Abstract
The sec6/8 complex or exocyst is an octameric protein complex that functions during cell polarization by regulating the site of exocytic vesicle docking to the plasma membrane, in concert with small GTP-binding proteins. The Sec5 subunit of the mammalian sec6/8 complex binds Ral in a GTP-dependent manner. Here we report the crystal structure of the complex between the Ral-binding domain of Sec5 and RalA bound to a non-hydrolyzable GTP analog (GppNHp) at 2.1 Å resolution, providing the first structural insights into the mechanism and specificity of sec6/8 regulation. The Sec5 Ral-binding domain folds into an immunoglobulin-like β-sandwich structure, which represents a novel fold for an effector of a GTP-binding protein. The interface between the two proteins involves a continuous antiparallel β-sheet, similar to that found in other effector/G-protein complexes, such as Ras and Rap1A. Specific interactions unique to the RalA·Sec5 complex include Sec5 Thr11 and Arg27, and RalA Glu38, which we show are required for complex formation by isothermal titration calorimetry. Comparison of the structures of GppNHp- and GDP-bound RalA suggests a nucleotide-dependent switch mechanism for Sec5 binding.
Keywords: exocyst/exocytosis/GTP-binding protein/ Ral effector/sec6/8
Introduction
Vesicular transport mediates the transfer of membrane components and cargo between organelles in eukaryotic cells. Transport vesicles bud from a donor compartment, are translocated through the cytoplasm and finally fuse with a target membrane. Prior to the final fusion reaction, two distinct and biochemically separable stages have been postulated: (i) a loose, reversible and Rab GTP-binding protein-dependent tethering of the vesicle to the target membrane, and then, (ii) engagement of Q- and R-SNAREs (soluble N-ethylmaleimide-sensitive factor [NSF] attachment protein receptor) (Fasshauer et al., 1998), which leads to full SNARE complex formation and membrane fusion. The tethering step is defined as the initial contact of the vesicle via a protein bridge with its target compartment and is the critical determinant in the specificity of membrane fusion. Essential for this tethering are multimeric protein complexes that have been shown to be involved in most, if not all, intracellular trafficking events (Whyte and Munro, 2002).
The tethering complex for exocytosis at the plasma membrane is referred to as the sec6/8 complex or exocyst in yeast (TerBush et al., 1996; Kee et al., 1997). This hetero-octameric protein complex is composed of the original SEC gene products (Sec3, Sec5, Sec6, Sec8, Sec10, Sec15), plus Exo70 and Exo84. The exocyst components were originally identified in a yeast genetic screen for mutants that are deficient in the fusion of secretory vesicles with the plasma membrane (TerBush et al., 1996, 2001). Later, the mammalian complex was purified and its components were characterized (TerBush et al., 1996; Kee et al., 1997; Hsu et al., 1998; Guo et al., 1999a; Matern et al., 2001; Terbush et al., 2001). The localization of the exocyst complex in yeast corresponds with sites of plasma membrane expansion, such as the bud tip early in the cell cycle and the neck separating mother and daughter cells (TerBush and Novick, 1995; Finger et al., 1998). The sec6/8 complex in higher eukaryotic cells is also believed to mediate fusion at a spatially restricted domain. In Madin–Darby canine kidney (MDCK) cells the complex is localized close to tight junctions and antibodies directed against Sec8, when introduced into permeabilized cells, block basal-lateral but not apical protein transport (Grindstaff et al., 1998). The precise role of the sec6/8 complex in neuronal cells remains to be elucidated, but it has been suggested that it functions in synapse formation and neurite outgrowth (Hazuka et al., 1999; Murthy et al., 2003).
Rho1, Rho3, Cdc42 (members of the Rho/Rac/Cdc42 family) and Sec4 (a member of the Rab family) interact with the yeast exocyst complex, suggesting that these small GTP-binding proteins regulate its function and/or its localization (Adamo et al., 1999; Guo et al., 1999b, 2001; Zhang et al., 2001; Novick and Guo, 2002). Although the overall function and composition of the yeast exocyst and mammalian sec6/8 complex might be very similar, the regulation of this complex is not well understood and it could be different for yeast and mammals. For example, Rab3A, a putative homolog of yeast Sec4, implicated in neurotransmitter release, does not interact with the sec6/8 complex. In addition, TC10, another member of the Cdc42 subfamily, binds Exo70 and plays a crucial role in the targeting of the glucose transporter, Glut4, to the plasma membrane in adipocytes (Inoue et al., 2003). Furthermore, Sec5 binds a Ral protein, which is not present in yeast (Brymora et al., 2001; Moskalenko et al., 2002; Sugihara et al., 2002).
Ral is a member of the Ras superfamily of GTP-binding proteins. It is likely that this ubiquitously expressed protein plays a role in cell transformation, differentiation and gene transcription; Ral may also function in the regulation of the cytoskeleton and in cell migration (Takai et al., 2001). In addition, Ral is localized to the plasma membrane, as well as to transport vesicles, suggesting that it is involved in membrane trafficking (Nakashima et al., 1999; Brymora et al., 2001; de Leeuw et al., 2001). Ral potentially regulates Rac/Cdc42 through a GTP-dependent association with RalBP1/RLIP76, a GTPase activating protein (GAP) for Rac/Cdc42 (Jullien-Flores et al., 1995). It has been suggested that the Ral–sec6/8 interaction is involved in the assembly of the sec6/8 complex and regulation of the basal-lateral distribution of proteins, such as epidermal growth factor receptor (EGFR) in MDCK cells (Moskalenko et al., 2002). The Ral–sec6/8 interaction may also regulate Cdc42-dependent filopodia formation in fibroblasts, which is induced by either interleukin 1 or tumor necrosis factor α (Sugihara et al., 2002). However, this regulation appears to be independent of vesicular transport, since brefeldin A, a vesicular transport inhibitor, does not affect Ral-dependent filopodia formation (Sugihara et al., 2002).
In this work, we report the 2.1 Å crystal structure of the complex formed between the Ral-binding domain of Sec5 and RalA bound to a non-hydrolysable GTP ana log {guanosine-5′-[β,γ-imido]triphosphate (GppNHp)}. Structure and thermodynamic binding studies using structure-based site-directed mutagenesis provide the first insights into the specificity of the interactions between the sec6/8 complex and one of its effectors.
Results and discussion
Structure determination
Human RalA is a member of the small GTP-binding protein family and comprises 206 amino acid residues. The C-terminal 23 amino acids and the N-terminal eight amino acids were truncated for soluble expression in Escherichia coli and improvement of crystal quality, respectively. This truncation of RalA did not affect the Kd for Sec5 (Table I). Here we refer to the RalA (9–183) fragment simply as RalA.
Table I. Thermodynamic quantities for the binding of RalA with the Sec5 Ral-binding domain and their variants in the presence of GppNHp.
| RalA·Sec5 complex | Ka × 10–4 (M–1) | –ΔH° (kcal mol–1) | ΔS (cal mol–1 K–1) | Kd (nM) |
|---|---|---|---|---|
| RalA (wt)–Sec5 (wt) | 726 ± 69 | 10.9 ± 0.63 | –4.58 ± 1.0 | 137 ± 13 |
| RalA (wt) full length–Sec5 (wt) | 848 ± 72 | 10.0 ± 0.21 | –1.22 ± 0.5 | 118 ± 10 |
| RalA (Glu38Ala)–Sec5 (wt) | 25 ± 6 | 6.2 ± 0.99 | 4.3 ± 1.0 | 4000 ± 960 |
| RalA (wt)–Sec5 (Thr11Ala) | 1.2 ± 0.25 | ND | ND | >8000 |
Each value is an average of three independent experiments, carried out as described in the legend to Figure 6. RalA refers to the RalA (9–183) fragment, except where specified. Data were fitted to a single-site model to obtain n, ΔH°, ΔS and Ka. In the case of the Sec5 (Thr11Ala) mutant, the Kd derived from the fit was a rough estimation as it was not possible to perfectly fit the data, since RalA binding to Sec5 (Thr11Ala) did not reach saturation (Figure 6D). Therefore, the thermodynamic quantities could not be determined for this mutant. The number of binding sites n was in the range of 0.9–1.1 for all other interactions listed in the table.
Sec5 is a 107 kDa subunit of the hetero-octameric sec6/8 complex. The 120 residue N-terminal fragment of Sec5 is structured as determined by a thermal melting experiment using circular dichroism spectroscopy (data not shown). This Sec5 fragment was thus used for the initial crystallization trials. Mass spectroscopic analysis of the resulting crystals showed that they contained only the N-terminal 99 residues of Sec5 (data not shown). Thus, we subcloned the N-terminal 99 residue fragment of Sec5 (simply referred to as the Ral-binding domain of Sec5) and used it for the subsequent crystallization.
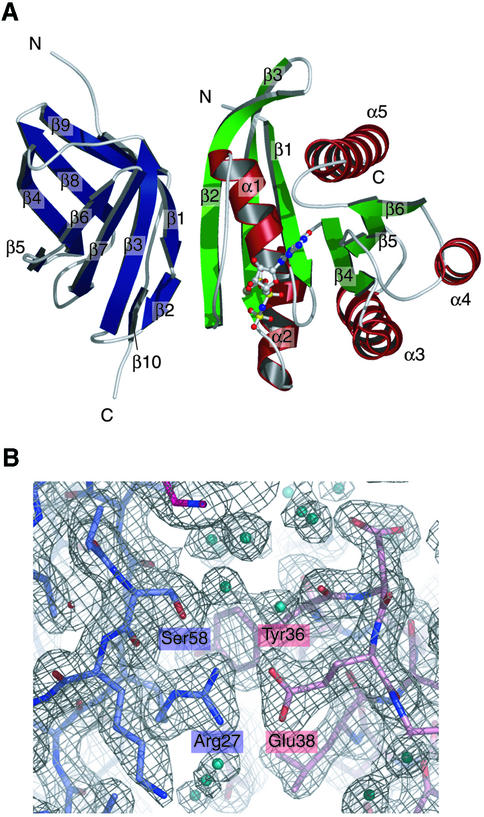
The complex of the Sec5 Ral-binding domain and GppNHp-bound RalA was purified using size-exclusion chromatography as the final purification step, and crystallized by hanging drop vapor diffusion. The complex crystallized in space group I4, with unit cell parameters of a = b = 117.42 Å, c = 102.68 Å, and contained two complexes per asymmetric unit. Initial phases were determined by molecular replacement using the crystal structure of GppNHp-bound Ras (PDB code 1CTQ), as the search model. Density modification with solvent flipping and NCS averaging produced an unambiguous electron density map. We could readily interpret the electron density of the Sec5 Ral-binding domain, as well as that of RalA. The model of the RalA·GppNHp·Sec5 Ral-binding domain complex was built and refined to a resolution of 2.1 Å with an Rfree value of 0.254. One of the two complexes in the asymmetric unit is shown in Figure 1A and a representative portion of a σA-weighted 2Fo – Fc electron density map is shown in Figure 1B. The model is complete except for a few regions with weak electron density: the N-terminal four and five residues of RalA and the Sec5 Ral-binding domain, respectively, and residues 72–76 of one RalA molecule, as well as residues 73–75 of the other RalA molecule.
Fig. 1. Structure of the complex of the Sec5 Ral-binding domain and GppNHp-bound RalA and a representative σA-weighted 2Fo –Fc electron density map. (A) Ribbon representation. The Sec5 Ral-binding domain consists of only β-strands and loops. The β-strands of the Sec5 Ral-binding domain are colored blue. The α-helices and β-strands of the GppNHp-bound RalA are colored red and green, respectively. The GppNHp molecule and the coordinated Mg2+ are shown as ball-and-stick models. For each protein, the α-helices and β-strands are numbered in sequential order. (B) The final refined model (shown as sticks) is superposed on the electron density map. The carbon atoms of the Sec5 Ral-binding domain and GppNHp-bound RalA are colored slate blue and pink, respectively. Water molecules are shown as balls colored cyan. The electron density map is contoured at the 1.3σ level around Sec5 Arg27 and RalA Glu38, showing a portion of the interface between the Sec5 Ral-binding domain and GppNHp-bound RalA.
In an asymmetric unit, two RalA·Sec5 complexes form an interface with a buried surface area of 925 Å2. However, 46 water molecules were found at the interface between the two complexes. Furthermore, in solution, the apparent molecular weight of the RalA·Sec5 complex corresponds to a quantitative 1:1 complex as determined by size exclusion chromatography and multi-angle light scattering spectroscopy (data not shown). We thus concluded that this interface represents a non-functional association induced by crystallization. This interface does not affect the nucleotide binding domains and the interaction between the Ral-binding domain and RalA, and therefore does not affect the conclusions drawn from our structure. The Cα atom root mean square (r.m.s.) deviation between the two RalA molecules and between the two Sec5 Ral-binding domains are 0.21 Å for residues 11–179 and 0.20 Å for residues 4–89, respectively. Residues 180–183 of RalA in one of the two complexes form an extended strand due to crystal packing contacts, whereas those in the other complex form an α-helix. The side chain conformations are also similar for the two complexes in the asymmetric unit, except for several solvent exposed residues.
Ral-binding domain of Sec5
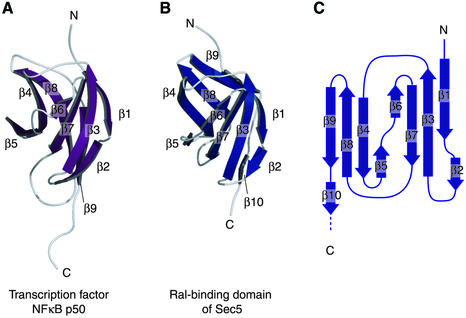
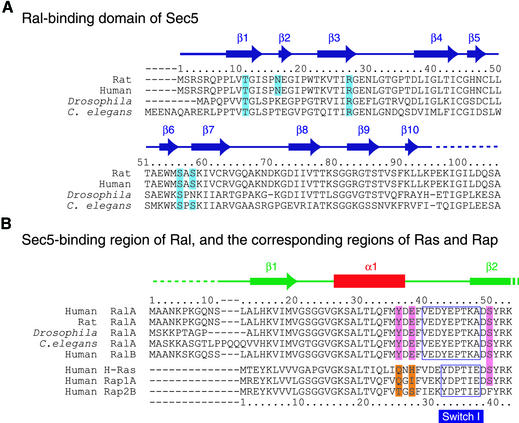
The Sec5 Ral-binding domain is conserved among higher eukaryotic cells. A BLAST search (Altschul et al., 1990; Madden et al., 1996) using Sec5 as the target sequence did not find any homologous proteins, although the BLAST Conserved Domain option (Altschul et al., 1997) predicted that the structure of the Sec5 N-terminal region might be similar to an IPT domain, an immunoglobulin (Ig)-like fold shared by plexins and transcription factors. An example of an IPT domain of known structure is the transcription factor NFκB p50 dimerization domain (Figure 2A), whose amino acid sequence is 12% identical to that of the Sec5 Ral-binding domain. The crystal structure of the Sec5 Ral-binding domain in complex with the GppNHp-bound RalA shows that the Sec5 Ral-binding domain consists of an Ig-like β-sandwich fold (Figure 2B). The antiparallel β-sheet comprised of strands β1, β3, β6 and β7 faces the other β-sheet comprised of strands β4, β5, β8 and β9. A hydrophobic core is formed between the two β-sheets. A characteristic kink occurs at the cis-peptide bond Ser14–Pro15 between β-strands β1 and β2 (Figure 1A). The topology of the domain is the same as that of the NFκB p50 dimerization domain, except that the Sec5 Ral-binding domain has one additional β-strand, β10, at the C-terminus (Figure 2A, B and C). The Cα atomic r.m.s. deviation is 2.4 Å between the Sec5 Ral-binding domain and the NFκB p50 dimerization domain. However, we could not find any similar modes of interaction when comparing all complexes with known structures involving the NFκB p50 dimerization domain and the Sec5·RalA complex. Although activated Ral is sufficient to induce NFκB transcription factor activity and accumulation of cyclin D1 protein (Henry et al., 2000), the structural similarity between NFκB and the Sec5 Ral-binding domain does not necessarily suggest a direct interaction between NFκB and Ral, since the Sec5 residues interacting with RalA are not conserved in NFκB.
Fig. 2. Comparison between the Sec5 Ral-binding domain and the NFκB p50 dimerization domain. (A) Structure of the NFκB p50 dimerization domain (PDB code 1IKN). (B) Structure of the Sec5 Ral-binding domain. (C) Schematic drawing of the topology of the Sec5 Ral-binding domain. β-strands are shown as blue arrows.
A structural similarity search by DALI (Holm and Sander, 1998) using the Ral-binding domain of Sec5 as the reference structure showed that a domain comprising residues 497–584 of cyclodextrin glycosyltransferase resembles the Sec5 Ral-binding domain with a Cα atom r.m.s. deviation of 2.2 Å over 86 residues, despite low sequence identity and no functional similarity between the proteins. Interestingly, the kink at the cis-proline residue between strands β1 and β2 was found in both structures.
An Ig-like fold has also been observed in the Rho/Rac-binding domain of Rho guanine nucleotide dissociation inhibitor (RhoGDI), although this domain was not among the top solutions found by the DALI search. The Rho/Rac-binding domain of RhoGDI covers the farnesylated C-terminus of Cdc42 in the complex of RhoGDI and GDP-bound Cdc42 (Hoffman et al., 2000). However, the interface between RhoGDI and GDP-bound Cdc42 is entirely different from that of the Sec5 Ral-binding domain and GppNHp-bound RalA. Thus, the Sec5 Ral-binding domain represents a novel fold among GTP-binding protein effectors.
Interactions between RalA and Sec5
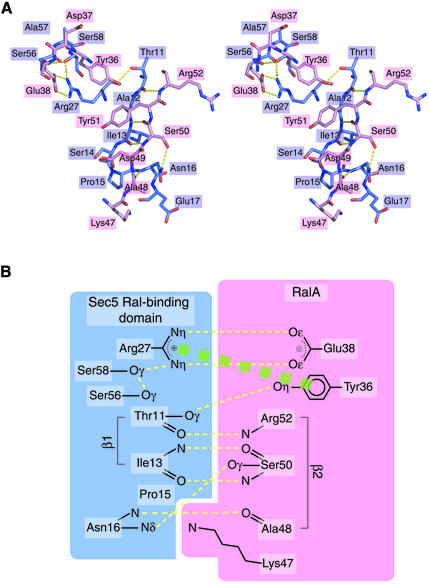
One prominent feature of the interaction between the Sec5 Ral-binding domain and GppNHp-bound RalA is an intermolecular antiparallel β-sheet formed by Sec5 β1 and RalA β2 (Figures 1A and 3A). Several conserved interactions were found. The main chain oxygen atom of Sec5 Thr11 hydrogen bonds with the main chain nitrogen atom of RalA Arg52 (Figure 3A). The main chain nitrogen and oxygen atoms of Sec5 Ile13 hydrogen bond with the main chain oxygen and nitrogen atoms of RalA Ser50, respectively. The main chain nitrogen atom of Sec5 Asn16 hydrogen bonds with the main chain oxygen atom of RalA Ala48. Another conserved interaction occurs between the Nη atoms of Sec5 Arg27 and the Oε atoms of RalA Glu38, which form two hydrogen bonds (Figure 3B). The side chain of Sec5 Arg27 is appropriately positioned by virtue of a hydrogen bond network comprising Sec5 Arg27 Nη, Ser58 Oγ and Ser56 Oγ. In addition, Sec5 Arg27 forms a conserved stacking interaction with RalA Tyr36, whose Oη atom hydrogen bonds with the Oγ atom of Sec5 Thr11.
Fig. 3. The interaction between the Sec5 Ral-binding domain and GppNHp-bound RalA. (A) Stereo view of a stick representation. The carbon atoms of the Sec5 Ral-binding domain and GppNHp-bound RalA are colored slate blue and pink, respectively. Labels within slate blue and pink rectangles correspond to Sec5 and RalA residues, respectively. Hydrogen bonds are shown as dotted yellow lines. (B) Schematic drawing of the interaction between the Sec5 Ral-binding domain and GppNHp-bound RalA. Hydrogen bonds are shown as dotted yellow lines. A stacking interaction is shown as a dotted green bold line.
The Nδ atom of Sec5 Asn16 hydrogen bonds with the Oγ atom of RalA Ser50 (Figure 3). However, this interaction is probably unimportant for the specificity of RalA and Sec5 complex formation, since Asn16 of rat Sec5 is replaced by Lys and Thr in Drosophila and Caenorhabditis elegans Sec5, respectively, in contrast to the invariant Ser50 of RalA (Figure 4).
Fig. 4. Sequence alignments of the Ral-binding domain of Sec5 and the Sec5-interacting region of Ral. (A) Sequence alignment of the Ral-binding domain of Sec5. The secondary structure is shown at the top. The amino acid residues whose side chains interact with RalA are highlighted with cyan rectangles. (B) Sequence alignment of the Sec5-interacting region of RalA. The secondary structure is shown at the top. The RalA amino acid residues whose side chains interact with the Sec5 Ral-binding domain are highlighted with pink rectangles. The corresponding amino acid residues in Ras/Rap are highlighted with orange rectangles.
Comparison between the Ral-binding domain of Sec5 and the canonical Ras-binding domain
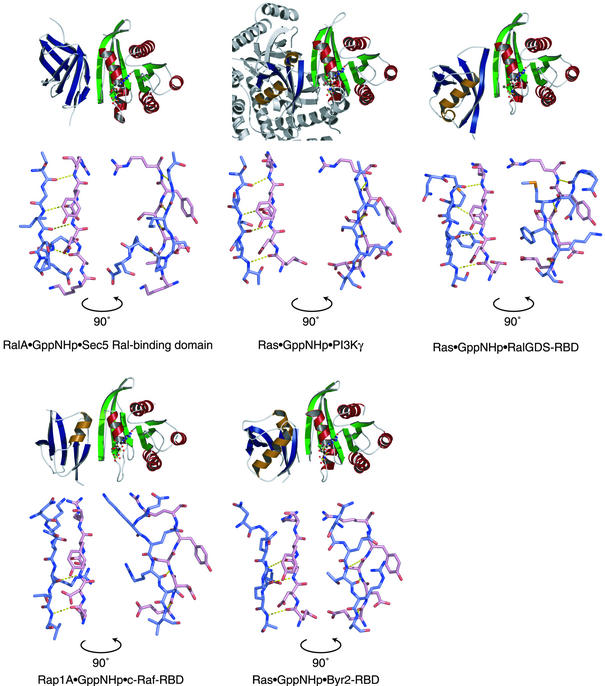
Ral is closely related to Ras and Rap (Reuther and Der, 2000). Rap antagonizes Ras function in the cell, and Ras and Rap share the same binding domain of several effector proteins, including Raf and phosphoinositide 3-kinase (PI3K). This shared effector domain, termed the Ras binding domain (RBD), consists of a ubiquitin fold (Figure 5). Ral does not bind any known RBDs, and the β-sandwich fold of the Sec5 Ral-binding domain is clearly different from that of the ubiquitin fold of a RBD. Nonetheless, the intermolecular antiparallel β-sheet found in the RalA·Sec5 complex is similar to that found between Ras/Rap and their RBDs (Nassar et al., 1995; Huang et al., 1998; Pacold et al., 2000; Scheffzek et al., 2001). The intermolecular antiparallel β-sheet of the Sec5·RalA complex appears to be particularly similar to that of the complex between PI3Kγ and GppNHp-bound Ras, both in terms of the number of hydrogen bonds and the alignment of the two β-strands (Figure 5).
Fig. 5. Comparison of the interface between the Sec5 Ral-binding domain and GppNHp-bound RalA with that between GppNHp-bound Ras/Rap and their corresponding Ras binding domains (RBDs). A ribbon representation is shown at the top of each panel. The α-helices and β-strands of the Sec5 Ral-binding domain/RBDs are colored brown and blue, respectively, while those of GppNHp-bound Ral/Ras/Rap are colored red and green, respectively. The GppNHp molecule and the coordinated Mg2+ are shown as ball-and-stick models. At the bottom of each panel, the amino acid residues involved in the intermolecular β-sheet are shown as sticks in two orientations differing by 90°. The carbon atoms of the Sec5 Ral-binding domain/RBDs and the GppNHp-bound Ral/Ras/Rap are colored slate blue and pink, respectively. The view angle of the left diagram is nearly the same as that of the ribbon representation drawn at the top of each panel.
Specificity
The structural similarity between RalA and Ras/Rap suggests that RalA could potentially bind to an RBD. Indeed, a RalA K47I/A48E double mutant, which has a ‘Ras-type’ switch I (Figure 4B), is able to bind with the RBDs of RalGDS, Rlf and Raf (Bauer et al., 1999). Even the K47I single mutant is able to bind with the RBDs of RalGDS and Rlf (Bauer et al., 1999). Thus, Lys47 prevents Ral from binding with the RBDs; that is, it negatively determines the specificity between the RBD and Ral. A superposition of RalA on the RBD-bound Ras/Rap reveals that the side chain of Ral Lys47 would clash with the Ras-interacting β-strand of the RBD, regardless of the rotamer of the side chain (data not shown). Indeed, in the complex of the Sec5 Ral-binding domain and GppNHp-bound RalA, the Sec5 Ral-binding domain appears to avoid this steric hindrance by the aforementioned kink at Sec5 Ser14–Pro15 (Figure 3).
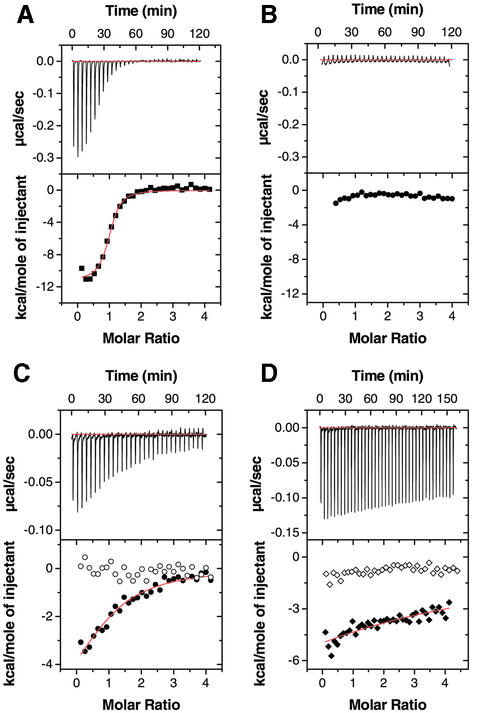
Our structure thus explains why Ral binds Sec5, but not the RBDs of RalGDS, Rlf and Raf. Next, we discuss why Sec5 does not bind small GTP-binding proteins other than Ral, such as Ras or Rap. As described above, conserved interactions occur between Sec5 Thr11 and Arg27 and RalA Tyr36 and Glu38, which are unique to RalA (Figure 4B). These interactions are specific for complex formation of Sec5 and GppNHp-bound RalA. This is in contrast to Ras/Rap and their RBDs where specific interactions occur primarily in the switch I region and the following β-strand of Ras/Rap (Nassar et al., 1995; Huang et al., 1998; Pacold et al., 2000; Scheffzek et al., 2001). To further assess the conserved interactions between RalA and Sec5, we introduced site-directed mutations into RalA (Glu38Ala and Glu38Arg) and Sec5 (Thr11Ala and Arg27Ala) so as to partially disrupt the interactions (Figure 3). Their influence on binding of RalA with Sec5 was tested by isothermal titration calorimetry. As shown in Figure 6A, the wild-type RalA and Sec5 Ral-binding domain titration in the presence of GppNHp exhibited a gradual decrease in the exothermic heat of binding with each successive injection until saturation was achieved. On the other hand, there was no exothermic heat response found for the GDP-bound RalA (Figure 6B) or GppNHp-bound RalA (Glu38Arg) titrated with wild-type Sec5, and for the wild-type GppNHp-bound RalA titrated with Sec5 (Arg27Ala) (Figure 6C and D, open symbols), indicating that these mutations disrupt the RalA–Sec5 interaction. The interaction between RalA (Glu38Ala) and wild-type Sec5 and that between wild-type RalA and Sec5 (Thr11Ala) is rather weak (Figure 6C and D) and their affinities are decreased 29- and 58-fold, respectively, as compared with that of the wild-type RalA–Sec5 interaction (Table I).
Fig. 6. Isothermal calorimetric titration of RalA with the Sec5 Ral-binding domain. The top panels show raw data obtained after 5 µl injections of 150 µM Sec5 RalA-binding domain into 4 µM RalA in 50 mM PBS containing 5 mM MgCl2 and 0.5 mM GppNHp or GDP. The bottom panels show a non-linear least-squares fit (–) of the heat released as a function of the added ligand. (A) RalA titration with Sec5 in the presence of GppNHp. (B) RalA titration with Sec5 in the presence of GDP. (C) RalA (Glu38Ala) titration with Sec5 in the presence of GppNHp. Bottom panel: solid circles for RalA (Glu38Ala) and open circles for RalA (Glu38Arg). (D) RalA titration with 110 µM Sec5 (Thr11Ala) in the presence of GppNHp. Bottom panel: solid diamonds for Sec5 (Thr11Ala) titration and open diamonds for Sec5 (Arg27Ala) titration. Note that the raw data for RalA (Glu38Arg)–Sec5 and RalA–Sec5 (Arg27Ala) in the presence of GppNHp were similar to that seen for RalA–Sec5 in the presence of GDP.
Conformational switch of RalA
The structure of the GppNHp-bound RalA complexed with Sec5 can be superposed on the effector-free GppNHp-bound H-Ras structure (PDB code 1CTQ) with a Cα atom r.m.s. deviation of 0.799 Å computed over all 166 residues. The main chain, and most of the side chains, are in very similar conformations (data not shown). Thus, it is unlikely that a significant conformational change of RalA occurs upon binding with Sec5. For the Sec5 Ral-binding domain, the topology of our high resolution structure agrees with that of the solution NMR structure of the isolated Sec5 Ral-binding domain (PDB code 1HK6; Mott et al., 2003). Thus, no gross conformational change of Sec5 occurs upon RalA binding, with the possible exception of the cis-peptide bond between Ser14 and Pro15, that is absent in the NMR structure. Therefore, we will consider the differences between the GppNHp-bound RalA structure complexed with Sec5 and the GDP-bound RalA structure to be primarily caused by the difference in nucleotide state rather than caused by binding of Sec5 to RalA.
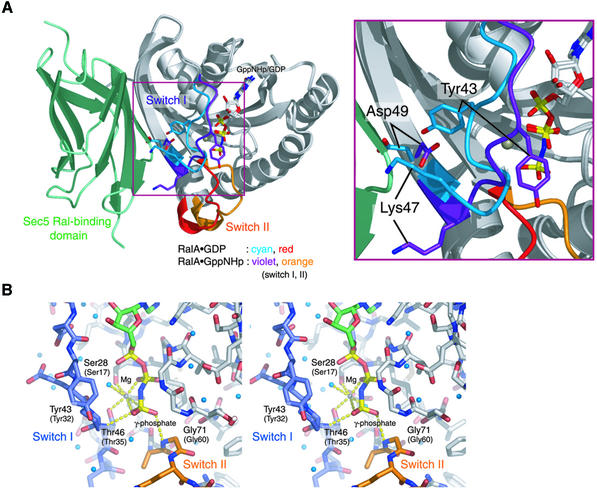
In the RalA·Sec5 complex, the structure of GppNHp-bound RalA is quite similar to that of GDP-bound RalA (see Materials and methods for refinement statistics) (Bauer et al., 1999; I.R.Vetter, personal communication), except for residues 40–48 and 70–78 (Figure 7A). The Cα atomic r.m.s. deviation over residues 12–39, 49–69 and 79–178 is 0.419 Å between the GppNHp-bound and GDP-bound RalA structures. Here, we define residues 40–48 and 70–78 of RalA as switch I and II, respectively. The length of switch I in RalA is longer by three residues than that of Ras (residues 32–38), whereas the length of switch II matches with that of Ras (residues 59–67). We were unable to precisely determine the nucleotide-dependent conformational differences for switch II, since this region is partially disordered in the crystal structure of the RalA·GppNHp·Sec5 complex (i.e. residues 73–75 in one RalA monomer and residues 72–76 in the other RalA monomer exhibit weak electron density). In contrast to the two switch regions, we did not find any large nucleotide-dependent conformational changes in the vicinity of residues 36 and 38, which specifically interact with the Sec5 Ral-binding domain. The Glu38 side chain of the GppNHp-bound RalA shifts slightly so as to form hydrogen bonds with Sec5 Arg27, as compared with that in the GDP-bound RalA.
Fig. 7. Guanine-nucleotide-dependent conformational change in RalA. (A) The GDP-bound RalA is superposed on the GppNHp-bound RalA in complex with the Sec5 Ral-binding domain. A ribbon representation is shown. The Sec5 Ral-binding domain is colored in green. The GDP- and GppNHp-bound RalA molecules are colored in white, except for the switch I (residues 40–48) and II (residues 70–78) regions. The coloring schemes for switch I and II are described in the figure. (B) Interactions of the γ-phosphate of GppNHp with switch I and II (stereo view). The amino acid residues around the γ-phosphate are shown as sticks. The carbon atoms of switch I, switch II and the other regions of RalA are colored in slate blue, orange and white, respectively. The carbon and phosphorus atoms of the GppNHp molecule are colored in green and yellow, respectively. The coordinated Mg2+ and water molecules are shown as gray and cyan balls, respectively. Dotted yellow lines indicate hydrogen bonds.
In GppNHp-bound RalA, a Mg2+ is coordinated with octahedral geometry by the side chains of Ser28 and Thr46, two water molecules and the β- and γ-phosphate groups of GppNHp (Figure 7B). The O2γ and O3γ atoms of the γ-phosphate group form hydrogen bonds with the main chain nitrogen atoms of Thr46, located in switch I, and Gly71, located in switch II, respectively. Thr46 and Gly71 correspond to Thr35 and Gly60 in human H-Ras. These invariant threonine and glycine residues arrest the conformations of switches I and II when GTP binds to the small GTP-binding protein. Then release of the γ-phosphate after GTP hydrolysis allows the switch regions to relax into the other conformation.
A striking difference between the GDP-bound and GppNHp-bound RalA is the position of Tyr43 (Figure 7A). In GDP-bound RalA, the aromatic side chain of Tyr43 is located in a hydrophobic pocket formed by the side chains of Leu32 and Val40, and the Oη atom of Tyr43 hydrogen bonds with the Oη atom of Tyr51 (data not shown). The Tyr43 side chain points towards Asp49, resulting in the exposure of the Asp49 side chain to solvent. In contrast, in GppNHp-bound RalA, the Tyr43 side chain flips to an entirely different site. This conformational switch creates a void near Asp49, allowing the Asp49 side chain to point inward. Superposition of the GDP-bound RalA structure on the structure of the Sec5·RalA shows that the side chain of Asp49 with the conformation found in GDP-bound RalA would clash with the loop connecting the β1 and β2 strands of the Sec5 Ral-binding domain. Therefore, the GDP-bound RalA would be expected to bind to Sec5 with much lower affinity than the GTP-bound RalA. Indeed, the replacement of RalA Asp49 with the isosteric Asn residue does not affect binding of Ral to Sec5, while a mutation to a larger glutamic acid residue disrupts binding (Moskalenko et al., 2002). The Tyr43/Asp49 conformational switch is the most probable explanation for the GTP-dependent binding of Sec5 to RalA.
Implications of the Sec5–Ral interaction
An initial tethering step has been postulated as the critical determinant in the specificity of membrane fusion. The best candidate for mediating the tethering to the plasma membrane is the sec6/8 complex functioning in concert with one or more small GTP-binding proteins (Lipschutz and Mostov, 2002; Novick and Guo, 2002). It has been proposed that RalA on the vesicle interacts with the plasma membrane sec6/8 complex, and that this interaction may also regulate aspects of the assembly of the sec6/8 complex. The RalA·Sec5 structure we present here might be the crucial initial contact between vesicle and target membrane that determines the recognition of membrane fusion partners.
The structure of the RalA·Sec5 complex indicates that a protein–protein interaction via an intermolecular antiparallel β-sheet is utilized for the regulation of the sec6/8 complex. This interaction is reminiscent of that between Ras/Rap and their effectors. Raf kinase, PI3Kγ and Byr2 kinase (Nassar et al., 1995; Pacold et al., 2000; Scheffzek et al., 2001), which are Ras effectors of known RBD structure, have catalytic domains following their N-terminal RBDs. For the Ras·PI3Kγ complex, the switch I region of Ras interacts with the RBD, whereas the switch II region contacts the catalytic domain along with a change in domain orientation of PI3Kγ, which presumably affects phospholipid binding and catalytic activity (Pacold et al., 2000). In this case, the switch I region contributes to the interaction with the effector via the RBD, whereas the switch II region is involved in effector regulation. If a similar principle applies to sec6/8 regulation by RalA, switch II and the surrounding residues of RalA might be involved in sec6/8 regulation. Another possibility is an interaction similar to that found in the Ran–importin/β–karyopherin-β complex (Chook and Blobel, 1999; Vetter et al., 1999a), where most of the binding interface is located on the opposite side of the switch I region and involves several α-helices of importin-β/karyopherin-β. Interestingly, Sec5 and other sec6/8 components are predicted to have a significant amount of α-helical structure (Rost, 1996).
The initial tethering of the vesicle to the target membrane is transient and likely to be regulated by GTP hydrolysis as would be required for the downstream fusion event to be mediated by the SNARE proteins. This regulation should require a conformational change in small GTP-binding proteins during GTP hydrolysis. For RalA, significant conformational differences between the GppNHp- and GDP-bound states were found exclusively in switches I and II (Figure 7A). RalA does not have any additional switch, such as the N-terminal region of Arf (Goldberg, 1998) or the C-terminal switch of Ran (Vetter et al., 1999b). As the switch I region of RalA is occupied by Sec5, the switch II region potentially plays an important role for the guanine nucleotide-dependent regulation of membrane tethering. In spite of the plausible hypothesis that RalA–sec6/8 interactions mediate the initial vesicle docking interaction and then lead to SNARE complex formation, it is unknown how RalA and the sec6/8 complex are themselves localized to their respective compartments. Furthermore, the complex might have additional functions in protein trafficking, cell polarization or organization of the actin cytoskeleton (Sugihara et al., 2002). This first glimpse into the structural basis of membrane trafficking specificity is only one of several aspects of the trafficking process that remains to be understood at a molecular level. The fidelity of membrane trafficking is the result of a large number of specific protein–protein and protein–lipid interactions that together build the complexity of the secretory pathway. It is the sum of all of these processes that leads to the exquisite architecture of the cellular membrane network.
Materials and methods
Cloning, expression and purification
In this section, we use the terms RalA (9–183) and Sec5 (1–99) to explicitly and more accurately describe the protocol. The genes encoding RalA (9–183) and Sec5 (1–99) were cloned into the pGEX2T vector (Amersham, Arlington Heights, IL), using BamHI and EcoRI restriction sites. Each protein was overproduced at 20°C for 12–17 h in the E.coli strain BL21-RIL (Stratagene, La Jolla, CA) as a GST fusion construct, after induction with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). For both proteins, cells were harvested and suspended in phosphate-buffered saline (PBS) containing 1 mM dithiothreitol (DTT) and 0.1 mM PMSF, then lysed using a microfluidizer at 17 000 p.s.i. The lysate was centrifuged for 1 h at 28 000 g. The supernatant was loaded into a glutathione–Sepharose 4B (Amersham) column at 4°C, and the column was washed with PBS containing 1 mM DTT. GST-fused RalA (9–183) and GST-fused Sec5(1–99) were cleaved with thrombin (Hematologic Technology) on the column, after replacement of PBS with 50 mM Tris–HCl buffer pH 7.5 containing 150 mM NaCl and 1 mM DTT. RalA (9–183) and Sec5 (1–99) were eluted after the cleavage. Fractions containing RalA (9–183) were directly loaded onto a Mono Q column (Amersham), which was pre-equilibrated with 50 mM Tris–HCl pH 7.5 containing 100 mM NaCl and 5 mM β-mercaptoethanol. RalA (9–183) was eluted with 50 mM Tris–HCl pH 7.5 buffer containing 5 mM β-mercaptoethanol using a linear gradient of 100–600 mM NaCl. Fractions containing Sec5 (1–99) were dialyzed against 50 mM Na-HEPES buffer pH 6.8 containing 100 mM NaCl and 5 mM β-mercaptoethanol, and loaded onto a Mono S column (Amersham), which was pre-equilibrated with the same buffer as that used for dialysis. Sec5 (1–99) was eluted with 50 mM Na-HEPES pH 6.8 containing 5 mM β-mercaptoethanol using a linear gradient of 100–350 mM NaCl.
To form the RalA (9–183)·GppNHp complex, we treated purified RalA (9–183) with 8 U/mg calf intestinal alkaline phosphatase (CIP) (New England Biolabs, Beverly, MA), an excess molar concentration (∼500 µM) of GppNHp (Calbiochem, San Diego, CA) and 2 mM MgCl2 at 4°C for 16–48 h. The RalA (9–183)·GppNHp·Sec5 (1–99) complex was formed by mixing the RalA (9–183)·GppNHp complex and purified Sec5 (1–99), and subsequent buffer exchange using Ultrafree-15 (Millipore). The buffer used for complex formation was 20 mM Tris–HCl pH 7.5 containing 50 mM NaCl, 5 mM β-mercaptoethanol, 300 µM GppNHp and 2 mM MgCl2. The complex was purified by using a HiLoad Superdex 75 (Amersham) size-exclusion column with 20 mM Tris–HCl pH 7.5 containing 50 mM NaCl, 5 mM β-mercaptoethanol and 2 mM MgCl2.
Crystallization, data collection and processing
Crystals of the complex grew at 20°C by hanging drop vapor diffusion. The protein solution for crystallization was prepared at a final concentration of 13–17 mg/ml. A 1–2 µl aliquot of protein solution was mixed with an equal volume of mother liquor, consisting of 50 mM Na acetate pH 5.5 and 7.5–8.5% polyethylene glycol (PEG) 8000. The drop solution was slowly equilibrated against the mother liquor. Thick rod-shaped crystals grew after 1–3 days. Crystals were flash frozen directly in liquid nitrogen, after soaking sequentially with mother liquor containing between 14 and 28% of PEG400. A diffraction data set for the final structure refinement was collected at 100 K at beamline ID9-2, SSRL (Stanford, CA) using an ADSC Q315 CCD detector. The diffraction data were processed with a HKL2000 (HKL Research) (Otwinowski and Minor, 1997). Statistics of the data processing are shown in Table II.
Table II. Crystallographic and refinement statistics.
| Space group | I4 |
| Cell dimensions | a = b = 117.42 Å, c = 102.68 Å |
| X-ray source | SSRL ID9-2 |
| dmin | 2.1 Å |
| No. of total reflections | 249 726 |
| No. of unique reflections | 40 357 |
| Completeness | 99.4% (98.6%) |
| I/σ | 40.4 (3.75) |
|
Rsyma |
0.044 (0.274) |
| Luzzati coordinate error | 0.28 Å |
| Cross-validated Luzzati coordinate error | 0.32 Å |
| R.m.s.d. bond length | 0.006 Å |
| R.m.s.d. bond angle | 1.143° |
| R.m.s.d. dihedral angle | 23.37° |
| R.m.s.d. improper angle | 0.759° |
| B factors: minimum, average, maximum | 18.3, 45.2, 92.1 |
| Residues in core regions | 90.3% |
| Residues in additionally allowed regions | 9.2% |
| Residues in generously allowed regions | 0.5% |
| Rwork, Rfreeb | 0.219, 0.254 |
Values in parentheses are for the high resolution bin, 2.14–2.10 Å.
aRsym = ΣhΣi|Ihi – <Ih>|/ΣhΣi|Ihi|, where h and i are unique reflection indices and symmetry-equivalent indices, respectively.
bRwork = Σ|Fobs – Fcalc|/Σ|Fobs| for the work set, and Rfree = Σ|Fobs – Fcalc|/Σ|Fobs| for the test set, which comprises randomly selected 10% of the total reflections.
Phasing by molecular replacement
Searches for peaks of the rotation and translation functions were performed with AMoRe (CCP4, 1994; Navaza, 1994). The 1.5 Å resolution structure of the GppNHp-bound Ras (PDB code 1CTQ) was used as a search model. At first, we used a diffraction data set collected at 100 K using a Rigaku RU-300 rotating anode and RaxisIV++ imaging plate detector (data not shown). A rotation search at 12–7.0 Å resolution showed a pronounced single peak producing a correlation coefficient of 0.217. A subsequent translation search using the 12–8.0 Å resolution data also showed a single peak producing a correlation coefficient of 0.323 and an R value of 0.483. Since the Matthews coefficient [5.75, 2.88 and 1.92 for one, two and three complex(es) per asymmetric unit] suggested the presence of two complexes in the asymmetric unit, we searched for another peak of the translation function, using the highest 20 peaks of the previous rotation function and including the fixed partial model obtained from the first translation search. This second translation search, using diffraction data at 12–7.5 Å resolution, clearly indicated another single peak producing a correlation coefficient of 0.402 and an R value of 0.462. After rigid-body refinement using 15–4.0 Å resolution data, the correlation coefficient and the R value improved to 0.448 and 0.449, respectively. Based on this final solution, we made a model of two RalA molecules in the asymmetric unit using the program O (Jones et al., 1991), and then refined it with conjugate gradient minimization. The resulting model was used for the molecular replacement phasing of the aforementioned data set collected at SSRL, beamline ID9-2. After initial rigid-body refinement, the correlation coefficient and R value were 0.522 and 0.434 in the resolution range of 15–4.0 Å. Application of density modification including solvent flipping (Abrahams and Leslie, 1996), histogram matching (Lunin, 1988) and NCS-related averaging (Bricogne, 1976) improved the phases, and produced an excellent quality electron density map. All phasing calculations and density modification were performed using the Crystallography & NMR System (CNS) (Brunger et al., 1998).
Model building and refinement
The model of the RalA·GppNHp·Sec5 Ral-binding domain complex was built using the program O (Jones et al., 1991). The refinement of the model was carried out by conjugate gradient minimization and torsion angle dynamics simulated annealing (Rice and Brunger, 1994) using the MLF target function (Adams et al., 1997). Individual restrained B factor refinement (Hendrickson, 1985) was performed after every positional refinement cycle. Ten per cent of the total unique reflections were randomly selected for the Rfree (Brunger et al., 1993) calculation to monitor the refinement and model building cycles. σA-weighted 2Fo – Fc, Fo – Fc and simulated-annealed 2Fo – Fc omit maps were calculated to improve the model by inspection, using the program O (Jones et al., 1991). Iterative model building and refinement improved the model, resulting in final Rwork and Rfree values of 0.219 and 0.254, respectively. The final model includes 222 water molecules and two Mg2+. The atomic occupancies of residues 72–76 in one RalA model and residues 73–75 in the other RalA model were set to 0, because of discontinuous electron density. All refinement calculations were carried out with CNS (Brunger et al., 1998). Refinement statistics are shown in Table II. The coordinates and structure factors have been deposited with the Protein Data Bank (PDB code 1UAD).
Site-directed mutagenesis and isothermal titration calorimetry (ITC)
Single site mutations were introduced into the genes encoding RalA and the Ral-binding domain of Sec5 by PCR using suitable primers. Nucleotide sequences were confirmed by dideoxy termination sequencing. ITC experiments were performed using a VP-ITC calorimeter from Microcal, LLC (Northampton, MA), as described in the instrument user’s manual. RalA (9–183) and full-length RalA were purified as described above except for after glutathione–Sepharose 4B (Amersham) column chromatography, the bound GST–RalA beads were processed through a series of nucleotide (GppNHp or GDP) exchange reactions in the presence of EDTA to purify protein to either GppNHp- or GDP-bound RalA, as described previously for Rab GTP-binding proteins (Christoforidis and Zerial, 2000). The thrombin-cleaved proteins were used for ITC studies. RalA (4 µM) was loaded in the sample cell (in PBS containing 5 mM MgCl2 and 0.5 mM GppNHp or GDP), using a volume of 1.42 ml, and titrated with the Sec5 Ral binding domain (150 µM) in the same buffer with each 5 µl injection for a total of 30 injections. The numbers of 5 µl injections for Sec5 (Thr11Ala) and Sec5 (Arg27Ala) were 40 and 35 as their concentrations in the syringe were 110 and 135 µM, respectively. The titrations were performed while samples were being stirred at 300 r.p.m. at 30°C. An interval of 4 min between each injection was used in order for the baseline to stabilize. The data were fitted with a single site model to calculate the number of binding sites (n), the binding constant (Ka) and the change in enthalpy (ΔH°) using Origin software, provided by Microcal, LLC.
GDP-bound RalA structure
The R and Rfree values of the GDP-bound RalA structure (Bauer et al., 1999) are 0.212 and 0.235, respectively, at 1.8 Å resolution (I.R.Vetter, personal communication). The r.m.s. deviations of bond lengths, bond, dihedral and improper angles are 0.006 Å, 1.2°, 21.6° and 0.7°, respectively. In the Ramachandran plot, 92.2 and 7.8% of the residues are in the core and additional allowed regions, respectively.
Figure preparation
All figures were prepared using MOLSCRIPT (Kraulis, 1991) and PyMOL (DeLano, 2001).
Acknowledgments
Acknowledgements
We thank Dr Ingrid R.Vetter (Max-Planck-Institut für Molekulare Physiologie) for providing us with the unpublished coordinates of the GDP-bound RalA, and M.Breidenbach and S.Kaiser for help with data collection at SSRL, beamline ID9-2. Portions of this research were carried out at the Stanford Synchrotron Radiation Laboratory (SSRL), a national user facility operated by Stanford University on behalf of the US Department of Energy, Office of Basic Energy Sciences. The SSRL Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program and the National Institute of General Medical Sciences. S.F. was supported by JSPS Research Fellowships for Research Abroad.
References
- Abrahams J.P. and Leslie,A.G.W. (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D, 52, 30–42. [DOI] [PubMed] [Google Scholar]
- Adamo J.E., Rossi,G. and Brennwald,P. (1999) The Rho GTPase Rho3 has a direct role in exocytosis that is distinct from its role in actin polarity. Mol. Biol. Cell, 10, 4121–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams P.D., Pannu,N.S., Read,R.J. and Brunger,A.T. (1997) Cross-validated maximum likelihood enhances crystallographic simulated annealing refinement. Proc. Natl Acad. Sci. USA, 94, 5018–5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer B., Mirey,G., Vetter,I.R., Garcia-Ranea,J.A., Valencia,A., Wittinghofer,A., Camonis,J.H. and Cool,R.H. (1999) Effector recognition by the small GTP-binding proteins Ras and Ral. J. Biol. Chem., 274, 17763–17770. [DOI] [PubMed] [Google Scholar]
- Bricogne G. (1976) Methods and programs for direct-space exploitation of geometric redundancies. Acta Crystallogr. A, 32, 832–847. [Google Scholar]
- Brunger A.T., Clore,G.M., Gronenborn,A.M., Saffrich,R. and Nilges,M. (1993) Assessing the quality of solution nuclear magnetic resonance structures by complete cross-validation. Science, 261, 328–331. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Brymora A., Valova,V.A., Larsen,M.R., Roufogalis,B.D. and Robinson,P.J. (2001) The brain exocyst complex interacts with RalA in a GTP-dependent manner: identification of a novel mammalian Sec3 gene and a second Sec15 gene. J. Biol. Chem., 276, 29792–29797. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chook Y.M. and Blobel,G. (1999) Structure of the nuclear transport complex karyopherin-β2–Ran–GppNHp. Nature, 399, 230–237. [DOI] [PubMed] [Google Scholar]
- Christoforidis S. and Zerial,M. (2000) Purification and identification of novel Rab effectors using affinity chromatography. Methods, 20, 403–410. [DOI] [PubMed] [Google Scholar]
- de Leeuw H.P., Fernandez-Borja,M., Reits,E.A., Romani de Wit,T., Wijers-Koster,P.M., Hordijk,P.L., Neefjes,J., van Mourik,J.A. and Voorberg,J. (2001) Small GTP-binding protein Ral modulates regulated exocytosis of von Willebrand factor by endothelial cells. Arterioscler. Thromb. Vasc. Biol., 21, 899–904. [DOI] [PubMed] [Google Scholar]
- DeLano W.L. (2001) The PyMOL Molecular Graphics System. http://www.pymol.org
- Fasshauer D., Sutton,R.B., Brunger,A.T. and Jahn,R. (1998) Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl Acad. Sci. USA, 95, 15781–15786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger F.P., Hughes,T.E. and Novick,P. (1998) Sec3p is a spatial landmark for polarized secretion in budding yeast. Cell, 92, 559–571. [DOI] [PubMed] [Google Scholar]
- Goldberg J. (1998) Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell, 95, 237–248. [DOI] [PubMed] [Google Scholar]
- Grindstaff K.K., Yeaman,C., Anandasabapathy,N., Hsu,S.-C., Rodriguez-Boulan,E., Scheller,R.H. and Nelson,W.J. (1998) Sec6/8 complex is recruited to cell–cell contacts and specifies transport vesicle delivery to the basal-lateral membrane in epitherial cells. Cell, 93, 731–740. [DOI] [PubMed] [Google Scholar]
- Guo W., Grant,A. and Novick,P. (1999a) Exo84p is an exocyst protein essential for secretion. J. Biol. Chem., 274, 23558–23564. [DOI] [PubMed] [Google Scholar]
- Guo W., Roth,D., Walch-Solimena,C. and Novick,P. (1999b) The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J., 18, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Tamanoi,F. and Novick,P. (2001) Spatial regulation of the exocyst complex by Rho1 GTPase. Nat. Cell Biol., 3, 353–360. [DOI] [PubMed] [Google Scholar]
- Hazuka C.D., Foletti,D.L., Hsu,S.-C., Kee,Y., Hopf,F.W. and Scheller,R.H. (1999) The sec6/8 complex is located at neurite outgrowth and axonal synapse assembly domains. J. Neurosci., 19, 1324–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson W.A. (1985) Stereochemically restrained refinement of macromolecular structures. Methods Enzymol., 115, 252–270. [DOI] [PubMed] [Google Scholar]
- Henry D.O., Moskalenko,S.A., Kaur,K.J., Fu,M., Pestell,R.G., Camonis,J.H. and White,M.A. (2000) Ral GTPases contribute to regulation of cyclin D1 through activation of NF-κB. Mol. Cell. Biol., 20, 8084–8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman G.R., Nassar,N. and Cerione,R.A. (2000) Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell, 100, 345–356. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1998) Touring protein fold space with DALI/FSSP. Nucleic Acids Res., 26, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S.-C., Hazuka,C.D., Roth,R., Foletti,D.L., Heuser,J. and Scheller,R.H. (1998) Subunit composition, protein interactions and structures of the mammalian brain sec6/8 complex and septin filaments. Neuron, 20, 1111–1122. [DOI] [PubMed] [Google Scholar]
- Huang L., Hofer,F., Martin,G.S. and Kim,S.H. (1998) Structural basis for the interaction of Ras with RalGDS. Nat. Struct. Biol., 5, 422–426. [DOI] [PubMed] [Google Scholar]
- Inoue M., Chang,L., Hwang,J., Chiang,S.H. and Saltiel,A.R. (2003) The exocyst complex is required for targeting of Glut4 to the plasma membrane by insulin. Nature, 422, 629–633. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Jullien-Flores V., Dorseuil,O., Romero,F., Letourneur,F., Saragosti,S., Berger,R., Tavitian,A., Gacon,G. and Camonis,J.H. (1995) Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J. Biol. Chem., 270, 22473–22477. [DOI] [PubMed] [Google Scholar]
- Kee Y., Yoo,J.S., Hazuka,C.D., Peterson,K.E., Hsu,S.C. and Scheller,R.H. (1997) Subunit structure of the mammalian exocyst complex. Proc. Natl Acad. Sci. USA, 94, 14438–14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: A Program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Lipschutz J.H. and Mostov,K.E. (2002) Exocytosis: the many masters of the exocyst. Curr. Biol., 12, R212–R214. [DOI] [PubMed] [Google Scholar]
- Lunin V.Y. (1988) Use of the information on electron density distribution in macromolecules. Acta Crystallogr. A, 44, 144–150. [Google Scholar]
- Madden T.L., Tatusov,R.L. and Zhang,J. (1996) Applications of network BLAST server. Methods Enzymol., 266, 131–141. [DOI] [PubMed] [Google Scholar]
- Matern H.T., Yeaman,C., Nelson,W.J. and Scheller,R.H. (2001) The Sec6/8 complex in mammalian cells: characterization of mammalian Sec3, subunit interactions and expression of subunits in polarized cells. Proc. Natl Acad. Sci. USA, 98, 9648–9653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskalenko S., Henry,D.O., Rosse,C., Mirey,G., Camonis,J.H. and White,M.A. (2002) The exocyst is a Ral effector complex. Nat. Cell Biol., 4, 66–72. [DOI] [PubMed] [Google Scholar]
- Mott H.R., Nietlispach,D., Hopkins,L.J., Mirey,G., Camonis,J.H. and Owen,D. (2003) Structure of the GTPase-binding domain of Sec5 and elucidation of its Ral binding site. J. Biol. Chem., 278, 17053–17059. [DOI] [PubMed] [Google Scholar]
- Murthy M., Garza,D., Scheller,R.H. and Schwarz,T.L. (2003) Mutations in the exocyst component sec5 disrupt neuronal membrane traffic, but neurotransmitter release persists. Neuron, 37, 433–447. [DOI] [PubMed] [Google Scholar]
- Nakashima S., Morinaka,K., Koyama,S., Ikeda,M., Kishida,M., Okawa,K., Iwamatsu,A., Kishida,S. and Kikuchi,A. (1999) Small G protein Ral and its downstream molecules regulate endocytosis of EGF and insulin receptors. EMBO J., 18, 3629–3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassar N., Horn,G., Herrmann,C., Scherer,A., McCormick,F. and Wittinghofer,A. (1995) The 2.2 Å crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature, 375, 554–560. [DOI] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Novick P. and Guo,W. (2002) Ras family therapy: Rab, Rho and Ral talk to the exocyst. Trends Cell Biol., 12, 247–249. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Pacold M.E. et al. (2000) Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase γ. Cell, 103, 931–943. [DOI] [PubMed] [Google Scholar]
- Reuther G.W. and Der,C.J. (2000) The Ras branch of small GTPases: Ras family members don’t fall far from the tree. Curr. Opin. Cell Biol., 12, 157–165. [DOI] [PubMed] [Google Scholar]
- Rice L.M. and Brunger,A.T. (1994) Torsion angle dynamics: reduced variable conformational sampling enhances crystallographic structure refinement. Proteins, 19, 277–290. [DOI] [PubMed] [Google Scholar]
- Rost B. (1996) PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol., 266, 525–539. [DOI] [PubMed] [Google Scholar]
- Scheffzek K., Grunewald,P., Wohlgemuth,S., Kabsch,W., Tu,H., Wigler,M., Wittinghofer,A. and Herrmann,C. (2001) The Ras–Byr2RBD complex: structural basis for Ras effector recognition in yeast. Structure, 9, 1043–1050. [DOI] [PubMed] [Google Scholar]
- Sugihara K., Asano,S., Tanaka,K., Iwamatsu,A., Okawa,K. and Ohta,Y. (2002) The exocyst complex binds the small GTPase RalA to mediate filopodia formation. Nat. Cell Biol., 4, 73–78. [DOI] [PubMed] [Google Scholar]
- Takai Y., Sasaki,T. and Matozaki,T. (2001) Small GTP-binding proteins. Physiol. Rev., 81, 153–208. [DOI] [PubMed] [Google Scholar]
- Terbush D.R. and Novick,P. (1995) Sec6, Sec8 and Sec15 are components of a multisubunit complex which localizes to small bud tips in Saccharomyces cerevisiae. J. Cell Biol., 130, 299–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terbush D.R., Maurice,T., Roth,D. and Novick,P. (1996) The Exocyst is a multiprotein complex required for exocytosis in Saccharomyces cerevisiae. EMBO J., 15, 6483–6494. [PMC free article] [PubMed] [Google Scholar]
- Terbush D.R., Guo,W., Dunkelbarger,S. and Novick,P. (2001) Purification and characterization of yeast exocyst complex. Methods Enzymol., 329, 100–110. [DOI] [PubMed] [Google Scholar]
- Vetter I.R., Arndt,A., Kutay,U., Gorlich,D. and Wittinghofer,A. (1999a) Structural view of the Ran–Importinβ interaction at 2.3 Å resolution. Cell, 97, 635–646. [DOI] [PubMed] [Google Scholar]
- Vetter I.R., Nowak,C., Nishimoto,T., Kuhlmann,J. and Wittinghofer,A. (1999b) Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature, 398, 39–46. [DOI] [PubMed] [Google Scholar]
- Whyte J.R.C. and Munro,S. (2002) Vesicle tethering complexes in membrane traffic. J. Cell Sci., 115, 2627–2637. [DOI] [PubMed] [Google Scholar]
- Zhang X., Bi,E., Novick,P., Du,L., Kozminski,K.G., Lipschutz,J.H. and Guo,W. (2001) Cdc42 interacts with the exocyst and regulates polarized secretion. J. Biol. Chem., 276, 46745–46750. [DOI] [PubMed] [Google Scholar]