of recombinant epitope-tagged NgR, NgR2 and NgR3 proteins is detected by immunostaining with the indicated anti-epitope-tag antibodies, anti-Myc or anti-His. None of the proteins binds 20 nM AP protein (bottom row).
Determination of the NgR structure and general protein architecture
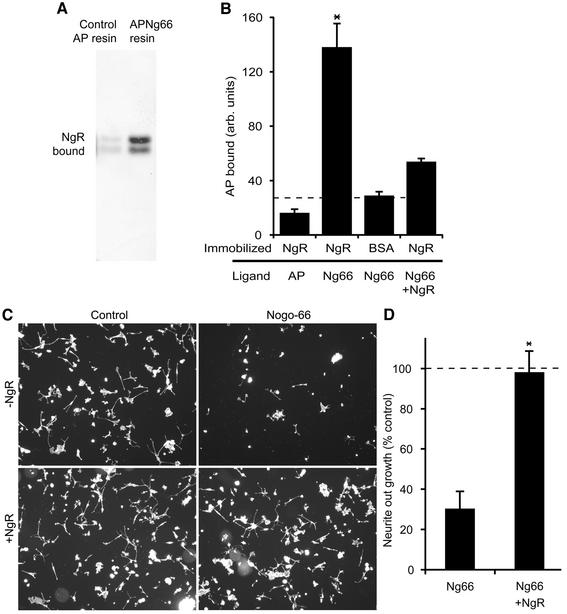
NgR residues 1–311, containing the LRRNT/LRR/LRRCT region, were expressed as a soluble secreted protein in HEK293 cells. Several assays were employed to verify that the purified recombinant receptor was active in binding its Nogo-66 ligand (Figure 4). Physical interaction of the two proteins was demonstrated by specific retention of NgR on a Nogo-66 affinity resin and by specific binding of AP-Nogo-66 to an NgR-coated surface. Furthermore, excess NgR protein completely reversed the DRG axon outgrowth-inhibiting effect of Nogo-66 (Figure 4C and D).
Fig. 4. Biological activity of the purified recombinant NgR protein. (A) Control AP protein or AP-Nogo-66 were bound to resin and then incubated with NgR. After washing, bound protein was examined by NgR immunoblot. The two bands on the gel correspond to differently glycosylated forms of NgR. (B) Microtiter wells were coated with the indicated proteins and then probed with AP-Nogo-66 or AP protein in the presence or absence of excess soluble NgR. (C) Rat P4-6 DRG neurons were plated on surfaces coated with or without GST–Nogo-66 and with or without the addition of excess NgR as indicated. Rhodamine-phalloidin staining is illustrated. (D) Neurite outgrowth from an experiment as in (C) is reported as a percentage of the value without GST–Nogo-66. Data are means ± SEM for three or more measurements. *Values with NgR are significantly different from those without NgR (p ≤ 0.02, Student’s t-test). Dissociated rat P4-6 DRG neurons were plated on surfaces coated with 90 ng of control GST or 90 ng of GST–Nogo-66 with the addition of 450 ng of NgR protein or 450 ng of control GST protein. After 6 h, cells were fixed and scored for neurite outgrowth per neuron as described previously (Fournier et al., 2002; Liu et al., 2002).
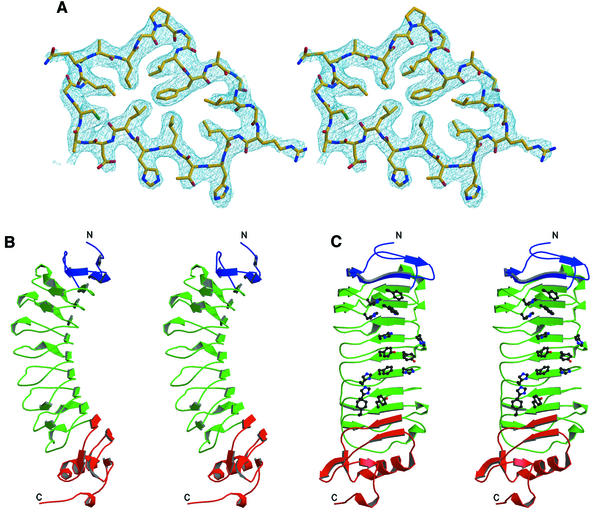
Soluble NgR was crystallized by hanging-drop vapor diffusion with 3.7 M NaCl as a precipitant. The initial crystals had poor diffraction quality (∼5.0 Å) and solvent content of 90%. Crystal dehydration (Heras et al., 2003) performed by transferring them into a stabilizing solution containing 4.5 M NaCl improved the diffraction resolution to 3.2 Å. The structure was determined using the multiple-wavelength anomalous dispersion (MAD) phasing method (Hendrickson, 1991) with a 12-tungsten-cluster derivative. The resulting electron density is of excellent quality (Figure 5A), and the final model is refined at 3.2 Å resolution to an R-factor of 26.5% (Rfree of 29.2%) with good stereochemistry.
Fig. 5. Structure of NgR. (A) Representative region of the density-modified experimental electron density map contoured at 1.5 σ. The refined NgR model is shown with carbons in yellow, nitrogens in blue and oxygens in red. (B and C) Orthogonal stereoviews of the NgR structure, the LRRNT subdomain is in blue, the central LRR subdomain in green and the LRRCT subdomain in red. The protein N- and C-termini are indicated. The side chains of the exposed aromatic and histidine residues lining the concave molecular surface are shown only in (C).
The structure of NgR (Figure 5B) reveals an elongated banana-shaped molecule with approximate dimensions of 80 Å by 35 Å by 35 Å. It has low secondary structure content, consisting mostly of short β strands that generate a long parallel β sheet spanning the concave surface of the molecule. The convex side is composed of the loops connecting the β strands and of several small helices. Eight LRRs make up the large central region of the molecule (green in Figure 5B), with each repeat contributing one strand to the concave-surface β sheet and one extended loop to the convex surface. The flanking LRRNT (blue) and LRRCT (red) sequences fold into smaller β and α/β structures, respectively, which cap the LRR region. NgR is a glycosylated protein, and N-linked N-actyl-β-d-glycosamine moieties, modifying Asn179 and Asn82, were clearly identifiable in the electron density map and were built into the model. These residues are located on the flat side of the molecule, away from the concave β sheet.
Homology of NgR to other LRR proteins
Comparison of the NgR structure with the contents of the FSSP structure database (Holm and Sander, 1998) reveals that the overall fold resembles that of other LRR- containing proteins. The closest structural homologs of NgR are the internalin B protein (InlB) of Listeria monocytogenes (Marino et al., 1999), the platelet glycoprotein Ibα (Huizinga et al., 2002) and the human ribonuclease inhibitor (RI) (Kobe and Deisenhofer, 1993; Kobe and Deisenhofer, 1995; Papageorgiou et al., 1997). The LRR domains of these proteins can be superimposed on the NgR LRR domain with root-mean-square deviations between α-carbon positions of 2.5 Å for InlB (166 atoms), 2.6 Å for platelet glycoprotein Ibα (234 atoms) and 4.9 Å for RI (216 atoms). However, only the model for the platelet glycoprotein Ibα contains LRRNT and LRRCT subdomains homologous to those in NgR.
The NgR β strands, although relatively short, are within the usual LRR range (Kobe and Kajava, 2001), with three residues in each strand engaging in characteristic backbone–backbone hydrogen bonding interactions, which are the hallmark of β-sheet structure. Figure 2B shows the alignment of the individual NgR LRR repeats. Certain positions of the LRRs in NgR are occupied by residues, which are highly conserved between repeats, whereas others can accommodate a wide variety of amino acids. Positions 3, 6, 8, 13, 16, 21 and 24 contain structural leucine, isoleucine, valine and phenylalanine residues, and the van der Waals interactions between them appear to be the main factor that stabilizes the overall fold of the molecule.
The NgR LRRs
NgR was recognized as an LRR family protein based on the fact that it contains the LRR-signature sequence (LxxLxLN/CxL), which corresponds to the segment surrounding the β strand (Figure 2B). LRRs vary in their length and pattern of conserved residues and are grouped into seven different subfamilies (Kobe and Kajava, 2001). The LRRs in NgR are either 24 or 25 aa long, which is midway between the long repeats (28–29 residues) of RI and the short (20 residues) repeats of the Yersinia outer protein YopM (Evdokimov et al., 2001). As such, the NgR repeats are classified as ‘typical’ (Kobe and Kajava, 2001). Each NgR LRR is composed of a single β strand followed by an extended loop. In some of the repeats, there is a small (one turn) α helix preceding the β strands. Most LRRs in other proteins are composed of a β-loop–helix–loop structure such as that observed in RI or InlB. NgR, on the other hand, does not adopt a regular helical structure on the convex side of the molecule. Instead, in NgR, this region contains a proline in seven of the eight LRRs (Figure 2B) and thus adopts an extended structure. In this respect, the LRRs of NgR most closely resemble those observed in YopM.
The LRRCT and LRRNT subdomains
In a regular LRR structure, the hydrophobic core would be exposed to solvent at the ends. Therefore, NgR, like many other LRR proteins, contain flanking regions or ‘caps’. Indeed, the NgR LRRNT and LRRCT subdomains represent more of an extension of the overall LRR region then separate domains (Figure 5A). The N-terminal NgR subdomain (residues 27–61) is composed of three short β strands forming a hydrophilic cap on the buried leucines and isoleucines of the first LRR. The NgR LRRNT forms a compact structure and, although is involved in ligand binding (Figure 1), does not extend a ligand-interacting β finger, such as the one observed in the glycoprotein Ibα structure (Huizinga et al., 2002). Instead, the corresponding loop is smaller and packs against the protein surface. The C-terminal NgR subdomain (residues 260–311) is composed of two helices and five β strands. Its hydrophobic core is continuous with the hydrophobic core of the LRR subdomain, and LRRCT residues form hydrogen bonds to LRR residues. The only other reported LRR structure containing an LRRCT subdomain is the structure of the glycoprotein Ibα. The NgR LRRCT subdomain is slightly larger than its Ibα counterpart but lacks the protruding ligand-binding loop observed in Ibα (Huizinga et al., 2002).
The LRRCT subdomain contains four cysteines forming two disulfide bonds (Cys264–Cys287 and Cys266–Cys309), which stabilize the NgR structure. This disulfide bond arrangement is the one most commonly observed in the sequences of LRR C-flanking regions and is referred to as a CF1-type domain (Kobe and Kajava, 2001). The LRRNT subdomain also contains four cysteines in two disulfide bridges (Cys27–Cys33 and Cys31–Cys43. Interestingly, NgR contains two free cysteines at positions 80 and 140. Cys80 is located next to an N-linked glycosylation site (Asn82), which explains its lack of reactivity, whereas Cys140 is not surface exposed.
The molecular surface of NgR
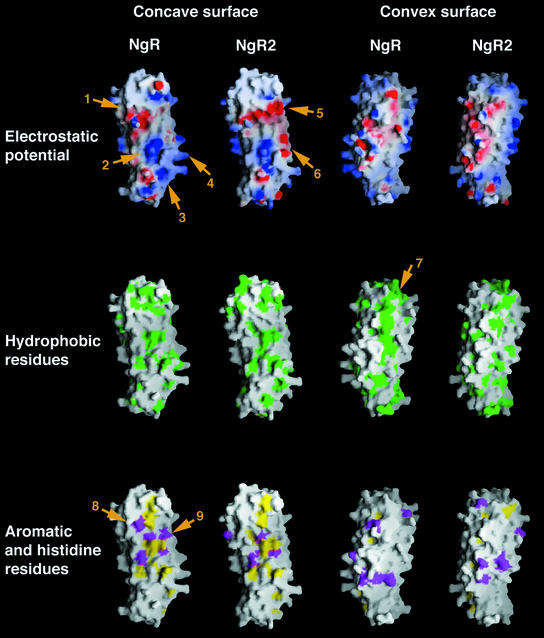
NgR, with its elongated form, presents an extensive molecular surface (∼12 500 Å2) for interactions with a large number of molecular partners. A schematic representation of this surface, colored in accordance with its electrostatic potential and the hydrophobicity of the exposed amino acid side chains, is presented in Figure 6. The figure also displays the predicted molecular surface characteristics of the related NgR2. Both the concave and the convex sides of NgR expose a variety of amino acid side chains. Interestingly, despite the abundance of exposed aromatic side chains, the concave NgR surface is somewhat less hydrophobic than the convex one. This is partly due to the presence of a chain of conserved proline residues on the convex side (arrow 7 in Figure 6).
Fig. 6. The molecular surface of NgR and NgR2 colored according to electrostatic potential (top row), hydrophobicity (middle row) or with colored exposed aromatic residues in yellow and histidines in magenta (bottom row). The model for the NgR2 structure was generated using the known structure of NgR (the two proteins share a 60% aa identity in the modeled region). Important surface regions, discussed in the text, are indicated with numbered arrows. The molecules are positioned with their N-termini pointing up and their C-termini pointing down.
NgR has a slight positive overall charge. From the individual NgR subdomains, the LRRCT is the most basic, with a theoretical pI (if isolated alone in solution) of 9.4, compared with a pI of 8.6 for the rest of the protein. There are two large positively charged areas on the NgR surface, one of which has a more localized round shape and is positioned at the C-terminal region of the LRR convex β sheet (arrow 2 in Figure 6), whereas the other is larger and more diffuse, comprising a significant portion of the LRRCT subdomain and some of the loops of the C-terminal LRR repeats (arrows 3 and 4 in Figure 6). In addition, a large acidic region is located on the concave side of the molecule at the N-terminal region of the central LRR subdomain (arrow 1 in Figure 6).
Implications for the interaction of NgR with ligands and co-receptors
The major biological function of LRRs is to provide a structural framework for the formation of specific protein–protein interactions (Kobe and Kajava, 2001). Several structures of macromolecular complexes involving LRR family members have been reported. They share the common theme that the LRR-containing proteins utilize large areas of exposed molecular surface to recognize and bind other proteins with high affinity. The binding interface usually resides within the LRR concave β sheet. In some cases, though, such as in the Ibα/Willebrand factor complex structure, most of the interactions involve the LRRCT and LRRNT subdomains.
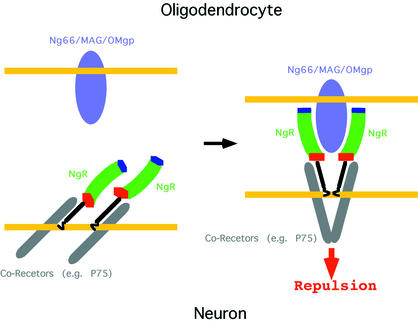
Our deletion analysis (Figure 1) documents that the NgR LRR, LRRCT and LRRNT subdomains are all involved in ligand binding. In order to gain insight into the precise molecular regions within these subdomains, which might mediate specific interactions, we generated a model of the structure of NgR2, a close sequence homolog of NgR (Figure 2A), which nevertheless displays completely distinct ligand preferences (Figure 3). Figure 6 illustrates the molecular surface characteristics of NgR and NgR2. The surface properties of mouse NgR and of human and mouse NgR3 (Figure 2A) are very similar to those of human NgR and NgR2, respectively, and are not presented in Figure 6. The concave faces of both NgR and NgR2 are dominated by aromatic residues such as tyrosines and phenylalanines and by histidines. This is a common feature of proteins belonging to the LRR family; and, although high-affinity ligand interactions involving these conserved residues are expected to provide the bulk of the binding energy, their strict conservation implies that they cannot be responsible for defining the ligand specificity of NgR. However, Figure 6 shows that a number of strategically positioned charged residues are also available for interaction in this area. Several basic and acidic patches, unique only to one of the two receptors, suggest possible regions involved in ligand recognition and binding. For example, as indicated in Figure 6, the NgR2 surface region (arrow 5) is very acidic, whereas the corresponding region in NgR is uncharged. In addition, the large basic NgR surface region, comprising part of the LRRCT subdomain and nearby LRR regions (arrows 3 and 4), is mostly uncharged in NgR2 and even partially acidic (arrow 6). This NgR positively charged region might be responsible for interactions with acidic regions of its ligands. Indeed, Nogo-66 has an overall negative charge and, furthermore, contains two consecutive acidic residues (Glu31 and Glu32), which are necessary for NgR binding (GrandPre et al., 2002). Interestingly, in our crystal, the large acidic patch on the NgR concave face (arrow 1) is packing against the basic LRRCT subdomain of another NgR molecule. This interaction might correspond to lower-affinity ligand-independent receptor/receptor association, which is observed experimentally at the cell surface (Fournier et al., 2002). Upon ligand approximation, the electrostatic receptor/receptor interaction would be disrupted in favor of the higher-affinity van der Waals ligand/receptor contacts, leading to the reorientation of receptor/co-receptor complexes and to the transduction of the repulsive signal to the inside of the cell. Figure 7 represents such a model for the initiation of NgR-mediated signaling.
Fig. 7. A model for initiation of the repulsive signaling mediated by NgR. Although recombinant NgR (NT/LRR/CT) is monomeric in solution, it has been shown to associate at the cell surface with itself (Fournier et al., 2002) and with its co-receptor p75 (Wang et al., 2002b). NgR interacts with a variety of structurally unrelated ligands, some of which may bind with 2:2 and some with 2:1 stoichiometry.
Another intriguing and unique feature of the NgR molecular surface is the abundance of exposed histidine residues (magenta in Figure 6). Other known LRR proteins have a much smaller number of exposed histidines, and even NgR2 has five fewer than NgR. Comparison of the location of the exposed histidine residues in NgR and NgR2 (Figure 6, bottom row) identifies a region at the interface between the LRRNT and the LRR domains of NgR (arrows 8 and 9) as another potentially important ligand-recognition surface. Finally, it should be noted that NgR interacts with multiple and structurally distinct ligands, and each of them is likely to utilize different structural features on the large NgR surface to recognize and bind the receptor. Indeed, the three currently known NgR ligands have very distinct molecular architectures: OMgp is a highly glycosylated LRR protein somewhat similar to NgR; MAG is composed of several immunoglobulin domains; and Nogo-A is a short (66 aa) domain of unknown structure or, even more likely, unstructured in its unbound form.
Conclusions and perspectives
The NgR plays a central role in mediating myelin-dependent restriction of axon outgrowth by three distinct ligands. We identified two proteins with a high degree of similarity to NgR. Despite high levels of amino acid identity with NgR in the ligand-binding LRRNT/LRR/LRRCT domain, these proteins do not bind MAG, Nogo-66, OMgp or NgR. Therefore, they do not appear to function in this pathway. The identity of the ligands for these proteins is yet to be determined. The lack of binding of the NgR ligands to NgR2 and NgR3 emphasizes the unique nature of NgR, rendering the possibility of redundant binding proteins for myelin-derived inhibitors much less probable. If there are any additional receptors for myelin-derived inhibitors of axon regeneration, they seem very unlikely to be structurally related to NgR.
We report the structure of the ligand-binding LRRNT/LRR/LRRCT domain of NgR. It shares some similarity with, but has numerous distinctions from, previously described LRR proteins. Amongst the unique features is the presence of a proline strip on the concave face of the molecule, preventing the formation of secondary structure in this region. There are numerous large basic patches on the surface of the NgR, especially near the LRRCT region. It is particularly tempting to speculate that the essential di-acidic motif in the region of Nogo-66 (Asp 31, Asp 32) most distant from the oligodendrocyte membrane might interact with this region. Comparison of NgR with closely related proteins highlights those surface residues and regions most likely to participate in interaction with different ligands. Such areas include two histidine residues on the convex face and a selected group of charged patches near the edges of the convex surface. The results presented here should lead to a greater understanding of the molecular basis for the inhibition of axon regeneration by CNS myelin.
Materials and methods
NgR2 and NgR3 expression
The mouse NgR3 cDNA was amplified by PCR from mouse adult brain cDNA from the signal sequence to the stop codon. The amplified product was ligated into the pSecTag2 vector such that the vector encoded a signal sequence (from the vector), followed by a Myc tag and the mature NgR3 sequence. The human NgR2 cDNA was derived from two human EST clones, AW293195.1 and BE222737.1. A His6 tag was inserted between the signal sequence and the mature polypeptide in a eukaryotic expression vector. After transfection of DNA into COS-7 cells, the expression of His-NgR2 or Myc-NgR3 protein of the appropriate size was verified by epitope-tag immunoblot. AP-Nogo-66, AP-MAG, AP-NgR and AP-OMgp binding studies and anti-epitope-tag immunohistology were conducted as described previously (Fournier et al., 2001, 2002; Liu et al., 2002).
Protein expression and purification
NgR (aa 1–311) was cloned into the pcDNA5FRT vector and transfected into HEK293/FRT cells (Gibco). Hygromycin-resistant stable cell lines were selected that express ∼5–7 mg protein per liter of culture. Cells were grown in large scale in roller bottles and supplemented with serum-free media. NgR was purified from the media by cation exchange chromotography, followed by ammonium sulfate precipitation and gel filtration chromotography. NgR migrates as a monomer on gel filtration columns and is monomeric in solution as judged by analytical ultracentrifugation (data not shown). The two observed NgR bands on SDS–PAGE gels (Figure 4A) are the result of heterogeneous glycosylation and can be converted to a single band by enzymatic in vitro deglycosylation.
Crystallization and structure determination
Purified NgR was concentrated to 10 mg/ml in a buffer containing 250 mM NaCl and 10 mM HEPES (pH 7.2) and crystallized at room temperature against a reservoir containing 3.7 M NaCl and 100 mM MES (pH 6.5). The space group is P3121 with a = b = 123.96 Å, c = 120.17 Å, and one molecule in the asymmetric unit. The final solvent content, after crystal dehydration, was 85%. Heavy-atom soaks were performed in 4.5 M NaCl at pH 6.5. All data were processed using DENZO and SCALEPACK (Otwinowski and Minor, 1997). The tungsten-cluster (W12) positions were identified using the program TRAWL, a real-space patterson search program. A model of the W12 structure was input into autoSHARP (C.Vonrhein, personal communication) and used to calculate a solvent flattened map of excellent quality.
Model building was initially performed using Arp/Warp (CCP4, 1994) and later proceeded through an iterative process of building in O and refinement of the model in CNS (Jones et al., 1991; Brunger et al., 1998). Stereochemical analysis of the refined models using PROCHECK of the CCP4 package (CCP4, 1994) revealed main chain and side chain parameters better than or within the typical range of values for protein structures determined at corresponding resolutions. None of the NgR residues fell in the disallowed region of the Ramachandran plot, whereas 68% of the residues were in the most favored region. Molecular graphic figures were created with MolScript (Kraulis, 1991), Raster3D (Merritt and Bacon, 1997) and GRASP (Nicholls et al., 1991).
AP-NgR binding assays and DRG axon outgrowth-inhibition assays
For affinity chromatography, 20 µg of His6-tagged AP or AP-Nogo-66 were immobilized on a Ni+2-containing resin (100 µl). The resin was incubated with 100 µg of NgR in 200 µl of 50 mM NaHEPES, 200 mM NaCl, 0.1% BSA (pH 7.5). After washing, bound protein was analyzed by NgR immunoblot (Fournier et al., 2001). For AP-Nogo-66 binding experiments to purified NgR protein, Nunc Maxisorp 96-well plates were coated with or without 0.5 µg of NgR per well and then blocked with BSA. Solutions containing 100 nM AP-Nogo-66 or 100 nM AP in the presence or absence of 20 µM soluble NgR were incubated in the wells for 1 h. After washing, bound AP was detected spectrophotometrically with p-nitrophenol phosphate as substrate. To assess neurite outgrowth, dissociated rat P4-6 DRG neurons were plated on surfaces coated with 90 ng of control GST or 90 ng of GST–Nogo-66 with the addition of 450 ng of NgR protein or 450 ng of control GST protein. After 6 h, cells were fixed and scored for neurite outgrowth per neuron as described previously (Fournier et al., 2002; Liu et al., 2002).
Coordinates and sequences
Coordinates and sequences have been deposited in the Protein Data Bank and GenBank (PDB accession number, 1P8T; DDBJ/EMBL/GenBank accession Nos: BK001302 for human NgR2, BK001303 for human NgR3, BK001304 for mouse NgR2 and BK001305 for mouse NgR3)
Table I. Summary of crystallographic data.
| Crystal | Peak | Inflection | Remote |
|---|---|---|---|
| Resolution (Å) | 3.2 | 3.2 | 3.2 |
| Wavelength (Å) | 1.210 | 1.212 | 1.186 |
| Anom. completeness (%) | 99.6 (99.0) | 99.7 (99.6) | 99.9 (100) |
| Redundancy (fold) | 4.4 | 4.4 | 4.2 |
| I/σI | 12 | 12 | 12 |
| Rmerge | 7.1 | 6.2 | 5.7 |
| F.O.M. (pre/post-dm) | 0.38/0.62 | ||
| Space group | P3121 | ||
| Cell dimensions (Å) |
a = b = 123.96 c =120.17 |
|
|
| Refinement |
|
|
|
| Resolution (Å) | 8.0–3.2 | ||
| Reflections (working/test) | 29 483/3133 | ||
| Non-hydrogen atoms | 2136 | ||
| Rcrys/Rfree | 26.5/29.2 | ||
| R.m.s. deviations | |||
| Bonds (Å) | 0.017 | ||
| Angle (degrees) | 2.27 |
Rmerge = Σ|I – <I>|/ΣI, where I is the observed intensity and <I> is the average intensity obtained from multiple observations of symmetry related reflections.
R.m.s. deviations in bond lengths and angles are the respective root-mean-square deviations from ideal values. r.m.s. thermal parameter is the r.m.s. deviation between the B values of covalently bound atomic pairs.
Acknowledgments
Acknowledgements
We thank Drs Seth Darst and Gabby Rudenko for supplying W-cluster compounds; and Dr Craig Ogata for assistance with crystallographic data collection at APS, Chicago, IL (NE-CAT). We thank Drs Mi Sha and Colleen Mullen of Biogen, Inc. (Cambridge, MA) for cDNA plasmids encoding AP-OMgp and NgR2, respectively. This work was supported by grants to D.B.N. from the New York State Spinal Cord Injury Research Program and to S.M.S. from the NIH and the McKnight Foundation for Neuroscience. D.B.N. is a PEW fellow and a Bressler Scholar. S.M.S. is an Investigator of the Patrick and Catherine Weldon Donaghue Medical Research Foundation.
Note added in proof
While this manuscript was under review, X.L.He and colleagues reported the structure of soluble human NgR ectodomain (He et al., 2003). The two structures are essentially the same, with the exception of one surface-exposed loop (β finger) in the LRRNT subdomain, which in their structure adopts an extended conformation, whereas in ours it packs against the surface of the receptor and is involved in a crystal-packing contact. The discussions about the potential location of the ligand-binding interface differ somewhat due to the fact that in our study we use the additional information that NgR2 and NgR3 do not interact with the known NgR ligands. In addition, V.Pignot and colleagues independently reported the identification of NgR2 and NgR3, which they have named NgRH1 and NgRH2, respectively (Pignot et al., 2003).
References
- Bandtlow C., Zachleder,T. and Schwab,M.E. (1990) Oligodendrocytes arrest neurite growth by contact inhibition. J. Neurosci., 10, 3837–3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartsch U. et al. (1995) Lack of evidence that myelin-associated glycoprotein is a major inhibitor of axonal regeneration in the CNS. Neuron, 15, 1375–1381. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for X-ray crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- David S. and Aguayo,A.J. (1981) Axonal elongation into peripheral nervous system ‘bridges’ after central nervous system injury in adult rats. Science, 214, 931–933. [DOI] [PubMed] [Google Scholar]
- Davies S.J., Fitch,M.T., Memberg,S.P., Hall,A.K., Raisman,G. and Silver,J. (1997) Regeneration of adult axons in white matter tracts of the central nervous system. Nature, 390, 680–683. [DOI] [PubMed] [Google Scholar]
- Dechant G. and Barde,Y.A. (2002) The neurotrophin receptor p75NTR: novel functions and implications for diseases of the nervous system. Nat. Neurosci., 5, 1131–1136. [DOI] [PubMed] [Google Scholar]
- Domeniconi M. et al. (2002) Myelin-associated glycoprotein interacts with the Nogo66 receptor to inhibit neurite outgrowth. Neuron, 35, 283–290. [DOI] [PubMed] [Google Scholar]
- Dou C.L. and Levine,J.M. (1994) Inhibition of neurite growth by the NG2 chondroitin sulfate proteoglycan. J. Neurosci., 14, 7616–7628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimov A.G., Anderson,D.E., Routzahn,K.M. and Waugh,D.S. (2001) Unusual molecular architecture of thee Yersinia pestis cytotoxin M: a leucine rich repeat protein with the shortest repeating unit. J. Mol. Biol., 312, 807–821. [DOI] [PubMed] [Google Scholar]
- Fournier A.E., GrandPre,T. and Strittmatter,S.M. (2001) Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature, 409, 341–346. [DOI] [PubMed] [Google Scholar]
- Fournier A.E., Gould,G.C., Liu,B.P. and Strittmatter,S.M. (2002) Truncated soluble Nogo receptor binds Nogo-66 and blocks inhibition of axon growth by myelin. J. Neurosci., 22, 8876–8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier A.E., Takizawa,B.T. and Strittmatter,S.M. (2003) Rho kinase inhibition enhances axonal regeneration in the injured CNS. J. Neurosci., 23, 1416–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GrandPre T., Nakamura,F., Vartanian,T. and Strittmatter,S.M. (2000) Identification of the Nogo inhibitor of axon regeneration as a Reticulon protein. Nature, 403, 439–444. [DOI] [PubMed] [Google Scholar]
- GrandPre T., Li,S. and Strittmatter,S.M. (2002) Nogo-66 receptor antagonist peptide promotes axonal regeneration. Nature, 417, 547–551. [DOI] [PubMed] [Google Scholar]
- He X.L., Bazan,J.F., McDermott,G., Park,J.B., Wang,K., Tessier-Lavigne,M., He,Z. and Garcia,K.C. (2003) Structure of the nogo receptor ectodomain. A recognition module implicated in myelin inhibition. Neuron, 38, 177–185. [DOI] [PubMed] [Google Scholar]
- Hendrickson W.A. (1991) Determination of macromolecular structures from anomalous diffraction of synchrotron radiation. Science, 254, 51–58. [DOI] [PubMed] [Google Scholar]
- Heras B., Edeling,M.A., Byriel,K.A., Jones,A., Raina,S. and Martin,J.L. (2003) Dehydration converts DsbG crystal diffraction from low to high resolution. Structure, 11, 139–145. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1998) Touring protein fold space with DALI/FSSP. Nucleic Acids Res., 26, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber A.B. and Schwab,M.E. (2000) Nogo-A, a potent inhibitor of neurite outgrowth and regeneration. Biol. Chem., 381, 407–419. [DOI] [PubMed] [Google Scholar]
- Huizinga E.G., Tsuji,S., Romijn,R.A.P., Schiphorst,M.E., de Groot,P.G., Sixma,J.J. and Gros,P. (2002) Structures of the glycoprotein Ibα and its complex with von Willebrand factor A1 domain. Science, 297, 1176–1179. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kim J.E., Bonilla,I.E., Qiu,D. and Strittmatter,S.M. (2003a) Nogo-C is sufficient to delay nerve regeneration. Mol. Cell. Neurosci., in press. [DOI] [PubMed] [Google Scholar]
- Kim J.E., Li,S., GrandPre,T., Qiu,D. and Strittmatter,S.M. (2003b) Axon regeneration in young adult mice lacking Nogo-A/B. Neuron, 38, 187–199. [DOI] [PubMed] [Google Scholar]
- Kobe B. and Deisenhofer,J. (1993) Crystal structure of porcine ribonuclease inhibitor; a protein with leucine rich repeats. Nature, 366, 751–756. [DOI] [PubMed] [Google Scholar]
- Kobe B. and Deisenhofer,J. (1995) A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature, 374, 183–186. [DOI] [PubMed] [Google Scholar]
- Kobe B. and Kajava,A.V. (2001) The leucine rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol., 11, 725–732. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) Molscript: A program to produce both detailed and schematic plots of portein structures. J. Appl. Cryst., 24, 946–950. [Google Scholar]
- Li S. and Strittmatter,S.M. (2003) Delayed systemic nogo-66 receptor antagonist promotes recovery from spinal cord injury. J. Neurosci., 23, 4219–4127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.P., Fournier,A., GrandPre,T. and Strittmatter,S.M. (2002) Myelin-associated glycoprotein as a functional ligand for the Nogo-66 receptor. Science, 297, 1190–1193. [DOI] [PubMed] [Google Scholar]
- Marino M., Braun,L., Cossart,P. and Ghosh,P. (1999) Structure of the InlB leucine rich repeats, a domain that triggers host cell invasion by the pathogen L.monocytogenes.Mol. Cell, 4, 1063–1072. [DOI] [PubMed] [Google Scholar]
- McGee A.W. and Strittmatter,S.M. (2003) The Nogo-66 receptor: focusing myelin inhibition of axon regeneration. Trends Neurosci., 26, 193–198. [DOI] [PubMed] [Google Scholar]
- McKerracher L., David,S., Jackson,D.L., Kottis,V., Dunn,R.J. and Braun,P.E. (1994) Identification of myelin-associated glycoprotein as a major myelin-derived inhibitor of neurite growth. Neuron, 13, 805–811. [DOI] [PubMed] [Google Scholar]
- Merrit E.A. and Bacon,D.J. (1997) Raster3D Version 2: Photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay G., Doherty,P., Walsh,F.S., Crocker,P.R. and Filbin,M.T. (1994) A novel role for myelin-associated glycoprotein as an inhibitor of axonal regeneration. Neuron, 13, 757–767. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Papageorgiou A.C., Shapiro,R. and Acharya,K.R. (1997) Molecular recognition of human angiogenin by placental ribonuclease inhibitor—an x-ray crystallographic study at 2.0 Å resolution. EMBO J., 16, 5162–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignot V. et al. (2003) Characterization of two novel proteins, NgRH1 and NgRH2, structurally and biochemically homologous to the Nogo-66 receptor. J. Neurochem., 85, 717–728. [DOI] [PubMed] [Google Scholar]
- Pot C. et al. (2002) Nogo-A expressed in Schwann cells impairs axonal regeneration after peripheral nerve injury. J. Cell Biol., 159, 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinjha R., Moore,S.E., Vinson,M., Blake,S., Morrow,R., Christie,G., Michalovich,D., Simmons,D.L. and Walsh,F.S. (2000) Inhibitor of neurite outgrowth in humans. Nature, 403, 383–384. [DOI] [PubMed] [Google Scholar]
- Raineteau O., Z’Graggen,W.J., Thallmair,M. and Schwab,M.E. (1999) Sprouting and regeneration after pyramidotomy and blockade of the myelin-associated neurite growth inhibitors NI 35/250 in adult rats. Eur. J. Neurosci., 11, 1486–1490. [DOI] [PubMed] [Google Scholar]
- Richardson P.M., McGuinness,U.M. and Aguayo,A.J. (1980) Axons from CNS neurons regenerate into PNS grafts. Nature, 284, 264–265. [DOI] [PubMed] [Google Scholar]
- Roux P.P. and Barker,P.A. (2002) Neurotrophin signaling through the p75 neurotrophin receptor. Prog. Neurobiol., 67, 203–233. [DOI] [PubMed] [Google Scholar]
- Savio T. and Schwab,M.E. (1989) Rat CNS white matter, but not gray matter, is nonpermissive for neuronal cell adhesion and fiber outgrowth. J. Neurosci., 9, 1126–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer M., Fruttiger,M., Montag,D., Schachner,M. and Martini,R. (1996) Disruption of the gene for the myelin-associated glycoprotein improves axonal regrowth along myelin in C57BL/Wlds mice. Neuron, 16, 1107–1113. [DOI] [PubMed] [Google Scholar]
- Schnell L. and Schwab,M.E. (1990) Axonal regeneration in the rat spinal cord produced by an antibody against myelin-associated neurite growth inhibitors. Nature, 343, 269–272. [DOI] [PubMed] [Google Scholar]
- Schwab M.E. and Caroni,P. (1988) Oligodendrocytes and CNS myelin are nonpermissive substrates for neurite growth and fibroblast spreading in vitro. J. Neurosci., 8, 2381–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow D.M., Lemmon,V., Carrino,D.A., Caplan,A.I. and Silver,J. (1990) Sulfated proteoglycans in astroglial barriers inhibit neurite outgrowth in vitro. Exp. Neurol., 109, 111–130. [DOI] [PubMed] [Google Scholar]
- Thallmair M., Metz,G.A., Z’Graggen,W.J., Raineteau,O., Kartje,G.L. and Schwab,M.E. (1998) Neurite growth inhibitors restrict plasticity and functional recovery following corticospinal tract lesions. Nat. Neurosci., 1, 124–131. [DOI] [PubMed] [Google Scholar]
- Wang K.C., Koprivica,V., Kim,J.A., Sivasankaran,R., Guo,Y., Neve,R.L. and He,Z. (2002a) Oligodendrocyte-myelin glycoprotein is a Nogo receptor ligand that inhibits neurite outgrowth. Nature, 417, 941–944. [DOI] [PubMed] [Google Scholar]
- Wang K.C., Kim,J.A., Sivasankaran,R., Segal,R. and He,Z. (2002b) P75 interacts with the Nogo receptor as a co-receptor for Nogo, MAG and OMgp. Nature, 420, 74–78. [DOI] [PubMed] [Google Scholar]
- Wong S.T., Henley,J.R., Kanning,K.C., Huang,K.H., Bothwell,M. and Poo,M.M. (2002) A p75NTR and Nogo receptor complex mediates repulsive signaling by myelin-associated glycoprotein. Nat. Neurosci., 5, 1302–1308. [DOI] [PubMed] [Google Scholar]