Abstract
Nuclear receptor corepressors SMRT (silencing mediator of retinoid and thyroid receptors) and N-CoR (nuclear receptor corepressor) recruit histone deacetylase (HDAC) activity to targeted regions of chromatin. These corepressors contain a closely spaced pair of SANT motifs whose sequence and organization is highly conserved. The N-terminal SANT is a critical component of a deacetylase activation domain (DAD) that binds and activates HDAC3. Here, we show that the second SANT motif functions as part of a histone interaction domain (HID). The HID enhances repression by increasing the affinity of the DAD-HDAC3 enzyme for histone substrate. The two SANT motifs synergistically promote histone deacetylation and repression through unique functions. The HID contribution to repression is magnified by its ability to inhibit histone acetyltransferase enzyme activity. Remarkably, the SANT-containing HID preferentially binds to unacetylated histone tails. This implies that the SMRT HID participates in interpreting the histone code in a feed-forward mechanism that promotes and maintains histone deacetylation at genomic sites of SMRT recruitment.
Keywords: deacetylase activation/histone deacetylation/N-CoR/SANT motif/SMRT corepressor
Introduction
Gene expression is regulated by changes in chromatin structure that include DNA unwinding and covalent modification of nucleosomal histones (Kouzarides, 2000; Jenuwein and Allis, 2001; Schreiber and Bernstein, 2002). Numerous proteins involved in transcription contain a conserved SANT motif, first identified by sequence homology analysis (Aasland et al., 1996), whose function is not well understood. Among the SANT-containing proteins are the nuclear receptor corepressor (N-CoR) and silencing mediator of retinoid and thyroid receptors (SMRT), related proteins that facilitate repression by unliganded nuclear receptors (Chen and Evans, 1995; Horlein et al., 1995). SMRT and N-CoR both exist in core repression complexes with histone deacetylase 3 (HDAC3) (Guenther et al., 2000; Li et al., 2000; Zhang et al., 2002). We previously reported that the enzyme activity of HDAC3 requires SMRT or N-CoR, which interact with and activate HDAC3 via a region that we have termed the deacetylase activation domain (DAD) (Guenther et al., 2001). DAD functions require the N-terminal SANT1 motif, but the downstream SANT2 is dispensable both for HDAC3 binding and enzyme activation (Guenther et al., 2001; Zhang et al., 2002). Thus, the function of the SANT2 motif and its conserved pairing with SANT1 is unknown.
While numerous transcriptional regulatory factors have intrinsic histone-modifying activity, other components of transcriptionally active complexes have been found to bind histones. A subset of such proteins serves as histone chaperones (Tyler, 2002). Other histone-binding proteins have been implicated in the regulation of histone modification. For example, the INHAT complex blocks transcriptional activation by interfering with histone acetyl transferase (HAT) activity (Seo et al., 2001). Also, histone binding by the coactivator Ada2 was shown to augment the catalytic activity of the GCN5 HAT (Boyer et al., 2002). This activity involves the lone SANT domain of Ada2, which has been demonstrated to be important for transcriptional activation (Boyer et al., 2002; Sterner et al., 2002). A bromodomain present in coactivators such as GCN5, PCAF, Bdf1 and TAFII250 preferentially bind histones whose tails have been marked by acetylation (Dhalluin et al., 1999; Ornaghi et al., 1999; Hudson et al., 2000; Jacobson et al., 2000; Owen et al., 2000; Ladurner et al., 2003; Matangkasombut and Buratowski, 2003), which is an important discriminator of the histone code (Strahl and Allis, 2000).
Here, we show the SMRT corepressor contains a histone interaction domain (HID) that includes the SANT2 motif that is adjacent to the SANT1-containing DAD. The HID functions in cis with the DAD to increase the enzymatic activity of HDAC3 by lowering the apparent Km for histone substrate. Furthermore, the HID inhibits histone acetylation by HAT enzymes. Thus, the SANT-containing DAD and the SANT-containing HID synergistically promote and maintain histone deacetylation. The HID preferentially recognizes unacetylated histone tails and thus is likely to have a role in interpreting the histone code.
Results
A HID in SMRT and N-CoR contains a SANT motif
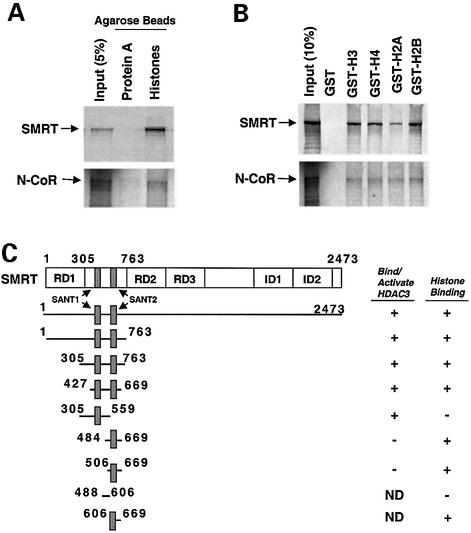
We previously noted that the SMRT/HDAC3/TBL1 complex associated with histones in nuclear extract (Guenther et al., 2000). Although WD40 proteins related to TBL1 such as Tup1 directly bind histones (Edmondson et al., 1996), recombinant TBL1 interacted poorly with histones in vitro (data not shown). In contrast, SMRT interacted strongly with histone agarose affinity matrix (Figure 1A) and with histone tails fused to GST (kindly provided by M.Grunstein) (Figure 1B). N-CoR also interacted specifically with histone tails (Figure 1A and B). A series of SMRT-derived polypeptides (Figure 1C) was used to determine the HID of SMRT. Histone interaction localized to a 243 amino acid region (427–669) that includes the DAD and contains both SANT motifs (Figure 1C). However, the DAD containing the upstream SANT1 motif (305–559) was insufficient for strong HID activity. This suggested that the downstream SANT2 was required for histone binding. Indeed, further mapping revealed that the upstream SANT1 was not required for HID activity, and a short polypeptide including SANT2 (606–669) was sufficient for histone binding (Figure 1C). In contrast to the SANT1-containing DAD, the HID was ineffective at binding to or activating HDAC3 (Guenther et al., 2001). Thus, HID function localized to the region containing the SANT2 motif immediately downstream of the SMRT DAD.
Fig. 1. SMRT and N-CoR interact with core histones. (A) SMRT and N-CoR bind to calf core histones coupled to agarose. 35S-labeled SMRT-Flag or N-CoR-Flag was incubated with protein A–agarose or histone-coupled protein A–agarose. Bound protein was eluted, resolved by SDS–PAGE and subjected to autoradiography. Input is 5% of total. (B) SMRT and N-CoR interact with core histone tails fused to GST. GST alone or GST fusions to histone H3, H4, H2A and H2B N-terminal tails were immobilized on glutathione beads and incubated with 35S-labeled SMRT-Flag or N-CoR-Flag. Bound protein was eluted, resolved by SDS–PAGE and subjected to autoradiography. Input is 10% of total. (C) Summary of results of pull-down assays of Gal4-SMRT polypeptides binding to GST–H3 and GST–H4. The gray bars indicate the two SANT motifs. Highly purified GST alone, GST–H3 and GST–H4 N-terminal tails were immobilized on glutathione beads and incubated with [35S]Gal4-SMRT derivatives. After extensive washing of the beads, bound proteins were eluted and separated by SDS–PAGE and visualized by autoradiography. Histone binding was considered positive (+) when binding of ≥10% of input SMRT polypeptide was observed. Results of HDAC3 binding/activation are also indicated (Guenther et al., 2001). ND, not done.
The SMRT HID interacts with endogenous histones
We next tested the ability of the SMRT HID to interact with endogenous histones in living cells. When transfected into 293T cells, Gal4-SMRT(1–763), which contains both the DAD and HID (Guenther et al., 2001), coimmunoprecipitated with endogenous histone H4, and deletion of the HID markedly reduced this interaction (Figure 2A). Gal4-HID, but not Gal4-DNA-binding domain (DBD) alone, was sufficient for cellular interaction with histone H4 (Figure 2B). Similar results were obtained with Flag-HID (Figure 2C). Thus, the HID mediates SMRT interaction with histones in living cells as well as in vitro.
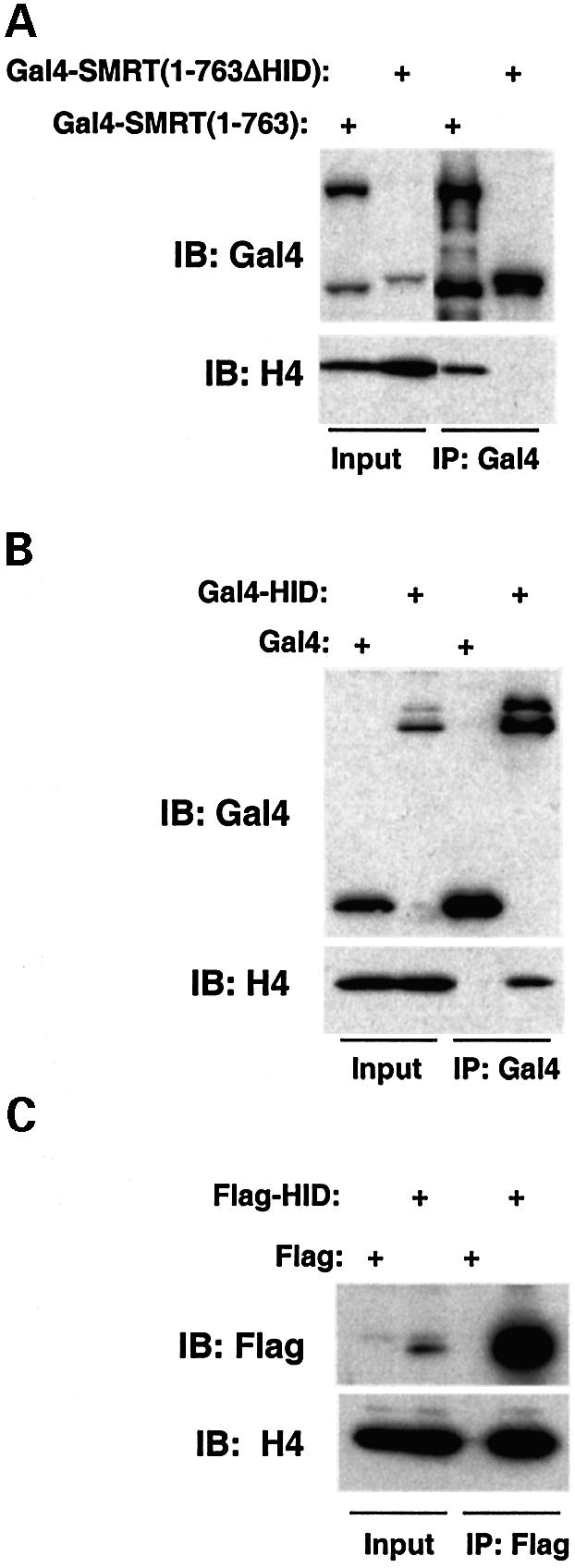
Fig. 2. The SMRT HID interacts with histones in living cells. 293T cells were transfected with (A) pCMX-Gal4-SMRT(1–763) or pCMX-Gal4-SMRT(1–763ΔHID) and (B) pCMX-Gal4-DBD or pCMX-Gal4- HID. Whole-cell lysates were immunoprecipitated with anti-Gal4 agarose beads. Bound proteins were separated by SDS–PAGE and immunoblotted with Gal4 antibody (top) or histone H4 antibody (bottom). Input is 5%. (C) 293T cells were transfected with pcDNA3-Flag or pcDNA3-Flag-HID. Whole-cell lysates were immunoprecipitated with anti-Flag agarose beads. Bound proteins were separated by SDS–PAGE and immunoblotted with Flag antibody (top) or histone H4 antibody (bottom). Input is 5%.
The two SMRT SANT motifs are not functionally interchangeable
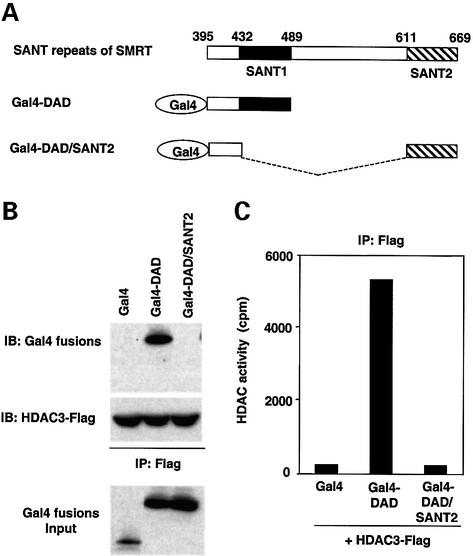
Because two SANT motifs of SMRT are quite homologous to one another, as well as to SANT motifs in other proteins, we next tested whether SANT2, implicated in histone binding, could substitute for SANT1 in the context of the DAD. Gal4 fusion proteins were constructed, containing the DAD with its own SANT1, or substituted by the downstream SANT2 (Gal4-DAD/SANT2, Figure 3A). As expected, Gal4-DAD interacted with HDAC3 (Figure 3B) and activated its enzyme activity (Figure 3C). In contrast, SANT2 was unable to substitute for SANT1 (Figure 3B and C). Thus, despite their structural similarities, the functions of the two SANT motifs in SMRT are quite unique.
Fig. 3. The SMRT SANT2 is unable to activate HDAC3 when substituted for SANT1. (A) Schematic diagram of the SMRT DAD chimera constructs. SMRT DAD comprises SANT1 (amino acids 432–489) and its N-terminal flank (amino acids 395–431). SANT2 (amino acids 611–669) was introduced into the DAD in the position of SANT1 (Gal4-DAD/SANT2). (B) The chimeric DAD containing SANT2 does not bind to HDAC3. (C) The chimeric DAD containing SANT2 does not activate HDAC3. HDAC3-Flag was incubated with Gal4-DAD or Gal4-DAD/SANT2, immunoprecipitated with anti-Flag agarose beads and assayed for binding by SDS–PAGE (B; input shown is 10%) and for HDAC activity (C).
The HID enhances the repression function of the SMRT DAD
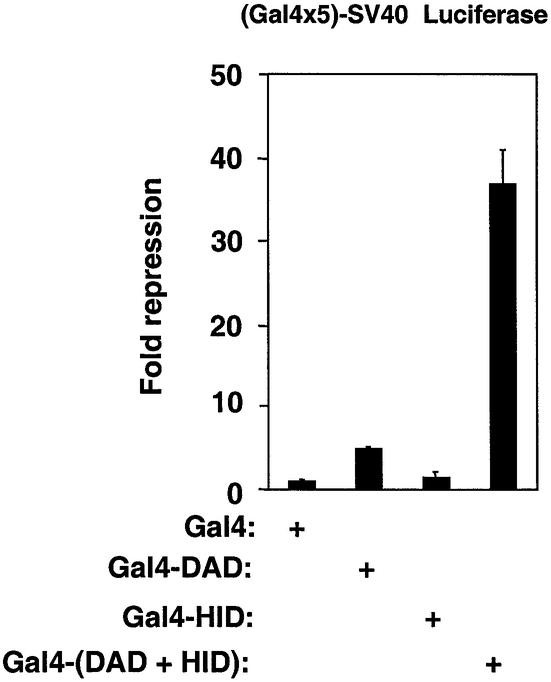
The proximity of the HID to the DAD suggested that the functions of the two SANT-containing domains might be linked. The SMRT DAD functions as a transrepression domain (Figure 4), due to recruitment of HDAC3 (Guenther et al., 2001). We next tested whether the ability of the HID to function in cis would lead to an enhancement of the DAD repression function. On its own, the repressive effect of Gal4-HID upon a luciferase reporter gene containing Gal4-DBD binding sites was not much different from the Gal4-DBD alone, whereas, as expected, Gal4-DAD is a repressor in this context (Figure 4). Notably, the presence of the HID in cis with the DAD as exists in SMRT (and N-CoR) led to marked augmentation of repression (Figure 4).
Fig. 4. The SMRT HID synergistically enhances repression by the SMRT DAD. (Gal4 × 5)-SV40 luciferase reporter was cotransfected with either Gal4-DAD (amino acids 305–559), Gal4-HID (amino acids 506–669) or Gal4-(DAD+HID) (amino acids 305–669) into 293T cells. Fold repression was measured relative to Gal4-DBD alone.
The HID reduces the Km for histone of the SMRT/HDAC3 enzyme
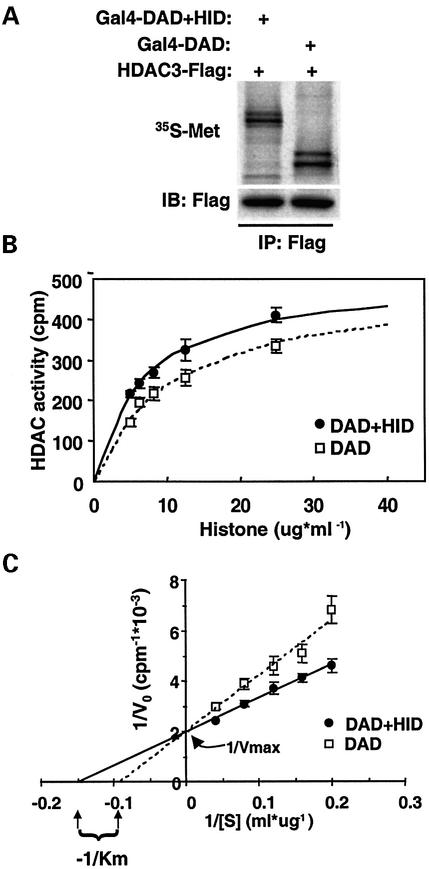
We next explored the mechanism by which the HID enhances DAD repression function. DAD repression function involves recruitment of HDAC3 (Guenther et al., 2001). As histone is the substrate of HDAC activity, we hypothesized that an SMRT-derived polypeptide containing both DAD and HID might lead to a more active HDAC3 enzyme than DAD alone. HDAC3-Flag was incubated with Gal4-(DAD+HID) or Gal4-DAD, and kinetic studies of HDAC activity were performed on Flag immunoprecipitates containing comparable amounts of the SMRT/HDAC3 complexes, whose formation was unaffected by the HID (Figure 5A). Although the HID has no ability to bind or activate HDAC3 on its own (Guenther et al., 2001; data not shown), the presence of the HID in cis with the DAD as normally found in SMRT synergistically activates the deacetylase activity of HDAC3 (Figure 5B). Lineweaver–Burk analysis of the data revealed that the presence of the HID in cis to the DAD lowered the apparent Km for histone (Figure 5C). The average reduction in four independent experiments was 2.11 ± 0.48-fold. Thus, in this context the HID functions to more effectively present histone to the DAD-HDAC3 enzyme.
Fig. 5. The SMRT HID activates the SMRT DAD-HDAC3 by reducing the apparent Km for histone substrate. (A) Formation of SMRT/HDAC3 complexes. HDAC3-Flag was incubated with 35S-labeled Gal4-DAD or Gal4-(DAD+HID), followed by immunoprecipitation and SDS–PAGE. (B) Histone substrate saturation curves for HDAC3 incubated with SMRT(1–763) or SMRT(1–763ΔHID). HDAC3 had no activity alone or incubated with the SMRT HID polypeptide that lacked the DAD. The curves shown were fitted to the Michaelis–Menten equation. (C) Lineweaver–Burk analysis of the experiment shown in (B). Data shown are representative of four independent experiments.
The HID synergizes with the DAD in promoting histone deacetylation in the context of chromatin
The above studies showed that, when present in cis as normally found in SMRT, the HID increased the repression and HDAC-activation functions of the DAD. Based on this, we hypothesized that adjoining the HID to the DAD polypeptide would lead to increased histone deacetylation at a promoter to which the polypeptide was targeted. This was tested using chromatin immunoprecipitation (ChIP). Consistent with its repression function, the Gal4-DAD itself led to modest reduction in local acetylation of histone H4 (Figure 6). Gal4-HID, which was transcriptionally inactive on its own, had little effect on histone acetylation. However, histone H4 deacetylation was dramatically increased when the DAD and HID were present in one polypeptide, as they are in full-length SMRT (Figure 6). In contrast, the acetylation of histone H4 in the constitutively expressed GAPDH gene was unaffected by the various Gal4 fusion proteins, which did not localize to this gene (Figure 6). Thus, the mechanism by which the HID synergizes with the DAD to increase repression likely involves enhancement of HDAC enzyme activity.
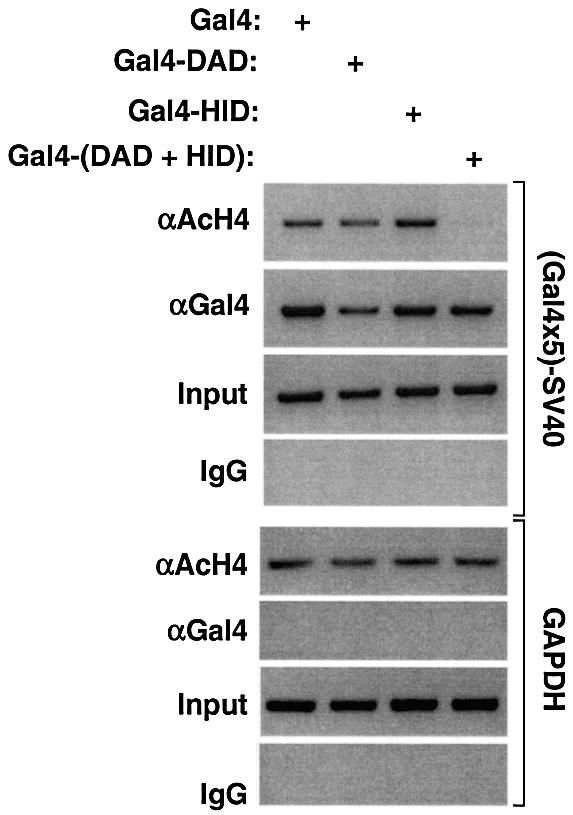
Fig. 6. The SMRT HID synergistically enhances local histone deacetylation by the SMRT DAD. ChIP analysis of 293T cells transfected with the (Gal4 × 5)-SV40 luciferase reporter and either Gal4-DAD (amino acids 305–559), Gal4-HID (amino acids 506–669) or Gal4-(DAD+HID) (amino acids 305–669). PCR was performed using primers flanking the Gal4 UAS or the endogenous GAPDH gene which served as control.
The SMRT HID inhibits HAT activity
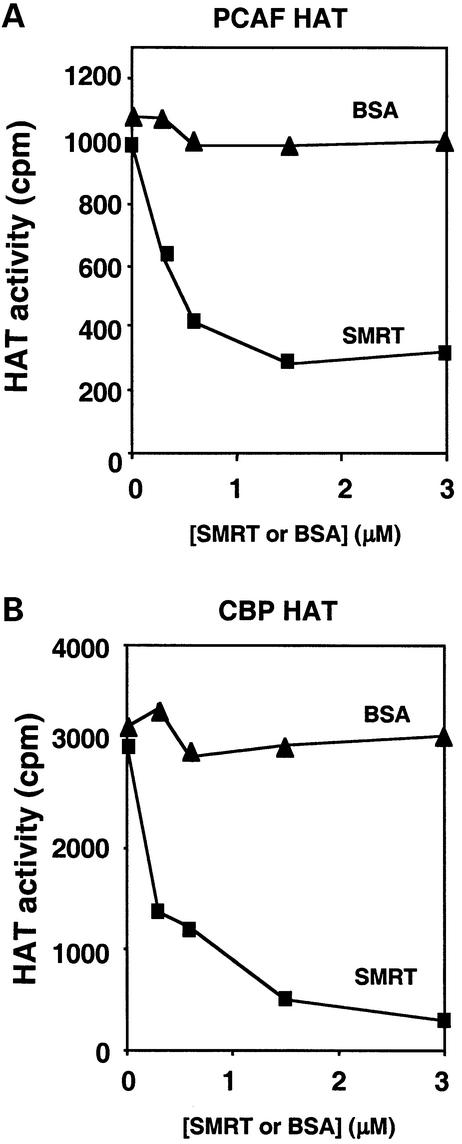
A subset of histone-binding proteins, including the INHAT complex (Seo et al., 2001), have the property of inhibiting HAT activity. We tested the SMRT HID for this activity in vitro using the PCAF HAT. A polypeptide containing the SMRT HID inhibited PCAF HAT activity in a dose-dependent manner (Figure 7A). Similar inhibition of HAT activity was noted for the CBP HAT (Figure 7B). Thus, binding of SMRT to histone inhibits HAT activity.
Fig. 7. The SMRT HID inhibits HAT activity. Increasing amounts of SMRT polypeptide or BSA control were added to core histone substrates prior to the addition of (A) PCAF HAT or (B) CBP HAT. Reaction samples were spotted on p81 paper and the 3H-labeled histones were measured by scintillation counting.
The SMRT HID binds selectively to unacetylated histone tails
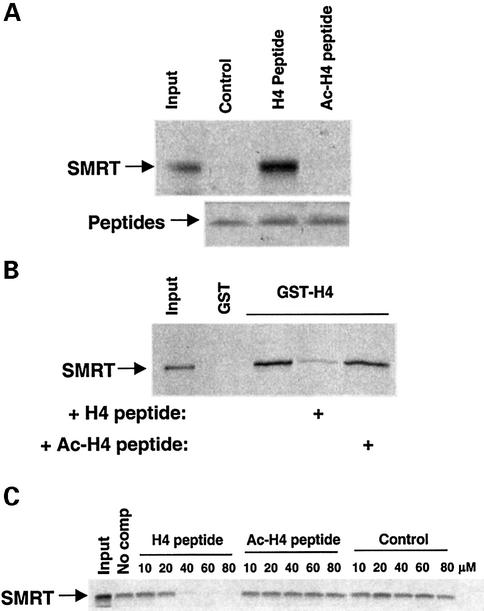
The histone code hypothesis postulates that the affinities of some histone-binding proteins will be regulated by post-translational modifications of histone, including acetylation. Indeed, the bromodomain found in coactivators has been shown to preferentially interact with acetylated histone tails (Dhalluin et al., 1999; Ornaghi et al., 1999; Hudson et al., 2000; Jacobson et al., 2000; Owen et al., 2000; Ladurner et al., 2003; Matangkasombut and Buratowski, 2003). Therefore, we hypothesized that the SANT motif found in the SMRT and N-CoR corepressors might direct the proteins to a subset of modified histones. To address this, we tested the ability of the SMRT HID to bind to unacetylated and tetra-acetylated (K5, 8, 12, 16) histone H4 tail polypeptides, as well as a control peptide which has a positive charge similar to that of the unacetylated H4 tail. Remarkably, the SMRT HID bound well to the unacetylated peptide but not to the tetra-acetylated or control peptide (Figure 8A). Consistent with the binding data, the unacetylated peptide competed well with GST–H4 tail for SMRT binding, whereas the tetra-acetylated histone H4 tail polypeptide was a poor competitor (Figure 8B). The EC50 for competition by the unacetylated peptide was ∼20–30 µM (Figure 8C), whereas up to a 10-fold greater concentration of the tetra-acetylated and control peptides did not measurably compete for binding (Figure 8C; data not shown). Together, these data strongly suggest that the SANT-containing SMRT HID specifically recognizes and binds to the unacetylated tail of histone H4.
Fig. 8. The SMRT HID preferentially binds to unacetylated histone tails. (A) Direct binding. The 35S-labeled Gal4-HID polypeptide was incubated with beads containing biotinylated control peptide (CHKtide), unacetylated histone H4 peptide (H4 peptide) or tetra- acetylated H4 peptide (Ac-H4 peptide). Beads were washed and then run on SDS–PAGE. Input is 10%. Silver staining of peptides used for binding is shown. (B) Competition experiment. In vitro-translated Gal4-HID polypeptide was incubated with GST or GST–H4 tail alone or in the presence of histone H4 tail peptide (80 µM) that was either unacetylated or tetra-acetylated. Input is 10%. (C) Dose response for peptide competition, using unacetylated histone H4 peptide, tetra- acetylated H4 peptide or control peptide (CHKtide).
Discussion
We have demonstrated that nuclear receptor corepressors SMRT and N-CoR bind directly to histones. The HID contains a SANT motif that is located just downstream from the previously characterized DAD and enhances the enzymatic activity of the DAD-HDAC3 by decreasing its apparent Km for histone. Peterson and colleagues have described an analogous role for the SANT domain of Ada2 (Boyer et al., 2002), and the similarities are remarkable. Like SMRT SANT2, the Ada2 SANT motif binds to histone (Boyer et al., 2002). Moreover, the Ada2 SANT motif is involved in enhancing the activity of the GCN5 HAT enzyme by reducing the apparent Km for histone (Boyer et al., 2002). Our work identifies a critical and general role of SANT motifs in the optimal delivery of histone substrates to chromatin-modifying enzymes.
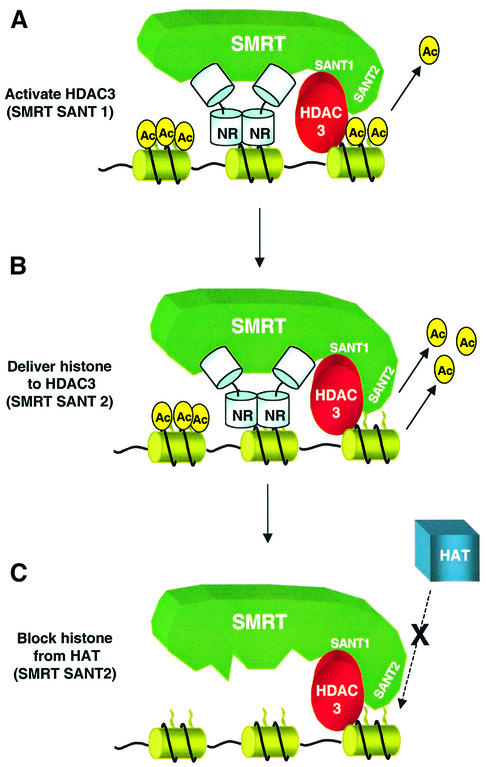
Although histone binding by Ada2 and SMRT HID have similar effects on the kinetics of HAT and HDAC function, respectively, the consequences for transcription are diametrically opposed. Activation of the GCN5 HAT favors histone acetylation, whereas enhancement of HDAC3 activity reverses the process. In addition, binding of the SMRT HID to deacetylated histone inhibits HAT activity. Histone binding by SMRT SANT2 thus has two independent effects, both of which favor histone deacetylation: delivery of substrate to HDAC3 actively deacetylates histone; and continued binding to histone prevents re-acetylation by local HATs. These mechanisms contribute to the repressive effects of SMRT/HDAC3 complex recruitment to regions of chromatin (Figure 9). It remains to be determined whether SMRT binding to deacetylated histones in chromatin continues after dissociation by nuclear receptor.
Fig. 9. Model of SMRT SANT functions in chromatin. (A) The SMRT DAD, requiring SANT1, binds and activates HDAC3. (B) The SMRT HID, requiring SANT2, binds histone and delivers substrate to DAD-HDAC3; this activity increases as the SMRT/HDAC3 deacetylates local histones, resulting in a feed-forward acceleration of deacetylation. (C) The SMRT HID, requiring SANT2, binds histone and blocks access of a HAT coactivator. All functions of SMRT SANT motifs promote and/or maintain deacetylation of histone. SMRT recruitment to chromatin is via unliganded nuclear receptor (NR) binding to target genes, although it is possible that in some cases histone binding by SMRT is sufficient for association with chromatin either before or after NR binding.
Our work links SANT motifs to chromatin structure and transcriptional regulation and also highlights intriguing differences among SANT motifs. SMRT SANT2 synergizes in cis with a SANT1, which is required for HDAC3 enzyme activity. Although SANT1 and SANT2 share 30% identity (Park et al., 1999; Tsai et al., 1999), SANT1 interacts only weakly with histone and SANT2 cannot substitute for SANT1 in the DAD. Thus, the paired SMRT SANT motifs have distinct functions in histone binding and in HDAC3 binding/activation.
Paired SANT motifs have also been found in other corepressors, including N-CoR, CoREST and yeast Snt1 (Aasland et al., 1996; Pijnappel et al., 2001; You et al., 2001). Interestingly, CoREST SANT1 is essential for HDAC1 activation, but CoREST SANT2 is not (You et al., 2001), suggesting a functional conservation with the paired SANT motifs in SMRT. Consistent with this, CoREST SANT1 is more similar to SMRT SANT1 (36.4% identity) than to SMRT SANT2 (26.1%). Moreover, CoREST SANT2 shares 43.5% identity with SMRT SANT2 but only 13.6% with SMRT SANT1. However, CoREST SANT1 cannot substitute for SMRT SANT1 in the function of the SMRT DAD (Y.Li and M.A.Lazar, unpublished results). Thus, SANT functions in corepressors are highly sequence specific, both within a class of SANT motifs involved in the activation of HDAC enzymes (e.g. SMRT and CoREST) and within a single transcriptional regulator containing two SANT motifs (e.g. SMRT SANT1 and SANT2). The coactivator Ada2 has only a single SANT motif which delivers histones to GCN5 as well as interacting directly with GCN5 and stimulating its HAT activity (Boyer et al., 2002). Thus, the Ada2 SANT appears to subserve both of the more specialized roles of the SANT pair in SMRT. Knowledge of the three-dimensional structure of SANT motifs will be required to understand the mechanism of SANT functions in histone binding and modification.
Of great interest is the observation that the SMRT HID binds much more avidly to unacetylated histone H4 tails than to hyperacetylated H4 tails. Specific recognition of hyperacetylated histone tails by the bromodomain present in several coactivators has been established (Dhalluin et al., 1999; Ornaghi et al., 1999; Hudson et al., 2000; Jacobson et al., 2000; Owen et al., 2000; Ladurner et al., 2003; Matangkasombut and Buratowski, 2003). Our work provides an example of a corepressor motif that recognizes hypoacetylated histone tails. This suggests that the SMRT SANT motif is involved in discrimination between different states of histone acetylation and therefore may play a role in interpretation of the histone code (Strahl and Allis, 2000). The existence of paired SANT motifs in other corepressors suggests that this may be a general mechanism. Future studies will be required to determine the precise code recognized by the SMRT SANT-containing HID, whether this is affected by nucleosomal architecture and whether SANT motifs from other proteins will differentially interpret the histone code.
The preference of the SMRT HID for deacetylated histone H4 tails suggests a feed-forward, processive function in repression. As the enzymatically active SMRT/HDAC3 complex is recruited to target genes by nuclear receptors (Figure 9A), local histone deacetylation would increase the affinity of the SMRT HID for histone. This would further promote deacetylation by increasing the activity of the SMRT/HDAC3 enzyme, either for partially deacetylated histone tails or, perhaps, for neighboring histones (Figure 9B). As histone tails become increasingly deacetylated, stronger binding of the SMRT HID prevents acetylation, thereby maintaining the deacetylated state (Figure 9C). This feed-forward role of SANT2 recognition of unacetylated histone is reminiscent of the processive roles proposed for binding of hyperacetylated histones by bromodomains (Hassan et al., 2002) and for binding of methylated histone by the chromodomain in corepressor HP1, which works together with its associated methyltransferase SUV39H1 (Bannister et al., 2001; Lachner et al., 2001; reviewed in Schreiber and Bernstein, 2002; Turner, 2002). Thus, the feed-forward regulation of chromatin modification that has been suggested for histone acetylation and histone methylation is likely to extend to histone deacetylation.
Materials and methods
Plasmids
The HDAC3-Flag construct has been described previously (Guenther et al., 2001). Full-length Flag-tagged mouse SMRT was a gift from Dr Ron Evans (Ordentlich et al., 1999), and full-length Flag-tagged mouse N-CoR was a gift from Dr Geoff Rosenfeld (Horlein et al., 1995). Plasmids for expression of GST fusions to core histone tails was a gift from Dr Michael Grunstein (Hecht et al., 1995). Gal4-mSMRT constructs containing Gal4-DBD fusion to mSMRT amino acids 1–2473, 1–763, 305–763, 305–669, 305–559, 427–669, 484–669, 506–669, 488–606 and 606–669 were produced by PCR amplification of the corresponding DNA sequences followed by insertion into pCMX-Gal4-DBD. Gal4-SMRT(1–763ΔHID) (deleted amino acids 507–693) was produced by PCR amplification of mSMRT sequences encoding amino acids 86–506 and insertion in frame into pCMX-Gal4-SMRT(1–763) with deletion of sequences encoding amino acids 86–693. Gal4-DAD/SANT2 was produced by overlapping PCR amplification of mSMRT sequence encoding amino acids 395–432 plus 611–669 followed by insertion into pCMX-Gal4-DBD. Flag-HID (amino acids 506–669) was produced by PCR amplification of the corresponding DNA sequence followed by insertion into pcDNA3-Flag.
In vitro interaction assays
For histone agarose pull-down assays, calf core histones coupled to agarose were obtained commercially (Sigma). [35S]methionine-labeled SMRT-Flag and N-CoR-Flag were produced using TNT T7 Quick coupled transcription–translation kit (Promega). The binding assays were carried out at 4°C for 12–16 h in HERR buffer (50 mM KCl, 20 mM HEPES pH 7.9, 2 mM EDTA, 0.1% NP-40, 10% glycerol, 0.5% nonfat dry milk). Bound proteins were eluted, resolved by SDS–PAGE and subjected to autoradiography. For the biotinylated histone tail binding assay, 1 µg of biotinylated histone H4 tail peptides (unacetylated or tetra-acetylated) were immobilized on streptavidin beads (Pierce). The pull-down assay was then performed as described above. A short peptide biotinylated CHKtide (Upstate), which contains three unmodified lysines, was used as control. For the GST pull-down assays, GST–histone tail fusion proteins were expressed in Escherichia coli as described previously (Zhang et al., 1997). The purity and concentration of the fusion proteins were determined by SDS–PAGE followed by Coomassie Blue. [35S]methionine-labeled in vitro-translated Gal4-DBD or Gal4-SMRT polypeptides were incubated with GST–histone fusion proteins bound to glutathione beads (Amersham) at 4°C for 1.5 h in HERR buffer. In peptide competition experiments, histone H4 amino acids 1–23 (MSGRGKGGKGLGKGGAKRHRKVC) were obtained unacetylated or tetra-acetylated (Upstate). For each competition, 80 µM peptide was used. For EC50 measurement, increasing amounts of CHKtide, unacetylated histone H4 or tetra-acetylated H4 peptides (0, 10, 20, 40, 60, 80 µM) were used.
Mammalian cell culture and transfection
293T cells were maintained in DMEM (high glucose) supplemented with 10% fetal bovine serum and l-glutamine (all from Gibco-BRL). Cells were grown at 37°C in 5% CO2. They were transfected with lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Cells were transfected for 24 h and washed with phosphate-buffered saline (PBS) before being harvested for immunoprecipitations, luciferase reporter assays or ChIP assays. In luciferase reporter assays, the Gal4 UAS × 5-SV40 luciferase reporter contains five copies of the Gal4 17mer binding site. A luciferase assay kit (Promega) was used to determine relative activity of the luciferase gene product. Light units were normalized to a cotransfected β-galactosidase expression plasmid. Fold repression is relative to the Gal4-DBD, and results of duplicate samples are plotted.
Immunoprecipitation
Cells were washed in PBS, resuspended in HERR buffer containing protease inhibitor cocktail (Roche) and then subjected to sonication. Lysates were clarified by centrifugation at 12 000 g for 10 min at 4°C. Supernatants were incubated with anti-Gal4 agarose-conjugated beads (Santa Cruz) or anti-Flag agarose beads (Sigma) at 4°C for 4 h. Immunoprecipitates were washed four times with HERR buffer. Bound proteins were eluted by boiling in SDS loading buffer and subjected to SDS–PAGE.
Immunoblot analysis and antibodies
Proteins were separated by SDS–PAGE and transferred to nitrocellulose membranes with HMW transfer buffer (50 mM Tris, 380 mM glycine, 0.05% SDS, 20% methanol). Blots were probed with primary antibodies in Tris-buffered saline containing 0.15% Tween 20 and 3.0% nonfat dry milk, followed by HRP-conjugated anti-rabbit antibodies (Pierce) at 1:5000. Alternatively, HRP-conjugated primary antibodies were directly used. All blots were visualized with the ECL kit (Amersham Pharmacia). Anti-Gal4-HRP mouse monoclonal antibody (Santa Cruz) was diluted 1:1000. Anti-Flag-HRP mouse monoclonal (Sigma) was diluted 1:5000. Anti-histone H4 rabbit polyclonal (Upstate) was used at a dilution of 1:1000.
HDAC assay
HDAC assays were performed essentially as described previously (Guenther et al., 2001) but with the following modifications. HDAC3 and SMRT polypeptides were translated individually with the TNT T7 Quick coupled transcription–translation kit (Promega). For kinetic studies, 10 µl of HDAC3-Flag were mixed with 25 µl of Gal4-SMRT(1–763) or Gal4-SMRT(1–763ΔHID) and added to 500 µl of buffer D-300 (300 mM KCl, 20 mM HEPES pH 7.9, 0.25 mM EDTA, 10% glycerol, 0.1% Tween 20). Complexes were purified by incubation with anti-Flag agarose beads (Sigma) in the presence of protease inhibitors (Roche) for 16–18 h at 4°C. Beads were washed three times in buffer D-300 and once in HD buffer (20 mM Tris–HCl pH 8.0, 150 mM NaCl, 10% glycerol). The beads were then incubated with 3H-labeled acetylated HeLa histones (0–25 µg/ml) in a total volume of 200 µl of HD buffer at 37°C for 5 min. To stop each reaction, 50 µl of Stop solution (1 M HCl, 0.16 M acetic acid) were added, and released 3H-labeled acetic acid was extracted with 600 µl of ethyl acetate and measured by scintillation counting. In SANT2 substitution experiments, 50 µl of HDAC3-Flag and 50 µl of Gal4-DAD or Gal4-DAD/SANT2 were used to form complexes as described above. The complexes were then incubated with 3H-labeled histone at 37°C for 2 h followed by scintillation counting.
ChIP assays
ChIP assays were performed essentially as described previously (Hartman et al., 2002). 293T cells were collected and cross-linked with 1% formaldehyde in PBS at room temperature for 10 min. They were then rinsed twice with ice-cold PBS and resuspended in SDS lysis buffer (1% SDS, 5 mM EDTA, 50 mM Tris–HCl pH 8.1). Following a 10 min incubation on ice, samples were sonicated at 15 s pulses four times at 4°C. The lysates were centrifuged at 14 000 g for 10 min and the supernatant precleared with 2 µg of sheared salmon sperm DNA and 50 µl of protein A–Sepharose beads at 4°C for 2 h in buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris–HCl pH 8.1) with protease inhibitors (Roche). Immunoprecipitation was performed at 4°C overnight with normal rabbit IgG (Santa Cruz), Gal4 antibody (Santa Cruz) or acetylated histone H4 antibody (Upstate). Samples were then incubated with 50 µl of protein A–Sepharose beads for 1 h followed by 10 min sequential washes in TSE I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl, 150 mM NaCl), TSE II (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl, 500 mM NaCl), buffer III (0.25 M LiCl, 1% NP-40, 1% deoxycholate, 1 mM EDTA, 10 mM Tris–HCl) and Tris–EDTA buffer; 150 µl elution buffer (1% SDS, 0.1 M NaHCO3) were added to pelleted beads to elute the immune complexes. Following reversal of cross-link by incubating the eluate in 5 M NaCl at 65°C for 6 h or overnight, DNA fragments were purified by using a PCR purification kit (Qiagen) and then subjected to PCR. For Gal4-reporter gene, primers were 5′-TGTATCTTATGGTACTGTAACTG-3′ and 5′-CTTTATGTTTTTGGCGTCTTCCA-3′. For GAPDH gene, primers were 5′-GACACCATGGGGAAGGTGAA-3′ and 5′-GAGTAGGGACCTCCTGT TTC-3′.
HAT inhibition assay
Active PCAF enzyme was obtained commercially (Upstate). Recombinant GST fusion CBP HAT domain (amino acids 1196–1718) was a gift from Dr Gerd Blobel (Hong et al., 2002). PCAF (500 ng) or CBP (50 ng) was incubated with 1 µg of core histones (Upstate) in the presence of [3H]acetyl CoA in HAT buffer (150 mM KCl, 20 mM HEPES pH 7.9, 0.25 mM EDTA, 10% glycerol, 0.1% Tween 20) for 30 min at 30°C. Reaction samples were then spotted on p81 paper and the 3H-labeled histones were measured by scintillation counting. To measure inhibition, increasing amounts of bovine serum albumin (BSA; Sigma) control or baculoviral SMRT(1–891)-Flag protein (0–3.0 µM) were added to reaction mixtures. Sodium butyrate (10 mM) was added to each reaction to block any possible contaminated HDAC activity. SMRT(1–891)-Flag was cloned into pBlue Bac4.5 (Invitrogen), and baculovirus expression and purification of SMRT(1–891)-Flag protein were carried out as described previously (Guenther et al., 2001).
Acknowledgments
Acknowledgements
We thank W.Hong and G.Blobel for providing the CBP HAT, A.Kimura and S.Liebhaber for GAPDH primers, and M.Grunstein for GST–histone constructs. This work was supported by NIH grants DK43806 and DK45586 (to M.A.L.) and the Penn Diabetes Center (DK19525).
References
- Aasland R., Steward,A.F. and Gibson,T. (1996) The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem. Sci., 21, 87–88. [PubMed] [Google Scholar]
- Bannister A.J., Zegerman,P., Partridge,J.F., Miska,E.A., Thomas,J.O., Allshire,R.C. and Kouzarides,T. (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature, 410, 120–124. [DOI] [PubMed] [Google Scholar]
- Boyer L.A., Langer,M.R., Crowley,K.A., Tan,S., Denu,J.M. and Peterson,C.L. (2002) Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol. Cell, 10, 935–942. [DOI] [PubMed] [Google Scholar]
- Chen J.D. and Evans,R.M. (1995) A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature, 377, 454–457. [DOI] [PubMed] [Google Scholar]
- Dhalluin C., Carlson,J.E., Zeng,L., He,C., Aggarwal,A.K. and Zhou,M.M. (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature, 399, 491–496. [DOI] [PubMed] [Google Scholar]
- Edmondson D.G., Smith,M.M. and Roth,S.Y. (1996) Repression domain of the yeast global repressor Tup1 interacts directly with histones H3 and H4. Genes Dev., 10, 1247–1259. [DOI] [PubMed] [Google Scholar]
- Guenther M.G., Lane,W.S., Fischle,W., Verdin,E., Lazar,M.A. and Shiekhattar,R. (2000) A core SMRT corepressor complex containing HDAC3 and a WD40 repeat protein linked to deafness. Genes Dev., 14, 1048–1057. [PMC free article] [PubMed] [Google Scholar]
- Guenther M.G., Barak,O. and Lazar,M.A. (2001) The SMRT and N-CoR corepressors are activating cofactors for histone deacetylase 3. Mol. Cell. Biol., 21, 6091–6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman H.B., Hu,X., Tyler,K.X., Dalal,C.K. and Lazar,M.A. (2002) Mechanisms regulating adipocyte expression of resistin. J. Biol. Chem., 277, 19754–19761. [DOI] [PubMed] [Google Scholar]
- Hassan A.H., Prochasson,P., Neely,K.E., Galasinski,S.C., Chandy,M., Carrozza,M.J. and Workman,J.L. (2002) Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell, 111, 369–379. [DOI] [PubMed] [Google Scholar]
- Hecht A., Laroche,T., Strahl-Bolsinger,S., Gasser,S.M. and Grunstein,M. (1995) Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell, 80, 583–592. [DOI] [PubMed] [Google Scholar]
- Hong W., Kim,A.Y., Ky,S., Rakowski,C., Seo,S.B., Chakravarti,D., Atchison,M. and Blobel,G.A. (2002) Inhibition of CBP-mediated protein acetylation by the Ets family oncoprotein PU.1. Mol. Cell. Biol., 22, 3729–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlein A.J. et al. (1995) Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature, 377, 397–404. [DOI] [PubMed] [Google Scholar]
- Hudson B.P., Martinez-Yamout,M.A., Dyson,H.J. and Wright,P.E. (2000) Solution structure and acetyl-lysine binding activity of the GCN5 bromodomain. J. Mol. Biol., 304, 355–370. [DOI] [PubMed] [Google Scholar]
- Jacobson R.H., Ladurner,A.G., King,D.S. and Tjian,R. (2000) Structure and function of a human TAFII250 double bromodomain module. Science, 288, 1422–1425. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. and Allis,C.D. (2001) Translating the histone code. Science, 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. (2000) Acetylation: a regulatory modification to rival phosphorylation? EMBO J., 19, 1176–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M., O’Carroll,D., Rea,S., Mechtler,K. and Jenuwein,T. (2001) Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature, 410, 116–120. [DOI] [PubMed] [Google Scholar]
- Ladurner A.G., Inouye,C., Jain,R. and Tjian,R. (2003) Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol. Cell, 11, 365–376. [DOI] [PubMed] [Google Scholar]
- Li J., Wang,J., Nawaz,Z., Liu,J.M., Qin,J. and Wong,J. (2000) Both corepressor proteins SMRT and N-CoR exist in large protein complexes containing HDAC3. EMBO J., 19, 4342–4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matangkasombut O. and Buratowski,S. (2003) Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol. Cell, 11, 353–363. [DOI] [PubMed] [Google Scholar]
- Ordentlich P., Downes,M., Xie,W., Genin,A., Spinner,N.B. and Evans,R.M. (1999) Unique forms of human and mouse nuclear receptor corepressor SMRT. Proc. Natl Acad. Sci. USA, 96, 2639–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornaghi P., Ballario,P., Lena,A.M., Gonzalez,A. and Filetici,P. (1999) The bromodomain of Gcn5p interacts in vitro with specific residues in the N terminus of histone H4. J. Mol. Biol., 287, 1–7. [DOI] [PubMed] [Google Scholar]
- Owen D.J., Ornaghi,P., Yang,J.C., Lowe,N., Evans,P.R., Ballario,P., Neuhaus,D., Filetici,P. and Travers,A.A. (2000) The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase gcn5p. EMBO J., 19, 6141–6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E.J., Schroen,D.J., Yang,M., Li,H., Li,L. and Chen,J.D. (1999) SMRTe, a silencing mediator for retinoid and thyroid hormone receptors-extended isoform that is more related to the nuclear receptor corepressor. Proc. Natl Acad. Sci. USA, 96, 3519–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijnappel W.W. et al. (2001) The S.cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1 and is a meiotic-specific repressor of the sporulation gene program. Genes Dev., 15, 2991–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber S.L. and Bernstein,B.E. (2002) Signaling network model of chromatin. Cell, 111, 771–778. [DOI] [PubMed] [Google Scholar]
- Seo S., McNamara,P., Heo,S., Turner,A., Lane,W.S. and Chakravarti,D. (2001) Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the Set oncoprotein. Cell, 104, 119–130. [DOI] [PubMed] [Google Scholar]
- Sterner D.E., Wang,X., Bloom,M.H., Simon,G.M. and Berger,S.L. (2002) The SANT domain of Ada2 is required for normal acetylation of histones by the yeast SAGA complex. J. Biol. Chem., 277, 8178–8186. [DOI] [PubMed] [Google Scholar]
- Strahl B.D. and Allis,C.D. (2000) The language of covalent histone modifications. Nature, 403, 41–45. [DOI] [PubMed] [Google Scholar]
- Tsai C.-C., Kao,H.-Y., Yao,T.-P., McKeown,M. and Evans,R.M. (1999) SMRTER, a Drosophila nuclear receptor coregulator, reveals that EcR-mediated repression is critical for development. Mol. Cell, 4, 175–186. [DOI] [PubMed] [Google Scholar]
- Turner B.M. (2002) Cellular memory and the histone code. Cell, 111, 285–291. [DOI] [PubMed] [Google Scholar]
- Tyler J.K. (2002) Chromatin assembly. Cooperation between histone chaperones and ATP-dependent nucleosome remodeling machines. Eur. J. Biochem., 269, 2268–2274. [DOI] [PubMed] [Google Scholar]
- You A., Tong,J.K., Grozinger,C.M. and Schreiber,S.L. (2001) CoREST is an integral component of the coREST–human histone deacetylase complex. Proc. Natl Acad. Sci. USA, 98, 1454–1458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Zamir,I. and Lazar,M.A. (1997) Differential recognition of liganded and unliganded thyroid hormone receptor by retinoid X receptor regulates transcriptional repression. Mol. Cell. Biol., 17, 6887–6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Kalkum,M., Chait,B.T. and Roeder,R.G. (2002) The N-CoR–HDAC3 nuclear receptor corepressor complex inhibits the JNK pathway through the integral subunit GPS2. Mol. Cell, 9, 611–623. [DOI] [PubMed] [Google Scholar]