Abstract
Formyl-CoA transferase catalyses transfer of CoA from formate to oxalate in the first step of oxalate degradation by Oxalobacter formigenes, a bacterium present in the intestinal flora which is implicated in oxalate catabolism in mammals. Formyl-CoA transferase is a member of a family of CoA-transferases for which no structural information is available. We now report the three-dimensional structure of O.formigenes formyl-CoA transferase, which reveals a novel fold and a very striking assembly of the homodimer. The subunit is composed of a large and a small domain where residues from both the N- and C-termini of the subunit are part of the large domain. The linkers between the domains give the subunit a circular shape with a hole in the middle. The enzyme monomers are tightly interacting and are interlocked. This fold requires drastic rearrangement of ∼75 residues at the C-terminus for formation of the dimer. The structure of a complex of formyl-CoA transferase with CoA is also reported and sets the scene for a mechanistic understanding of enzymes of this family of CoA-transferases.
Keywords: CAIB-BAIF family/CoA-transferase/crystal structure/knotted fold/oxalate degradation
Introduction
Oxalate is a common by-product of metabolism in virtually every life form, but in common with other mammals, human cells are not able to degrade this metabolite. Oxalate is maintained at a low level in tissues by secretion into the blood, from which it is eventually removed by the kidneys or released into the intestinal lumen. The accumulation of oxalate in the body leads to several disorders such as hyperoxaluria, pyridoxine deficiency, cardiomyopathy, cardiac conductance disorders, calcium oxalate stones and renal failure (Williams and Smith, 1968; Rodby et al., 1991). Hyperoxaluria is observed in diverse medical conditions, e.g. aspergillosis and cystic fibrosis, and also in patients who have undergone ileal-bypass surgery (Hylander et al., 1978; Clark et al., 1985). Moreover, hyperoxaluria is rather common in individuals who have received prolonged treatment with antibiotics, such as patients affected by cystic fibrosis. This interesting observation can be correlated with the disappearance of an obligate anaerobe, Oxalobacter formigenes, from the intestine of these patients. A direct correlation between the disappearance of O.formigenes from the intestinal flora and the appearance of hyperoxaluria symptoms has been established in humans with urolithiasis and in rats (Sidhu et al., 1998, 1999). These studies have therefore generated significant interest in the role of O.formigenes in maintaining oxalate homeostasis in humans, leading to the somewhat controversial hypothesis that O.formigenes plays an important role in the intestinal flora of mammals by degrading dietary and secreted oxalate within the lumen. Not only does this process prevent re-absorption of oxalate in the lower tract of the intestine (Sidhu et al., 1999), but it also maintains a favourable transepithelial gradient for further oxalate excretion.
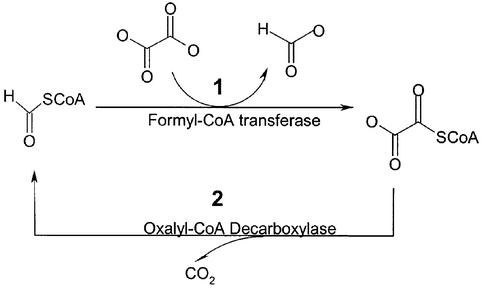
Oxalobacter formigenes is an unusual bacterium in that oxalate is of vital necessity as an energy source for its survival. Oxalate is anaerobically decarboxylated to give CO2 and formate in a two-step pathway that is mediated by the coupled action of the enzymes formyl-CoA transferase and oxalyl-CoA decarboxylase (Figure 1). Thus, as observed in Pseudomonas oxalaticus (Quayle, 1963), formyl-CoA transferase catalyses the transfer of CoA from formate to oxalate (Baetz and Allison, 1990), activating the oxalyl moiety for thiamine-dependent decarboxylation in a reaction that is mediated by oxalyl-CoA decarboxylase (Baetz and Allison, 1989). The metabolic importance of these two enzymes to O.formigenes is supported by the observation that they make up 20% of the total protein content of the cell (Baetz and Allison, 1989, 1990). It has been proposed that this oxalate degradation pathway, coupled with oxalate/formate transport by a oxalate/formate transmembrane transporter, is the origin of the transmembrane potential required for ATP production in the bacterium (Ruan et al., 1992).
Fig. 1. Oxalate degradation pathway in O.formigenes. Step 1: formyl-CoA reacts with oxalate to give formate and oxalyl-CoA; step 2: oxalyl-CoA is decarboxylated and regenerates formyl-CoA.
Native formyl-CoA transferase (FRC) has been purified from O.formigenes (Baetz and Allison, 1990), and the gene encoding the enzyme has been cloned and expressed in Escherichia coli (Sidhu et al., 1997). FRC consists of 428 amino acids with a mass of 44 kDa, and has been shown to be specific for oxalate or succinate as coenzyme A acceptors. No reaction is observed if these substrates are replaced by acetate or malonate. The optimal pH for enzyme activity is between 6.5 to 7.5. Detailed kinetic characterization of the recombinant, wild-type FRC used in these structural studies has shown that the apparent Km is 11.1 µM for formyl-CoA and 5.25 mM for oxalate; the Vmax is 6.49 µmol/min/mg (unpublished data), but the chemical mechanism employed by the enzyme remains to be established.
CoA transferases, ubiquitous enzymes that catalyse the transfer of coenzyme A from a donor to an acceptor, have traditionally been divided into two families. Enzymes belonging to family I are mostly involved in fatty acid metabolism, and function using a ping-pong mechanism involving enzyme-bound CoA as well as mixed substrate/product anhydrides (Heider, 2001). Family II include the homodimeric α-subunits of the citrate and citramalate lyases, which carry out CoA transfer through a ternary complex intermediate (Heider, 2001). Recent sequence alignments (Elssner et al., 2001) have, however, revealed the existence of a third family of CoA transferases to which FRC belongs (Heider, 2001). Most of the genes belonging to family III are present in anaerobic bacteria, but a few similar genes have also been found in archea and eukaria. Enzymes belonging to family III are involved in the anaerobic metabolism of toluene, carnitine and bile acids, and in Stickland fermentation and oxalate catabolism (Heider, 2001). These CoA transferases are active as homo- or heterodimers and have a similar mass, between 42 and 47 kDa. The characterized enzymes in this family are very substrate- and stereo-specific. Sequence comparison shows that some parts of these proteins are quite conserved, with a pairwise identity in the order of 18–37% (Heider, 2001). No knowledge on structure or catalytic mechanism is available for this class of CoA transferases. We now present the crystal structure of formyl-CoA transferase, as both its apoenzyme and in complex with coenzyme A. In addition to revealing an unusual new protein fold, our results have important implications for both the folding pathways that lead to the homodimer and the catalytic mechanism.
Results and discussion
Quality of electron density maps and resulting models
The experimental electron density obtained from seleno-methionine single wavelength anomalous dispersion (SAD) phasing to 2.8 Å was of excellent quality for both chains in the asymmetric unit, except for residues 239–240 and 300–309 in subunit B, which are all solvent exposed. The main chain of each subunit, consisting of residues 2–428, could be traced unequivocally, yielding high-quality models as judged by commonly accepted criteria (see Table I) after refinement to 2.2 and 2.5 Å for the apoenzyme and the CoA complex, respectively.
Table I. Data collection and refinement.
| Native | Semet-frc Peak | CoA complex | |
|---|---|---|---|
| Data collection | ESRF ID14–1 | DESY BW7A | ESRF ID 29 |
| Resolution (Å) | 25–2.2 | 25–2.8 | 25–2.5 |
| Unique reflections | 57 045 | 30 805 | 38 678 |
| Completeness (%)a | 99.0 (99.0) | 98.6 (98.0) | 99.7 (98.1) |
| Redundancya | 5.1 (4.6) | 3.2 (2.8) | 5.4 (3.1) |
| I/σa | 15.3 (4.9) | 13.3(2.4) | 16.4 (4.2) |
| Rsym (%)a | 9.8 (23.6) | 8.2 (43.5) | 8.5 (21.9) |
| Phasing powerb | 1.36 | ||
| Refinement | |||
| B-factor value from Wilson plot | 29.5 | 49.8 | |
| Mean B-factor for model | 32.6 | 32.0 | |
| R-factor (%) | 17.1 (19.0) | 19.4 (27.2) | |
| Rfree (%) | 20.9 (21.9) | 23.6 (34.1) | |
| Atoms in the model | 6624 protein atoms, 528 waters | 6624 protein atoms, 261 waters, 2 CoA molecules | |
| Ramachandran plot | |||
| Most allowed region (%) | 91.1 | 86.7 | |
| Additional allowed region (%) | 8.3 | 12.4 | |
| Generously allowed region (%) | 0.3 | 0.7 | |
| Disallowed region (%) | 0.3 | 0.3 |
aNumbers in parentheses are calculated using data from the highest resolution shell.
bPhasing power from SOLVE run using the peak dataset to 3.5 Å.
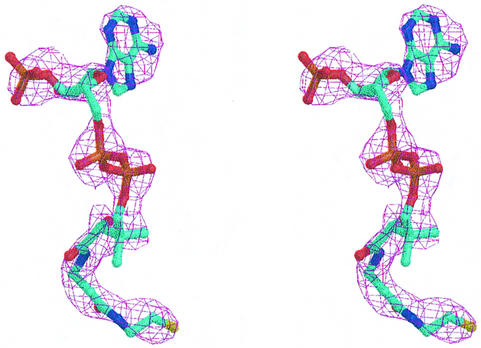
Both formyl-CoA and oxalyl-CoA are unstable in water and free CoA is already an inhibitor at a low concentration (40 µM; unpublished data). In order to elucidate the position and the organization of the active site and the amino acids involved in binding and catalysis, FRC was crystallized in the presence of oxalyl-CoA and free CoA as described previously (Ricagno et al., 2003). In the virtually identical structures of both complexes, only electron density for CoA is present, and it is very likely that hydrolysis of oxalyl-CoA occurred in the aqueous solution before the complex with the enzyme was formed. The structure from FRC co-crystallized with oxalyl-CoA is of slightly higher quality, with better electron density for the CoA moiety (Figure 2), and has been used in the following description.
Fig. 2. The initial difference Fourier electron density map at 2.5 σ for the CoA complex of formyl-CoA transferase.
Subunit structure
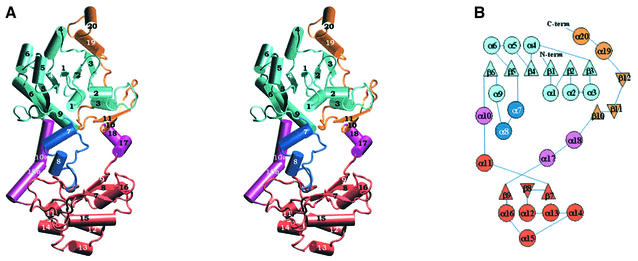
The subunit consists of two domains, the large domain and the small domain, and two linker regions (α10 and α17–α18) that connect the two domains. The large domain comprises both the N-terminal (residues 2–199, β1–β6) and C-terminal parts of the protein chain (residues 367–428, β10–α20), while the small domain contains residues 219–351, α11–β9 (Figure 3). The large domain contains an open α/β-twisted sheet structure with a Rossmann fold consisting of a central six-stranded parallel β-sheet with helices packed to both sides of it. It starts with a βαβαα motif, repeated twice, which forms the main part of the β-sheet. The connection between strands β5 and β6 is left-handed; after β5 the polypeptide chain, including two α helices (α7 and α8), makes a protrusion towards the small domain. A long loop leads back to the large domain where α9 and β6 complete the α/β structure. The linker α10, a long, very bent helix, then connects the large domain with the small domain.
Fig. 3. (A) Schematic stereo picture showing the monomer of formyl-CoA transferase. The large domain is in two colours: the N-terminal part in cyan and the C-terminal part in orange. The small domain is coloured red, the chain protruding from the large domain to the small domain is blue and the two linkers between the two domains are purple. (B) A topology diagram of formyl-CoA transferase.
The small domain starts with a short helix (α11) and a long loop (residues 232–268) connected to a three-stranded, anti-parallel β-sheet. The first two strands (β7 and β8) are joined by a short loop, while five helices (α12–α16) are inserted between the second and the third strand. After strand β9, the second linker (α17–α18) connects the small domain back to the large domain.
The C-terminal part of the large domain consists of a two-stranded β sheet (β10–β11), a long loop and two short helices (α19 and α20). These residues are not part of the central β-sheet of the domain but are packed tightly to it through, amongst other contacts, 15 hydrogen bonds mainly from the long loop between β11 and α19, three that are mediated by the first short helix and three by the last three C-terminal amino acids. In the dimer, however, an anti-parallel strand β12, comprising residues 385–389, from the other subunit extends the central parallel β-sheet of the large domain.
An unexpected feature of the subunit is the hole formed between the domains. The hole is at least 13 Å wide between the linkers and 22 Å long between the two domains.
Superimposition of the Cα atoms of the two subunits gives a r.m.s. deviation of 0.6 Å, while superposition of only the large domains gives 0.27 Å. The small domain shows larger deviations (r.m.s. 0.73 Å), some of which are due to local areas of different crystal contacts. However, a short loop of four glycine residues (258–261) in the interior of the protein shows very different conformations that are coupled to two different conformations for the side chain of Trp48 in the two subunits.
Dimer structure
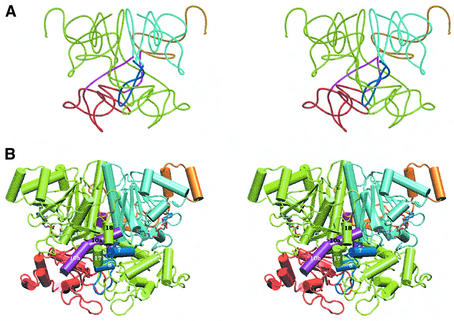
The 2-fold related subunits in the dimer embrace each other with their C-terminal parts, giving rise to an interlocked dimer (Figure 4). The linker helix α10 of molecule A is passing through the hole present in molecule B and vice versa. This puts the two domains of one subunit on top of and below the hole of the other subunit. The hole is filled further by the stretch α7–α8, but this does not give rise to a knot since this is only a protrusion from the large domain.
Fig. 4. (A) Simplified chain trace showing the dimer organization: the linkers α10 are passing through the hole of the other monomer, forming an interlocked dimer. Colour coding as in Figure 2A for one subunit while the other subunit is in green. (B) Stereo picture showing the interlocked dimer organization of formyl-CoA transferase and the bound CoA molecules.
Thus, the two subunits are interlocked and only a drastic rearrangement of ∼75 residues (α17–α20) at the C-terminus would allow the dimer to break apart; and the reverse process, dimerization, can not occur when the two subunits are completely folded. Perhaps the two polypeptides start to form dimers already before they are wholly synthesized; the large amount of FRC generated in the bacteria would ease the interaction between ribosomes producing FRC. Alternatively the subunits are not completely folded when leaving the ribosome and the last steps in folding occur when the two polypeptide chains interact. A chaperone might keep the polypeptides in a partially unfolded state until two monomers start to interact and to fold on each other. If so, this chaperone must also be present in E.coli since the structure is constructed from recombinant protein expressed in that organism. Due to this peculiar packing of subunits, the dimer interface is very extensive, comprising an area of 6200 Å2 with 38% of the amino acids involved, while the average buried area reported for 32 homodimers previously analysed is 1600 Å2 (see Materials and methods).
There are no strong indications that this dimer fold is caused by domain swapping (Rousseau et al., 2003). There is a short sequence where corresponding amino acids from the two subunits are adjacent in the structure, the loop protruding from the large domain into the hole of the other subunit (residues 152–165); domain swapping could have taken place here. Reconnecting the two chains at this position to mimic a non-swapped dimer removes the interlock. However, this results in a dimer where α9 and β6 are now in a separate subunit from the rest of the α/β-domain and must be inserted into the open α/β sheet between β5 and β12 of the other subunit upon formation of the dimer. Furthermore, this dimer would also require a similar drastic rearrangement of the C-terminus to separate into subunits.
A rather unusual feature of FRC is the abundance of water molecules buried in the interior of the protein; out of 528 water molecules located in the model, 100 are not accessible from bulk solvent. One hundred and sixty-nine water molecules reside in the dimer interface making direct or indirect hydrogen bonds with both subunits, and 13 of these are totally buried.
In one of the subunits there is a deep cavity between the large domain of one subunit and the small domain of the second subunit. However, the symmetric cavity is even deeper, making a sharp curve and almost reaching out to the surface. The reason is that loop 258–260 has a different conformation and opens the tunnel. This difference is coupled to a rotation of 90° of the side chain of Trp A48 away from loop B258–B260. Since all these interactions are in the interior of the dimer it is very unlikely that different crystal contacts are forcing the two different conformations observed in the dimer; they must represent two possible low energy states. The open tunnel contains 26 water molecules, while the corresponding closed cavity contains 22 water molecules in total.
Comparison with other enzymes
Structure alignment searches have been performed for the dimer of formyl-CoA transferase using the program TOP (Lu, 2000). The only structural similarities found are between the large domain and open α/β-sheet structures with the Rossmann fold. The two most similar protein structures are the NAD binding domain (dI) of transhydrogenase from Rhodospirillum rubrum (Buckley et al., 2000) and saccaropine reductase from Magnaporthe grisea (Johansson et al., 2000). When the dI component of transhydrogenase and formyl-CoA transferase was superimposed, 94 residues were spatially matching: 18 of these were identical amino acids and the r.m.s. Cα-difference was 1.85 Å. In the three-dimensional aligment with saccaropine reductase, 81 amino acids were structurally matched, 15 of these were identical and the r.m.s. deviation was 1.74 Å. In both cases the similarity is restricted to secondary structures in the open α/β-sheet of the NAD and NADPH binding domains, respectively.
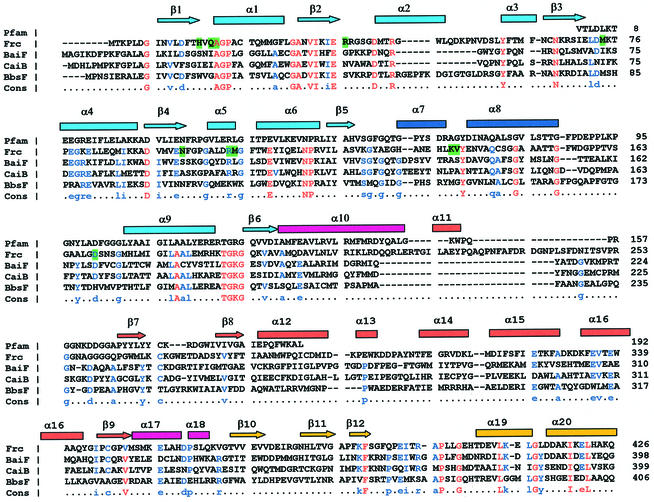
Sequence analysis (Figure 5) shows that the N-terminal part of formyl-CoA transferase belongs to pfam02515, the CaiB-BaiF family of enzymes with diverse functions including fatty acid racemases, carnitine dehydratase (CaiB) and bile acid inducible operon protein F (BaiF). The three-dimensional structure of this domain family can thus now be described in terms of the open α/β-domain of formyl-CoA transferase.
Fig. 5. Sequence alignment of formyl-CoA transferase to three enzymes in the CaiB-BaiF and CoA-transferase family III: Frc Formyl-CoA transferase (AAC45398), BaiF putative cholate CoA-transferase (AAC45415), CaiB (R)-carnitine CoA-transferase (CAA52112), and BbsF (R)-benzylsuccinate CoA-transferase (AAF89841). Secondary structures in formyl-CoA transferase are shown above the sequences. Identical residues are shown in red. CoA-contacting residues are marked green and residues suggested to be involved in catalysis in cyan.
Elssner et al. compared gene sequences encoding CoA-transferases belonging to family III (Elssner et al., 2001). In addition to the sequence conservation corresponding to the pfam-domain described above, the longest stretch of similarity for the family III CoA transferases is located in the C-terminus, comprising helices 19 and 20 (Figure 5). There are few other conserved amino acids, which makes it difficult to predict the degree of structure conservation within this family outside the open α/β-domain; in particular it is unclear whether the knotted dimer structure will be present in the other enzymes.
Coenzyme A binding
With one exception, binding of CoA to FRC does not provoke any significant domain movement or change in dimer interactions, and the dimer structure remains mostly unchanged, with a r.m.s. deviation to the Cα-atoms of the apoenzyme of 0.33 Å for subunit A and 0.62 Å for subunit B. The only large difference observed is in loop 258–261; in the complex with CoA, both loops are in the closed conformation.
The dimer of formyl-CoA transferase presents two equivalent CoA binding sites, located in the interface between the large domain of one subunit and the small domain of the other subunit (Figure 4B). Despite this, all interactions between the cofactor and the protein are established by the large domain, where it binds to the Rossmann fold.
Binding of CoA to a Rossmann fold has been reported previously for succinyl-CoA synthetase (Wolodko et al., 1994). In such a case, CoA is bound analogously to NADH/NADPH at the first βαβ-motif, as in the canonical nucleotide binding to the Rossmann fold. In FRC, however, CoA is displaced such that instead of the pyrophosphate, the SH-group of CoA is interacting with the N-terminus of the helix in the βαβ-motif (Figure 6). Intriguingly, a glutaconate CoA-transferase from Acidaminococcus fermentans (Jacob et al., 1997), belonging to family I of CoA-transferases, is also reported to have its active site at the N-terminus of the α-helix of a Rossmann fold, but seems to have a completely different binding of the CoA moiety.
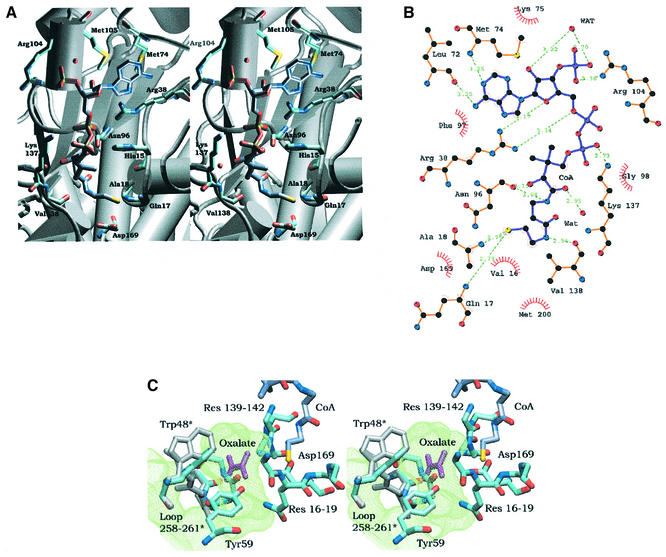
Fig. 6. (A) Stereo picture of coenzyme A in its binding site. Amino acids involved in hydrogen bonds with CoA are coloured according to atom type (C atoms cyan for FRC and ice blue for CoA). (B) Schematic drawing of the CoA interactions. Hydrogen bond distances are given and hydrophobic contacts are indicated. (C) Stereo picture of the end of the pantetheine chain of CoA and amino acids in the surrounding active site. Superimposed to loop 258–261 in the closed conformation is the same loop in the open conformation of the apoenzyme (grey); the two conformations correspond to different rotamer conformations of Trp48. The cavity formed when the loop is in the open conformation is shown in green. A model of bound oxalate, in magenta, is included but its orientation is unknown.
In FRC, the adenine part of CoA is wedged into a thin cleft and buried from solvent while the ribose, the ribose phosphate and the pyrophosphate are solvent exposed. The pantetheine chain is buried in a cleft formed mainly by the large domain. The small domain participates by closing off the cleft where the SH-group on the pantetheine arm of CoA is bound, through loop 258–261 in the closed conformation. The adenine moiety is positioned by a stacking interaction (Figure 6A) with the side chain of Arg38, and on the other side of the adenine plane by interactions with Met74 and Met105. The edge of the pyrimidine ring packs against main-chain atoms of residues 72–75. The phosphoribose is anchored to FRC through four hydrogen bonds: one between O4′ and the side chain of Arg38, another two as a result of O2′and one of the phosphate oxygen atoms interacting with the main chain carbonyl of Ala101 via a neighbouring water molecule, and the fourth by the hydrogen bonding of a phosphate oxygen atom to the side chain of Arg104. The pyrophosphate moiety interacts with the basic side-chains Lys137, Arg38 and His15. The dimethyl group of the pantetheine chain makes hydrophobic interactions with Val16, Met44 and Tyr139. The pantetheine hydroxyl group hydrogen bonds to the side chains of His15 and to the main chain carbonyl of Asn96. This carbonyl oxygen also interacts with one of the amino groups on the pantetheine chain, while the second amino group forms a hydrogen bond to the carbonyl oxygen of Val138. One of the pantetheine carbonyl oxygen atoms forms hydrogen bonds via a water molecule to the Leu136 main-chain carbonyl and Gly126 amino group. The SH group points towards the N-terminus of α-helix 1 at hydrogen bond distance from the main-chain amino group of Gln17 and Ala18, and is close to the side chain of Asp169.
In the apoenzyme structure, the loop composed of residues 258–261 adopts an open conformation in one subunit, but has a closed conformation in the other. In the FRC/CoA complex, this loop is in the closed conformation in both monomers, leaving insufficient space for an oxalate molecule to bind in the active site. However, there is a suitable binding site for oxalate in the vicinity of the SH group when the loop is in the open conformation (Figure 6C), this pocket being occupied by water molecules in the apoenzyme. In the open loop conformation, the deep cavity makes a turn and continues from the presumed binding pocket for oxalate, almost to the surface of the protein, which could be the entry point for oxalate to the active site.
The catalytic mechanism
Given their relatively recent identification, many questions remain concerning the catalytic mechanisms used by type III acyl-CoA transferases (Heider, 2001). Steady-state kinetic studies of succinyl-CoA:benzylsuccinate CoA-transferase (Leutwein and Heider, 2001) were not consistent with a ping-pong mechanism, as observed for type I acyl-CoA transferases (Selmer and Buckel, 1999). Kinetic experiments using FRC give similar results to those reported for succinyl-CoA:benzylsuccinate CoA-transferase (S.Jonsson and N.G.J.Richards, unpublished results). One explanation for the differences in the steady-state kinetics of type I and type III acyl-CoA-transferases could be that acyl transfer in the latter family proceeds via a ternary complex in which both substrates are bound within the active site as in family II (Heider, 2001). In this mechanism, the enzyme catalyses nucleophilic attack of the carboxylate on the thioester to yield an anhydride intermediate and free coenzyme A. Addition of coenzyme A at the other end of the anhydride then completes the acyl transfer, and the products are released from the active site. Note the absence of covalent intermediates formed by a reaction of the substrates with active site residues in such a mechanism, the function of the catalyst being primarily to bring the reactants together in the correct orientation.
On the other hand, this proposal for the mechanism of type III acyl-CoA transferases appears inconsistent with the observed structure of the FRC–CoA complex in which the side chain of Asp169 is positioned so as to preclude direct attack of oxalate on formyl-CoA in a ternary complex (Figure 6C). Asp169 is highly conserved in enzymes that have been assigned to be type III CoA-transferases (Figure 5), and its location within the putative active site of FRC suggests that it might play a functional role in catalysis.
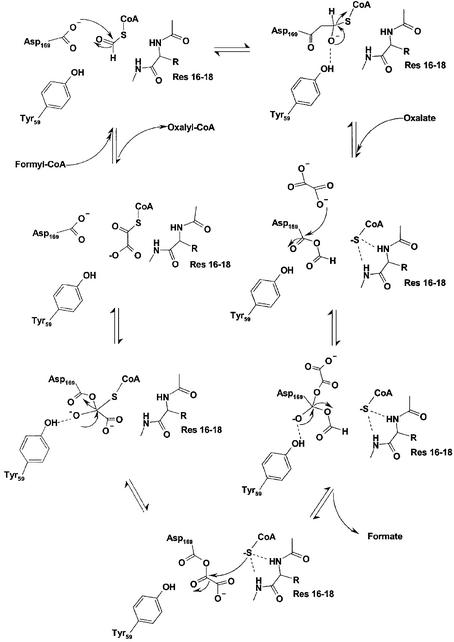
We therefore suggest an alternate hypothesis for the catalytic mechanism of FRC (Figure 7), which is quite analogous to that observed in type I CoA-transferases. Hence, formyl-CoA binds to the active site in the initial step and the substrate thioester reacts with the Asp169 side chain to form a covalent anhydride intermediate and coenzyme A. The negatively charged tetrahedral transition state could be stabilized by interaction with the side chain of the conserved residue Tyr59, while formation of the CoAS– anion is facilitated by the positive dipole at the N-terminus of α-helix 1 (Hol, 1985).
Fig. 7. Reaction mechanism for formyl-CoA transferase as suggested from the crystal structure and available kinetic data.
Oxalate could be bound through interaction with the main-chain amino groups of residues 139–142 at the N-terminus of helix 8 and/or the oxyanion site created by loop residues 259–261, then in the open conformation. The binding of oxalate requires the loop to be in its open conformation; in the absence of oxalate, the loop closes down to protect the enzyme-anhydride intermediate from hydrolysis.
Oxalate then reacts to generate a new enzyme anhydride intermediate and formate. Subsequent attack of bound coenzyme A on the mixed anhydride then yields oxalyl-CoA and regenerates the carboxylate moiety of the aspartate. Again, Tyr59 could stabilize the tetrahedral intermediate that is formed in this transfer reaction. Release of the products then yields the free apoenzyme. Since free formate is not produced until after oxalate is bound, kinetic plots cannot assume the form observed for classical ping-pong mechanisms.
The suggested mechanism will now be tested by site-directed mutagenesis of putative catalytic residues, kinetic studies and experiments aiming to trap catalytic intermediates for structure analysis.
Function of FRC in its physiological environment
The intrinsic instabilities of formyl-CoA and oxalyl-CoA suggest that the proteins involved in the oxalate degradation pathway might interact in order to prevent hydrolysis of the thioesters in the cytoplasm. Oxalyl-CoA decarboxylase could interact with FRC by binding at the cleft (Figure 4B) close to the CoA moiety. The CoA moiety is firmly anchored to FRC by its adenine group, while the ribose, ribose-phosphate and pyrophosphate are solvent accessible. A reasonable conjecture is that CoA is able to swing its pantetheine arm esterified with oxalate directly into the active site of oxalyl-CoA decarboxylase, and that the pantetheine arm with the formyl product then swings back into FRC. Preliminary in vitro experiments using native gel electrophoresis of a mixture of the two proteins and CoA support the formation of a complex between the two proteins.
The oxalate/formate antiporter might bind to FRC at the other end of the deep cavity, opposite to CoA, and transport of formate and oxalate could regulate the opening and closure of loop 258–261. The loosely packed structure at this end of FRC, as evidenced by relatively higher B-factors and many buried water molecules, and the concentration of aromatic and hydrophobic residues at this location is an indication that this could form a binding site for another protein at the membrane. The oxalate/formate antiporter, FRC and oxalyl-CoA decarboxylase should then form a large complex, a hypothesis which has to be tested by fluorescence labelling (for instance using GFP) and/or immunohistochemistry to reveal the localization and possible interaction of the proteins in O.formigenes.
Materials and methods
Expression and purification
FRC was expressed and purified as described previously (Ricagno et al., 2003). Selenomethionine-labelled formyl-CoA transferase was produced according to Doublie (1997) and purified as the native enzyme.
Oxalyl-CoA was prepared by the reaction of thiocresol oxalate (Stolle, 1914) with the sodium salt of coenzyme A following standard procedures (Sly and Stadman, 1963). Pure oxalyl-CoA was obtained by reverse-phase HPLC on a C18 column, using a gradient elution protocol with 10 mM sodium phosphate pH 5.0, and 10 mM sodium phosphate pH 5.0 containing 20% CH3CN. Under these conditions, oxalyl-CoA eluted with a retention time of 5 min. Oxalyl-CoA could then be isolated as a white powder after lyophilization.
Crystallization
FRC has been crystallized as described previously (Ricagno et al., 2003). Selenomethionine-substituted enzyme was crystallized under the same conditions, except at a lower protein concentration (4.75 mg/ml). The crystals grew to almost the same size as native protein in 2 weeks. The crystals revealed the same space group, I4, and unit cell (a = b = 151.4 Å, c = 99.6 Å) as crystals of the native protein. Mass spectrometry showed full incorporation of selenomethionine and that the first methionine in the protein chain was cleaved off (data not shown).
Data collection and structure determination
All data were collected in a nitrogen stream at 110 K. Data for native enzyme and for the CoA complex were collected as described previously (Table I; Ricagno et al., 2003). MAD data collection, to a resolution of 2.8 Å, was carried out on multipole wiggler beam line BW7A at the Deutsches Elektronen Synchrotron (DESY; Hamburg, Germany) equipped with a MAR CCD detector. Data were processed using DENZO/SCALEPACK (Otwinowski and Minor, 1996; Table I).
Structure determination and refinement
Phases were calculated to 3.5 Å from the peak data set using the program SOLVE (Terwilliger and Berendzen, 1999) which located 29 selenium sites out of a possible 36. The sites not found were later located in areas of high B-factors. The sites were then refined by SOLVE with ANALYSE_SOLVE to 2.8 Å. The phases were extended to 2.2 Å using non-crystallographic symmetry averaging and solvent flattening in RESOLVE, which also gave a preliminary chain tracing of 60% of residues. Model building was then performed using program O (Jones et al., 1991). Refinement using CNS (Brunger et al., 1998) consisted of anisotropic scaling, bulk-solvent correction, steps of conjugate gradient minimization and isotropic B-factor refinement against the maximum likelihood target, using amplitudes and the phase probability distribution. Final refinement was performed with REFMAC5 (Murshudov et al., 1997) using the maximum likelihood residual, anisotropic scaling, bulk-solvent correction, and atomic displacement parameter refinement using the ‘translation, libration, screw-rotation’ method with each monomer as a rigid group. Non-crystallographic symmetry averaging was not used during the refinement. To rule out the possibility of wrong chain tracing and to correct misplaced side-chains, a composite omit map (CNS) was calculated in the following way: for every step, 5% of the dimer model was excluded followed by annealing from 4000°C. The CoA model was built in O from difference Fourier electron density maps. The refinement of the model of the CoA complex has been performed in CNS using the same procedure as for the apoenzyme model, except that non-crystallographic symmetry was applied at the end of refinement of CoA and its binding site.
The analysis of the dimer surface was performed using the Protein–Protein interaction server (http://www.biochem.ucl.ac.uk/bsm/PP/server/), and analysis of the enzyme geometry was performed using PROCHECK (Laskowski et al., 1993). The real space correlation was calculated in CNS giving an average correlation coefficient of 0.94. Structure comparisons have been made using TOP with default parameters (Lu, 2000), and cavity and surface calculations with VOIDOO (Kleywegt and Jones, 1994). The pictures were prepared with Visual Molecular Dynamics (Humphrey et al., 1996), BobScript (Kraulis, 1991; Esnouf, 1997), POV-ray (Persistence of Vision Development Team), RASTER3D (Merritt and Bacon, 1997) and Ligplot (Wallace et al., 1995).
Structure factors and coordinates have been deposited in the Protein Data Bank under accession codes 1P5H (FRC) and 1P5R (FRC–CoA complex).
Acknowledgments
Acknowledgements
Drs Doreen Dobritzsch and Guido Pintacuda are acknowledged for helpful discussions and technical support, and Drs Kjell Ericsson and Harmeet Sidhu are acknowledged for help in developing the initial purification procedure. We also gratefully acknowledge the EMBL outstation, DESY (Hamburg), and the European synchotron Research Facility (Grenoble) for provision of synchrotron radiation. This work was supported by a grant to Ylva Lindqvist from the Swedish Research Council–Scientific Council for Natural and Engineering Sciences. We acknowledge partial support for Stefan Jonsson from the National Institutes of Health (DK53556).
References
- Baetz A.L. and Allison,M.J. (1989) Purification and characterization of oxalyl-coenzyme A decarboxylase from Oxalobacter formigenes. J. Bacteriol., 171, 2605–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baetz A.L. and Allison,M.J. (1990) Purification and characterization of formyl-coenzyme A transferase from Oxalobacter formigenes. J. Bacteriol., 172, 3537–3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Buckley P.A., Baz Jackson,J., Schneider,T., White,S.A., Rice,D.W. and Baker,P.J. (2000) Protein-protein recognition, hydride transfer and proton pumping in the transhydrogenase complex. Structure Fold. Des., 8, 809–815. [DOI] [PubMed] [Google Scholar]
- Clark J.H., Fitzgerald,J.F. and Bergstein,J.M. (1985) Nephrolithiasis in childhood inflammatory bowel disease. J. Pediatr. Gastroenterol. Nutr., 4, 829–834. [DOI] [PubMed] [Google Scholar]
- Doublie S. (1997) Preparation of selenomethionyl proteins for phase determination. Methods Enzymol., 276, 523–530. [PubMed] [Google Scholar]
- Elssner T., Engemann,C., Baumgart,K. and Kleber,H.P. (2001) Involvement of coenzyme A esters and two new enzymes, an enoyl-CoA hydratase and a CoA-transferase, in the hydration of crotonobetaine to l-carnitine by Escherichia coli. Biochemistry, 40, 11140–11148. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model., 15, 133–138. [DOI] [PubMed] [Google Scholar]
- Heider J. (2001) A new family of CoA-transferases. FEBS Lett., 509, 345–349. [DOI] [PubMed] [Google Scholar]
- Hol W.G. (1985) The role of the alpha-helix dipole in protein function and structure. Prog. Biophys. Mol. Biol., 45, 149–195. [DOI] [PubMed] [Google Scholar]
- Humphrey W., Dalke,A. and Schulten,K. (1996) VMD—Visual Molecular Dynamics. J. Mol. Graph., 14, 33–38. [DOI] [PubMed] [Google Scholar]
- Hylander E., Jarnum,S., Jensen,H.J. and Thale,M. (1978) Enteric hyperoxaluria: dependence on small intestinal resection, colectomy and steatorrhoea in chronic inflammatory bowel disease. Scand. J. Gastroenterol., 13, 577–588. [DOI] [PubMed] [Google Scholar]
- Jacob U., Mack,M., Clausen,T., Huber,R., Buckel,W. and Messerschmidt,A. (1997) Glutaconate CoA-transferase from Acidaminococcus fermentans: the crystal structure reveals homology with other CoA-transferases. Structure, 5, 415–426. [DOI] [PubMed] [Google Scholar]
- Johansson E., Steffens,J.J., Lindqvist,Y. and Schneider,G. (2000) Crystal structure of saccharopine reductase from Magnaporthe grisea, an enzyme of the α-aminoadipate pathway of lysine biosynthesis. Structure Fold. Des., 8, 1037–1047. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1994) Detection, delineation, measurement and display of cavities in macromolecular structures. Acta Crystallogr. D, 50, 178–185. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Laskowski R.A., Moss,D.S. and Thornton,J.M. (1993) Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol., 231, 1049–1067. [DOI] [PubMed] [Google Scholar]
- Leutwein C. and Heider,J. (2001) Succinyl-CoA:(R)-benzylsuccinate CoA-transferase: an enzyme of the anaerobic toluene catabolic pathway in denitrifying bacteria. J. Bacteriol., 183, 4288–4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu G. (2000) TOP: a new method for protein structure comparison and similarity searches. J. Appl. Crystallogr., 33, 176–183. [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D: Photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin,A.A. and Dodson,E.J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D, 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1996) Processing X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Quayle J.R. (1963) Carbon assimilation by Pseudomonas oxalaticus (OX1). Reactions of oxalyl-coenzyme A. Biochem. J., 87, 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricagno S., Jonsson,S., Richards,N. and Lindqvist,Y. (2003) Crystallisation and preliminary crystallographic analysis of formyl-CoA transferase from Oxalobacter formigenes. Acta Crystallogr. D, in press. [DOI] [PubMed] [Google Scholar]
- Rodby R.A., Tyszka,T.S. and Williams,J.W. (1991) Reversal of cardiac dysfunction secondary to type 1 primary hyperoxaluria after combined liver–kidney transplantation. Am. J. Med., 90, 498–504. [PubMed] [Google Scholar]
- Rousseau F., Schymkowitz,J.W.H. and Itzhaki,L.S. (2003) The unfolding story of three-dimensional domain swapping. Structure, 11, 243–251. [DOI] [PubMed] [Google Scholar]
- Ruan Z.S., Anantharam,V., Crawford,I.T., Ambudkar,S.V., Rhee,S.Y., Allison,M.J. and Maloney,P.C. (1992) Identification, purification and reconstitution of OxlT, the oxalate: formate antiport protein of Oxalobacter formigenes. J. Biol. Chem., 267, 10537–10543. [PubMed] [Google Scholar]
- Selmer T. and Buckel,W. (1999) Oxygen exchange between acetate and the catalytic glutamate residue in glutaconate CoA-transferase from Acidaminococcus fermentans. Implications for the mechanism of CoA-ester hydrolysis. J. Biol. Chem., 274, 20772–20778. [DOI] [PubMed] [Google Scholar]
- Sidhu H., Ogden,S.D., Lung,H.Y., Luttge,B.G., Baetz,A.L. and Peck,A.B. (1997) DNA sequencing and expression of the formyl coenzyme A transferase gene, frc, from Oxalobacter formigenes. J. Bacteriol., 179, 3378–3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidhu H., Hoppe,B., Hesse,A., Tenbrock,K., Brömme,S., Rietschel,E. and Peck,A.B. (1998) Absence of Oxalobacter formigenes in cystic fibrosis patients: a risk factor for hyperoxaluria. Lancet, 352, 1026–1029. [DOI] [PubMed] [Google Scholar]
- Sidhu H., Schmidt,M.E., Cornelius,J.G., Thamilselvan,S., Khan,S.R., Hesse,A. and Peck,A.B. (1999) Direct correlation between hyperoxaluria/oxalate stone disease and the absence of the gastrointestinal tract-dwelling bacterium Oxalobacter formigenes: possible prevention by gut recolonization or enzyme replacement therapy. J. Am. Soc. Nephrol., 10, S334–S340. [PubMed] [Google Scholar]
- Sly W.S. and Stadman,E.R. (1963) Formate metabolism. 1. formyl coenzyme A, an intermediate in formate-dependent decomposition of acetyl phosphate in Clostridium kluyveri. J. Biol. Chem., 238, 2632–2638. [PubMed] [Google Scholar]
- Stolle R. (1914) Uber methyl-thionaphthenchinon. Ber. Dtsch. Chem. Ges., 47, 1130–1132. [Google Scholar]
- Terwilliger T.C. and Berendzen,J. (1999) Automated MAD and MIR structure solution. Acta Crystallogr. D, 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace A.C., Laskowski,R.A. and Thornton,J.M. (1995). LIGPLOT: A program to generate schematic diagrams of protein–ligand interactions. Protein Eng., 8, 127–134. [DOI] [PubMed] [Google Scholar]
- Williams H.E. and Smith,L.H.,Jr. (1968) Disorders of oxalate metabolism. Am. J. Med., 45, 715–735. [DOI] [PubMed] [Google Scholar]
- Wolodko W.T., Fraser,M.E., James,M.N. and Bridger,W.A. (1994) The crystal structure of succinyl-CoA synthetase from Escherichia coli at 2.5-Å resolution. J. Biol. Chem., 269, 10883–10890. [DOI] [PubMed] [Google Scholar]