Abstract
The AXR6 gene is required for auxin signaling in the Arabidopsis embryo and during postembryonic development. One of the effects of auxin is to stimulate degradation of the Aux/IAA auxin response proteins through the action of the ubiquitin protein ligase SCFTIR1. Here we show that AXR6 encodes the SCF subunit CUL1. The axr6 mutations affect the ability of mutant CUL1 to assemble into stable SCF complexes resulting in reduced degradation of the SCFTIR1 substrate AXR2/IAA7. In addition, we show that CUL1 is required for lateral organ initiation in the shoot apical meristem and the inflorescence meristem. These results indicate that the embryonic axr6 phenotype is related to a defect in SCF function and accumulation of Aux/IAA proteins such as BDL/IAA12. In addition, we show that CUL1 has a role in auxin response throughout the life cycle of the plant.
Keywords: Arabidopsis/auxin/cullin/IAA/SCF
Introduction
The plant hormone auxin regulates diverse aspects of development throughout the plant life cycle (Davies, 1995). In Arabidopsis seedlings, auxin response depends on the ubiquitin protein ligase SCFTIR1 (Ruegger et al., 1998; Gray et al., 1999, 2001). SCF E3s consist of four subunits: an F-box protein (FBP) responsible for substrate binding, SKP1 (ASK in Arabidopsis), the RING protein RBX1/ROC1/HRT1 and CUL1. Biochemical and structural studies show that the CUL1 subunit functions as a scaffold, binding RBX1 and a SKP1–FBP dimer (Zheng et al., 2002). RBX1 also binds the ubiquitin-conjugating enzyme (E2) thus bringing E2 in close proximity to the substrate. There are over 700 FBPs encoded by the Arabidopsis genome suggesting that SCFs have a prominent role in cellular regulation in plants (Gagne et al., 2002; Kuroda et al., 2002).
SCF function requires modification of CUL1 by the ubiquitin-related protein RUB (Nedd8 in some species) (Yeh et al., 2000; Hellmann and Estelle, 2002). In Arabidopsis, RUB modification requires a heterodimeric E1 enzyme composed of AXR1 and ECR1, and a RUB1-conjugating enzyme called RCE1 (del Pozo et al., 1998, 2002). RUB is conjugated to a conserved lysine residue near the C-terminal region of CUL1 (del Pozo et al., 1998, 2002). Although the function of the modification is unknown, recent genetic results suggest that dynamic cycling of CUL1 between the unmodified and modified forms is required for normal SCF activity (Lyapina et al., 2001; Schwechheimer et al., 2001; Gray et al., 2002).
The only known substrates of SCFTIR1 are the Aux/IAA proteins (Gray et al., 2001). These short lived, nuclear proteins consist of four conserved domains. Domains III and IV are required for dimerization between Aux/IAA proteins and with members of a second family of transcriptional activators called the auxin response factors (ARFs) (Reed, 2001). The Aux/IAA proteins repress ARF function through this interaction (Reed, 2001). In Arabidopsis seedlings, auxin relieves this repression by promoting binding of the Aux/IAA proteins by SCFTIR1, resulting in their ubiquitination and degradation (Gray et al., 2001; Tiwari et al., 2001; Zenser et al., 2001). Domain II of the Aux/IAA protein is necessary and sufficient for interaction with the SCF (Ramos et al., 2001). Dominant mutations within domain II prevent the interaction and stabilize the affected protein resulting in a reduction in auxin response (Hellmann and Estelle, 2002).
Studies of the bodenlos (bdl), monopteros (mp) and hobbit (hbt) mutants suggest that auxin response in the embryo requires regulated protein degradation (Berleth and Jurgens, 1993; Hardtke and Berleth, 1998; Hamann et al., 1999, 2002). The mp and bdl mutants have a similar phenotype characterized by cell division defects as early as the two-cell stage of embryogenesis. Mutant embryos complete embryogenesis but lack basal embryonic structures including the hypocotyl and root meristem. The hbt mutants display early cell division defects that are similar to those of mp and bdl (Willemsen et al., 1998). However, hbt seedlings are relatively normal except for the absence of the root meristem (Willemsen et al., 1998). The MP gene encodes ARF5 while BDL encodes IAA12 (Hamann et al., 2002). The mp phenotype is caused by loss of ARF5 while bdl is a gain-of-function mutation in domain II similar to other mutations known to prevent interaction with SCFTIR1. Based on these results Hamann et al. (2002) have proposed that BDL/IAA12 represses MP/ARF5 during embryogenesis. The HBT gene encodes a subunit of the anaphase promoting complex (APC), another class of ubiquitin protein ligase (Blilou et al., 2002). The Aux/IAA protein IAA17 is stabilized in hbt mutants suggesting that Aux/IAA proteins are substrates of APC in the embryo. Thus it is possible that APC or SCF (or both) are responsible for degradation of BDL/IAA12 and other Aux/IAA proteins during embryogenesis.
The phenotype of the auxin-resistant mutant axr6 closely resembles that of mp and bdl (Hobbie et al., 2000). Here we show that AXR6 encodes the SCF subunit CUL1. The mutations result in stabilization of the AXR2/IAA7 protein suggesting that the axr6 embryonic phenotype is caused by accumulation of Aux/IAA proteins, probably including BDL/IAA12. Furthermore, a reduction in CUL1 levels in transgenic lines leads to defects in organ initiation similar to those observed in mutants deficient in auxin transport (Okada et al., 1991). These results suggest that SCF-mediated degradation of the Aux/IAA proteins is important for early events in embryogenesis and diverse growth process throughout plant development.
Results
AXR6 gene encodes SCF subunit CUL1
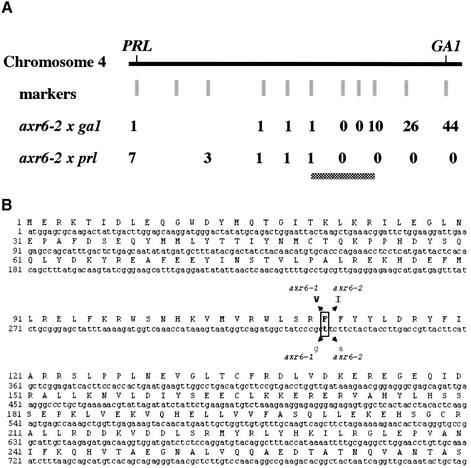
In a previous report, we showed that the AXR6 gene is required for auxin response throughout plant growth and development (Hobbie et al., 2000). To further explore the function of AXR6 we isolated the gene using a map-based approach. Earlier mapping studies had shown that AXR6 is located on chromosome 4 near PRL. Additional recombinants were generated by crossing axr6-2 (Col-0) to ga1 and prl (Ler) (Figure 1A). The AXR6 gene was localized to an ∼80 kilobase (kb) interval on chromosome 4. Among the genes in this region is CUL1. Since SCFTIR1 is required for auxin response, we sequenced CUL1 from axr6-1 and axr6-2 mutants. Both alleles carry a base substitution at the same position in the coding region of CUL1. The mutations result in replacement of phenylalanine 111 with valine (axr6-1) or isoleucine (axr6-2) (Figure 1B). Based on the crystal structure of human SCFSKP2, Phe111 is within a region of CUL1 that interacts with the SKP1–FBP module (Zheng et al., 2002).
Fig. 1. Mapping of the AXR6 gene. (A) AXR6 was mapped using crosses between axr6-2 (Col-0 background) and two mutants in the Ler background, ga1 and prl. The number of recombinants at each position for each cross are indicated. The mutation was localized to a region of ∼80 kb (indicated by the gray horizontal bar) on the short arm of chromosome 4 containing CUL1. (B) Sequencing of CUL1 in two independent alleles, axr6-1 and axr6-2, showed mutations in the same base at position 336. The mutations result in substitution of phenylalanine with valine and isoleucine in axr6-1 and axr6-2, respectively.
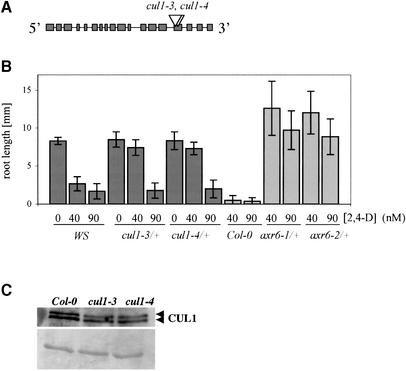
Shen et al. (2002) used two CUL1 T-DNA insertion mutants (cul1-1 and cul1-2) to show that loss of CUL1 results in lethality at the zygote stage of embryogenesis. We isolated two new T-DNA insertion lines from the Wisconsin collection. The new alleles, called cul1-3 and cul1-4, each have a T-DNA insert in exon 15 of the CUL1 gene (Figure 2A). Like cul1-1 and cul1-2, the new alleles are recessive embryo lethal mutations (data not shown). Heterozygous plants are normal in appearance but display a mild auxin-resistant phenotype (Figure 2B). This defect is associated with reduced levels of CUL1 (Figure 2C). Additional cross-reacting proteins were not observed in cul1-3 or cul1-4 plants indicating that auxin resistance was not related to accumulation of a truncated version of CUL1 (data not shown). This result indicates that auxin response is unusually sensitive to CUL1 levels. However, it is also important to note that cul1-3/+ and cul1-4/+ are much less auxin resistant than axr6/+ plants (Figure 2B).
Fig. 2. CUL1 T-DNA insertion mutants are auxin resistant. (A) Two independent T-DNA insertion lines, cul1-3 and cul1-4 [Wasselewskja (WS) background], were identified carrying insertions in exon 15 of the gene. (B) Response of mutant and wild-type seedlings to auxin. At least 15 seedlings were tested for each genotype. Error bars represent standard deviation. (C) Heterozygous cul1-3/+ and cul1-4/+ plants have reduced CUL1 levels. Ten-day-old seedlings were used. The loading control (lower panel) shows an unidentified protein visualized by Poinceau staining.
To confirm that the axr6 mutations are in the CUL1 gene, heterozygous axr6-1 and axr6-2 plants were crossed to heterozygous cul1-4 plants. In each case ∼25% of the progeny seed were not viable, showing that axr6 does not complement the embryo-lethal phenotype of cul1-4 (Table I). Of the viable seedlings, the expected 33% displayed the axr6/+ auxin resistant phenotype (data not shown).
Table I. Analysis of progeny from cross of axr6 × cul1-4.
| axr6-1/+ | axr6-2/+ | cul1-4/+ | Col-0 | axr6-1/+ × cul1-4/+ | axr6-2 × cul1-4/+ | |
|---|---|---|---|---|---|---|
| Viable seed | 100 | 100 | 228 | 100 | 95 | 221 |
| Inviable seed | 0 | 0 | 72 | 0 | 31 | 80 |
| Total | 100 | 100 | 300 | 100 | 126 | 301 |
Degradation of the AXR2/IAA7 protein is impaired in axr6 mutants
Heterozygous axr6-1/+ plants are deficient in auxin induction of the BA3–GUS reporter gene indicating that AXR6/CUL1 is required for this auxin response (Hobbie et al., 2000). We also examined transcript levels of the IAA5 and IAA7 primary auxin response genes. We found that accumulation of both in response to auxin is reduced in the mutants compared with wild type (Figure 3A). In the case of IAA7, RNA levels were also reduced in the absence of exogenous auxin. These results confirm that AXR6/CUL1 is required for auxin regulation of gene expression. We also show that the CUL1 RNA level does not change in response to auxin and is not affected in the axr6 mutants (Figure 3A).
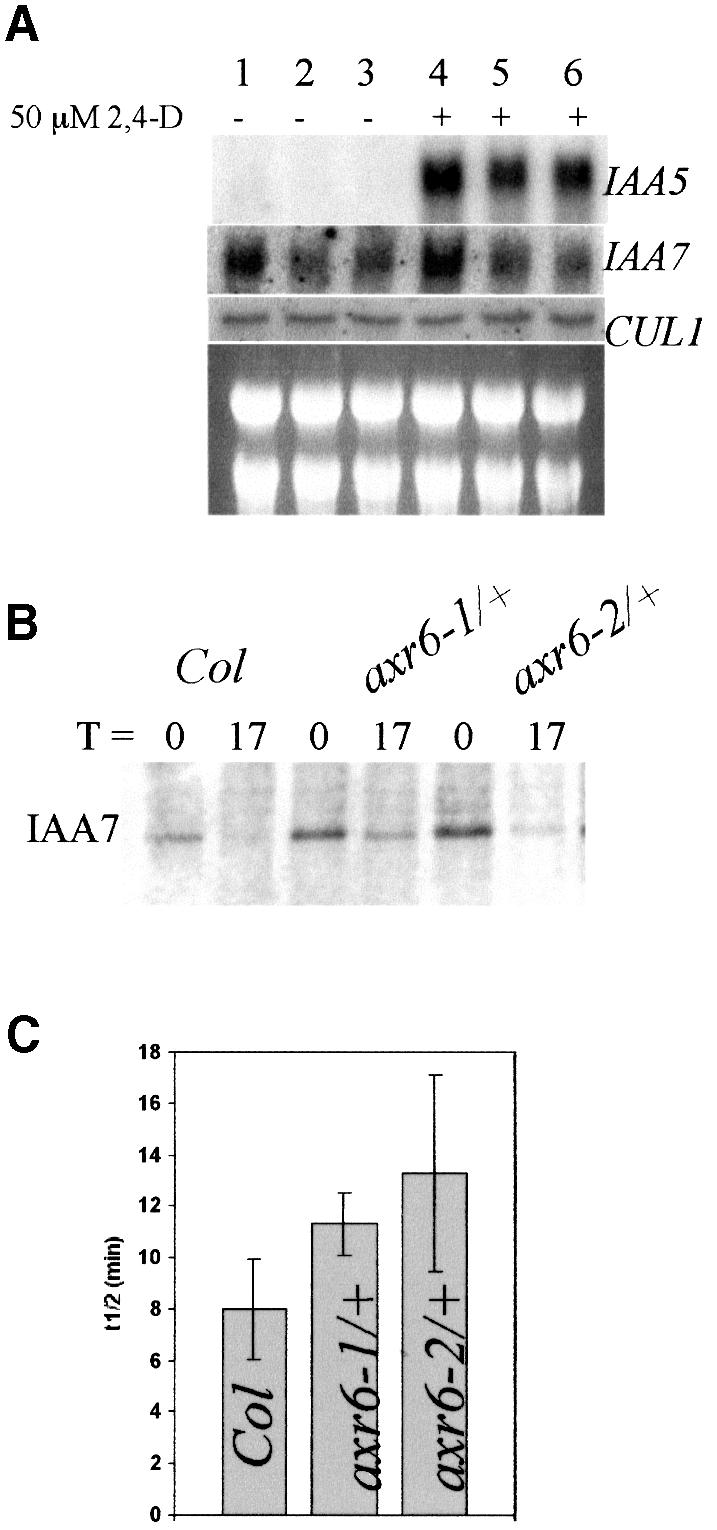
Fig. 3. The axr6 mutants are deficient in auxin-induced gene expression and degradation of AXR2/IAA7. (A) RNA blot analysis shows that auxin-regulated expression of IAA5 and IAA7 is reduced in axr6 mutants. Seven-day-old seedlings were treated with buffer or 50 µM 2,4-D for 1 h before RNA extraction. CUL1 expression is not regulated by auxin and is not altered in axr6-1 or axr6-2 plants. Col-0 (lanes 1 and 4), axr6-1/+ (lanes 2 and 4), axr6-2/+ (lanes 3 and 6). (B) Pulse–chase analysis in 7-day-old seedlings showed that stability of the IAA7 protein is increased in axr6-1 and axr6-2. (C) The half-life of IAA7 is higher in the axr6 mutants. Values presented are the averages of three independent experiments ± SD. (P <0.05 for each mutant compared with wild type; Student’s t-test.)
Since SCF-dependent degradation of Aux/IAA proteins is required for auxin response in seedlings, we speculated that this process might be impaired in the axr6 mutants (Gray et al., 2001). To test this possibility, the half-life of IAA7 was determined in wild-type and heterozygous axr6 seedlings. The results show that IAA7 is more stable in seedlings heterozygous for either axr6-1or axr6-2 compared with wild-type seedlings (Figure 3B and C). This difference is statistically significant (P <0.05 for each mutant compared with wild type; Student’s t-test).
CUL1 levels are increased in axr6 plants
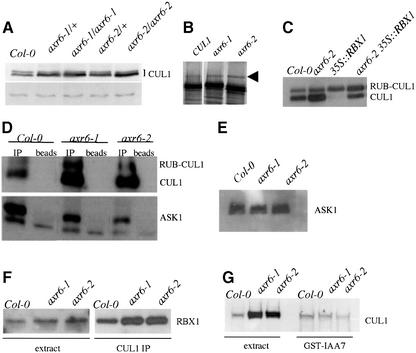
To explore the effects of the axr6 mutations on CUL1 function, we examined CUL1 in mutant plants by immunoblotting. Each mutation causes an increase in CUL1 protein levels (Figure 4A). Since CUL1 RNA level is not affected by the mutations (Figure 3A), this difference must reflect a post-transcriptional change. A fraction of the CUL1 protein is modified by the ubiquitin-related protein RUB1 (del Pozo et al., 2002). Despite the increase in total CUL1 levels in the axr6 mutants, the amount of RUB-modified CUL1 is similar to wild type. The relative level of RUB-CUL1 may be decreased because the mutant proteins are poor substrates for RUB modification. To test this possibility a rabbit reticulocyte lysate was used to perform in vitro RUB modification of mutant and wild-type CUL1. The results in Figure 4B show that the mutant proteins are effective substrates for RUB modification in this system. However, it is still possible that in the context of the plant cell, RUB modification of CUL1 is impaired. Alternatively, the mutant proteins may be subject to increased RUB de-conjugation, perhaps catalyzed by the COP9 signalosome (CSN) (Schwechheimer et al., 2001).
Fig. 4. The axr6-1 and axr6-2 mutations alter CUL1 levels and formation of stable SCF complexes. (A) CUL1 levels in wild-type and mutant seedlings determined by western blot with antibody against CUL1. The lower panel shows an unidentified cross-reacting protein used as a loading control. (B) In vitro translation and GST–RUB1 modification of CUL1, AXR6-1 and AXR6-2 proteins in rabbit reticulocyte lysates. Arrow indicates GST–RUB1 modified cullin. (C) Effect of RBX1 over-expression on CUL1 levels in axr6 plants determined by western blot. (D) Co-immunoprecipitation of CUL1 (upper panel) and ASK1 (lower panel) from Col-0 and homozygous axr6-1 or axr6-2 seedlings. Proteins were immunoprecipitated with CUL1 antibody and analyzed by western blot. The ASK1 antiserum detected an unknown protein that was recovered after incubation with beads alone. (E) ASK1 levels in Col-0 and homozygous axr6-1 or axr6-2 extracts determined by western blot using α-ASK1 antibody. (F) Co-immunoprecipitation of CUL1 and RBX1 in Col-0 and mutant seedlings. (G) CUL1 levels in GST-IAA7 pulldowns from Col-0 and homozygous axr6 seedlings determined by western blot.
Genetic studies indicate that decreased RUB-CUL1 levels result in reduced SCF function (del Pozo et al., 2002). Thus the axr6 phenotype may be caused by a decrease in the relative levels of RUB-CUL1. To test this model, we used a line that carries a 35S::RBX1 transgene. Over-expression of RBX1 results in increased levels of RUB-CUL1 (Gray et al., 2002). Surprisingly, the effects of this change are similar to those of decreased RUB-CUL1, including stabilization of Aux/IAA proteins. To examine the effects of increased RBX1 levels in an axr6 background, we crossed axr6-2 to the 35S:RBX1 line (Gray et al., 2002). F2 plants that were homozygous for the axr6-2 mutation and carrying the 35S:RBX1 transgene were recovered and analyzed by western blot (Figure 4C). These plants had increased levels of RUB-modified CUL1. However, no effect on the morphology of homozygous axr6-2 seedlings was observed, suggesting that decreased RUB-CUL1 levels do not cause this phenotype (data not shown).
axr6 mutations affect the interaction between CUL1 and ASK1
Based on the structure of mammalian SCFSKP2, phenyl alanine 111 of CUL1 lies adjacent to residues that contribute directly to SKP1-FBP binding (Zheng et al., 2002). To determine if the mutations affect the interaction between CUL1 and ASK1, we tested for co-immunoprecipitation of the two proteins (Figure 4D). As expected, more CUL1 was recovered from mutant extracts than wild-type extracts. However, we observed less ASK1 in the CUL1 immunoprecipitate compared with wild type despite the presence of similar levels of ASK1 in the extract (Figure 4D and E). We conclude that the mutations affect the ability of CUL1 to form a stable complex with ASK1.
RBX1 binds cullin near its C-terminus (Furukawa et al., 2000; Zheng et al., 2002). We used an RBX1 antibody to examine RBX1 levels in plant extracts and CUL1 immunoprecipitates. Surprisingly, RBX1 levels are higher in axr6 seedlings than in wild type (Figure 4F). Consistent with this difference, and the higher level of CUL1 in these extracts, we found increased levels of RBX1 in the CUL1 immunoprecipitate from mutant plants. Thus the mutations do not affect the interaction between CUL1 and RBX1. However, RBX1 appears to be more stable in mutant plants.
The IAA7 protein is a substrate for SCFTIR1 and interacts with the complex in plant extracts prepared from seedlings. As a further test of the effects of the axr6 mutations on CUL1 function, we performed a GST–IAA7 pulldown experiment from wild-type and axr6 extracts. Figure 4G shows that less CUL1 is pulled down from axr6 extracts than from wild-type extracts, despite an increase in CUL1 levels. Since IAA7 interacts with CUL1 indirectly through the ASK-TIR1 module, these results indicate that less CUL1 is assembled into SCFTIR1 complexes in axr6 plants.
Reduction in CUL1 levels results in severe shoot meristem defects
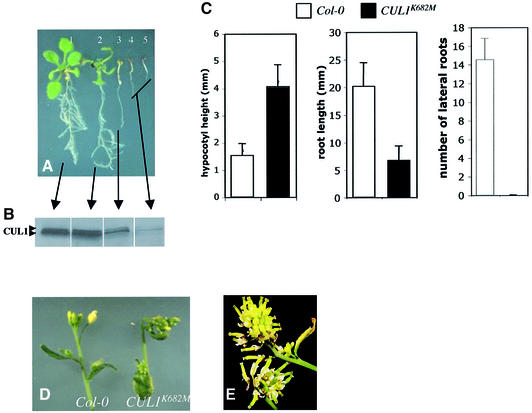
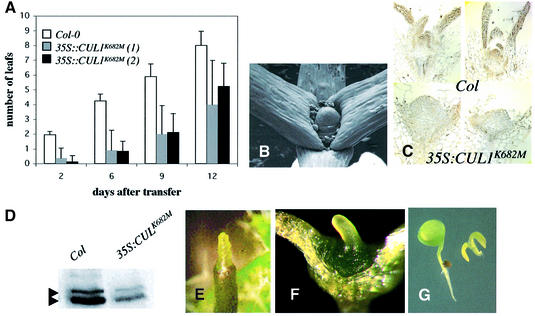
To learn more about the function of CUL1 throughout plant development, we attempted to overexpress the protein in Col-0 plants. Two 35S::CUL1 constructs were used containing either the wild-type cDNA or a mutant form in which lysine 682, the site of RUB attachment, is changed to methionine. Over 100 transformants were generated with each construct. No lines with increased expression of either gene were recovered suggesting that CUL1 overexpression is severely detrimental to the plant. However, 25–30% of the lines exhibited defects in development (Table II). Figure 5A illustrates the range of phenotypes observed in T1 plants. Most seedlings exhibited a severe phenotype similar to seedlings 3 through 5 in Figure 5A. The severity of the phenotype correlated with the level of CUL1 protein, suggesting that the phenotype is caused by co-suppression of CUL1 expression (Figure 5B). A variety of defects were observed. The hypocotyls of affected T1 seedlings were two to three times longer than Col-0 seedlings (Figure 5C), but had the same number of cells (data not shown), indicating an increase in cell size. Root growth was reduced and no lateral roots were observed in these seedlings (Figure 5C). Subsequent growth was slow and most seedlings died before they developed leaves. The occasional survivor produced a compact inflorescence (Figure 5D and E). These plants produced seed but only a small fraction of the T2 population (typically <2%) displayed the severe co-suppressor phenotype. These lines were not analyzed further.
Table II. Recovery of pin phenotype after transformation of Col-0 and tir1-1 plants with CUL1 and CULK682M constructs.
| Construct/genotype | No. of transformants | No. of co-suppressor [n (%)] | Plants with pin inflorescence [n (%)] |
|---|---|---|---|
| CUL1/Col-0 | 105 | 26 (24.8) | 2 (1.9) |
| CUL1K682M/Col-0 | 117 | 39 (33) | 2 (1.7) |
| CUL1/tir1-1 | 129 | 30 (23) | 11 (8.5) |
| CUL1K682M/tir1-1 | 135 | 61 (45) | 15 (11.1) |
Fig. 5. Reduced CUL1 levels lead to severe developmental defects. Arabidopsis Col-0 plants were transformed with a CUL1 or CUL1K682M cDNA under the control of the 35S promoter from cauliflower mosaic virus. (A) Approximately 25% of the transgenic lines displayed growth defects of varying severity. The plant labeled ‘1’ is untransformed Col-0. The others are transgenic for 35S::CUL1K682M. (B) CUL1 levels in transgenic lines from (A) determined by western blot; 30 µg total protein were loaded in each lane. (C) Three-day-old kanamycin-resistant 35S::CUL1K682M seedlings with elongated hypocotyls were transferred to minimal medium lacking antibiotics and compared with Col-0 seedlings 12 days after transfer. Transgenic seedlings also displayed reduced root elongation and almost no lateral roots. (D) Surviving plants developed strongly distorted and compact inflorescences. (E) Compact inflorescence from a mature 35S::CUL1K682M plant. The frequency and severity of defects observed with the 35S::CUL1 construct were similar to those shown here.
In addition to the severely affected T1 plants, ∼2% of the transformed lines displayed a less severe but completely heritable phenotype (Table II). Each of the lines had a similar phenotype regardless of the transgene. Leaf initiation was slower and irregular compared with Col-0 plants (Figure 6A). After producing a few leaves, the shoot apical meristem (SAM) ceased organogenesis and formed a short pin-shaped structure similar to the pin meristem observed in the mp, pinformed1 (pin1) and pinoid (pid) mutants (Figure 6B) (Okada et al., 1991; Bennett et al., 1995; Przemeck et al., 1996). In most seedlings, the SAM did not develop further. To determine if these defects were associated with changes in meristem structure we prepared paraffin sections from 9-day-old Col-0 and 35S:CUL1K682M seedlings and immunolocalized CUL1 (Figure 6C). The SAM of 35S:CUL1K682M seedlings was enlarged compared with Col-0. As expected, the levels of CUL1 were reduced in the transgenic lines compared with Col-0 (Figure 6C). This was confirmed by western blotting (Figure 6D). After SAM arrest, the axillary buds developed, resulting in the formation of a ‘normal looking’ rosette. When these plants flowered, many of the inflorescences also formed a pin-like structure (Figure 6E). These results show that CUL1 is required for normal organ initiation from the SAM and inflorescence meristem
Fig. 6. CUL1-deficient plants display defects in meristem growth and lateral organ initiation. (A) Stable CUL1-deficient plants exhibit slow and irregular leaf development. Leaf number was counted at intervals in Col-0 and two independent 35S::CUL1K682M lines. (B) Scanning electron micrograph of the SAM from a 35S::CUL1 seedling showing the pin phenotype. (C) Immunolocalization of CUL1 in Col-0 and 35S::CUL1K682M 9-day-old seedlings. Images from two different seedlings are shown for each genotype. The brown staining represents CUL1 protein. (D) Protein blot showing reduced CUL1 levels at the apex of 21-day-old plants. Thirty micrograms of total protein extract were loaded in each lane. (E) Pin-like meristem from inflorescences of 35S::CUL1K682 plants. (F) Homozygous axr1-13 carrying the 35S::CUL1K682 transgene generated by crossing a stable CUL1-deficient line(35S::CUL1K682) to axr1-13. All homozygous axr1-13 seedlings with at least one transgene had this phenotype. (G) Example of a T1 axr1-3 plant transformed with the 35S:CUL1 construct. Approximately 10% of the T1 seedlings had this phenotype.
Pin formation is enhanced by mutations in AXR1 or TIR1
Modification of CUL1 by the ubiquitin-related protein RUB is important for auxin response (Hellmann and Estelle, 2002). To determine if this is the case during organ initiation, we crossed the 35S:CUL1K682M line to the axr1-13 mutant, known to be deficient in RUB-CUL1 conjugation (del Pozo et al., 2002). All of the F1 plants displayed the pin phenotype. Strikingly, F2 seedlings that were homozygous for axr1-13 and either hetero- or homozygous for 35S:CUL1 developed a pin-shaped meristem immediately after germination and did not develop further (Figure 6F). Direct transformation of axr1-3 with 35S:CUL1 also produced dramatically affected seedlings. In this case, ∼10% of the transformants resemble homozygous axr6 or mp plants with severe defects in basal structures (Figure 6G). These results indicate that RUB modification of CUL1 is essential for organ initiation.
A reduction in auxin response is a striking feature of the axr1 phenotype. However, the RUB conjugation pathway is probably required for normal function of many different SCFs. To determine if the defects in organ initiation observed in the 35S:CUL1 lines are related to auxin response and SCFTIR1 in particular, we introduced the 35S:CUL1 transgenes into tir1 plants. The frequency of transformants with a pin inflorescence in the tir1 background was ∼5-fold higher than in wild type (Table II). Since TIR1 is required for the auxin-dependent degradation of the Aux/IAA proteins, the defect in organ initiation is probably related to stabilization of these proteins.
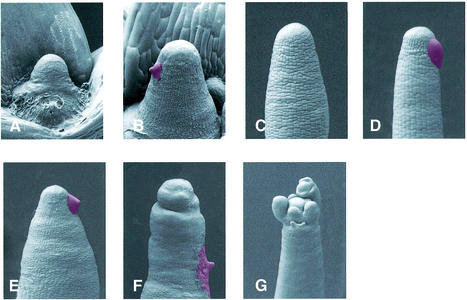
35S::CUL1 pin meristems are auxin insensitive
The failure to initiate lateral organs in pin1 is related to a defect in auxin transport. Auxin application to the flank of the pin1 meristem results in normal organ initiation suggesting that localized auxin accumulation is required for organ initiation (Reinhardt et al., 2000). To see if CUL1 deficient plants respond to auxin similarly, we applied IAA to the meristems of 35S::CUL1 plants. A total of 59 SAMs were examined. Of 32 pin SAMS that were not treated, nine recovered and formed new leaves. The other 23 SAMs terminated without further organogenesis. IAA was applied to the flanks of 27 defective SAMs. Twenty terminated without formation of additional leaves (Figure 7A and B). Seven SAMs recovered, but of these only two initiated an organ at the site of IAA application. We conclude from these results that pin SAMs on CUL1-deficient plants are insensitive to auxin. Similarly, application of IAA to the flanks of pin inflorescence meristems on 35S::CUL1 plants did not induce organ formation (n = 25) (Figure 7C–F). Occasionally a meristem resumed flower formation after prolonged growth (Figure 7G). These results indicate that the pin phenotype is related to a defect in auxin response rather than transport. This is consistent with a model in which accumulation of Aux/IAA proteins prevents organ formation in 35S::CUL1 lines.
Fig. 7. Response of CUL1-deficient pins to exogenous auxin. (A) Two-week-old 35S::CUL1K682 seedling with two cotyledons and two true leaves (one removed) and a central pin. (B) Arrested SAM 3 days after application of 1 mM IAA in lanolin paste (red). (C) Inflorescence pin on CUL1-deficient plant. (D) Inflorescence pin 3 days after local treatment with 1 mM IAA (red). (E) Inflorescences pin 3 days after local treatment with 1 mM IAA (red). (F) Floral pin 7 days after local treatment with 1 mM IAA (red). (G) Spontaneous flower formation on an inflorescence pin after prolonged growth.
Discussion
Recent advances in our understanding of auxin action have demonstrated that auxin regulation of transcription involves two families of proteins (Hagen and Guilfoyle, 2002; Liscum and Reed, 2002). The ARF transcription factors bind AuxRE promoter elements and either activate or repress transcription depending on the ARF. The short-lived Aux/IAA proteins negatively regulate ARFs, probably through a direct interaction (Hagen and Guilfoyle, 2002; Liscum and Reed, 2002). In Arabidopsis seedlings, auxin promotes degradation of Aux/IAA proteins by stimulating an interaction between the Aux/IAA proteins and the ubiquitin protein ligase SCFTIR1 (Gray et al., 2001; Ramos et al., 2001; Tiwari et al., 2001). In this study we show that decreased AXR6/CUL1 function results in auxin-related defects during embryogenesis and throughout plant development. These results indicate that auxin and SCF-dependent degradation of Aux/IAA proteins is a general aspect of auxin response.
CUL1 is required throughout the plant life cycle
Animal cullins function as subunits in several classes of ubiquitin protein ligases (Deshaies, 1999; Winston et al., 1999; Pickart, 2001). There are six known cullin proteins in humans (Winston et al., 1999). Cul1 interacts with SKP1–FBP to form SCFs while Cul2 and Cul5 assemble into a second large group of E3s all of which also contain the Elongin BC complex. Cul3 interacts with RBX1 (also called Roc1) to form an E3 that functions to regulate the level of cyclin E and possibly other proteins (Singer et al., 1999; Winston et al., 1999). In Arabidopsis there are at least five genes encoding canonical cullins called CUL1, CUL2, CUL3A, CUL3B and CUL4 (Shen et al., 2002). Because of sequence divergence, orthologous relationships between the plant and human cullins are difficult to discern. CUL1 is expressed throughout the life cycle of the plant from the zygote to the pollen (Farras et al., 2001; del Pozo et al., 2002; Shen et al., 2002). However, because null mutations in CUL1 result in embryonic arrest at the zygote stage, the function of CUL1 later in development has been impossible to evaluate (Shen et al., 2002). The phenotype of the axr6 mutants, and the CUL1-deficient lines described here, indicate that CUL1 is required for auxin-dependent patterning in the embryo and auxin response throughout the life of the plant. It is also important to note that CUL1 is very likely to be part of many different SCF complexes with diverse physiological roles (Xu et al., 2002).
axr6 mutations affect SCF complex formation
The axr6 mutations affect the same residue near the N-terminus of CUL1, a region known to interact with the SKP1–FBP module (Zheng et al., 2002). Co-immunoprecipitation experiments indicate that the mutations alter the interaction between the Arabidoipsis SKP1 ortholog, ASK1, and CUL1, but not between RBX1 and CUL1. Consistent with these findings, the results of GST–IAA7 pulldown experiments indicate that less SCFTIR1 is present in axr6 extracts.
The mutations also stabilize CUL1 and decrease the relative levels of RUB-modified CUL1. When the level of RUB-CUL1 is increased by introducing the 35S::RBX1 transgene, CUL1 continues to accumulate to higher levels than the wild-type control suggesting that increased stability of cullin is not directly related to decreased RUB modification. Rather, we suspect that increased CUL1 levels are related to reduced incorporation of CUL1 into active SCF complexes. Some cullin degradation may be a normal consequence of SCF activity. If this is the case, CUL1 that is not assembled into an active SCF may be more stable. This hypothesis may also explain why RBX1 is more abundant in axr6 plants. A similar effect on CUL1 levels is observed in plants that are severely deficient in the RUB/Nedd8 conjugation pathway (Dharmasiri et al., 2003). Seedlings that are deficient for the RUB-activating enzyme AXR1-ECR1, and the RUB-conjugating enzyme RCE1, accumulate high levels of CUL1 (Dharmasiri et al., 2003). In this case, the defect is related to SCF function rather than assembly. It is not clear why axr6 mutants also have decreased relative RUB-CUL1 levels. One possibility is that CUL1 modification requires ASK1–FBP binding. This would also explain why increased levels of RBX1 in the axr6 mutants are not associated with increased RUB-CUL1 formation as in the wild type (Figure 4C and F). Alternatively, the mutations cause increased removal of RUB, perhaps by the CSN (Schwechheimer et al., 2001).
The axr6 mutations are homozygous seedling lethals and result in auxin resistance in heterozygous plants. Because triploid plants with the genotype axr6-2/AXR6/AXR6 are auxin resistant, Hobbie et al. (2000) proposed that the axr6-2 allele has a gain-of-function quality. Our results confirm this hypothesis. Heterozygous cul1-3 and cul1-4 plants are slightly auxin resistant, but display no other defects, axr6-1/+ and axr6-2/+ have a higher level of auxin resistance and a pronounced post-embryonic phenotype (Figure 2) (Hobbie et al., 2000). It seems likely that this genetic behavior is related to accumulation of defective CUL1. Mutant CUL1 interacts normally with RBX1. Because RBX1 binds the ubiquitin E2, it is possible that both RBX1 and E2 become limiting in axr6/+ plants. In addition, CUL1 has been shown to interact with the CSN (Schwechheimer et al., 2001). The accumulation of defective CUL1 may disrupt the function of this complex.
CUL1 is required for auxin signaling in the embryo
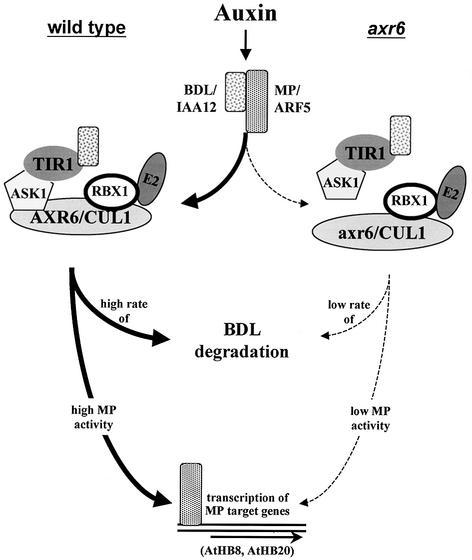
A striking aspect of the axr6 phenotype is its similarity to the mp and bdl phenotypes. All three mutations affect the orientation of cell division as early as the two-cell stage of the embryo and they each lack the SAM, hypocotyl and root meristem and display vascular defects (Berleth and Jurgens, 1993; Hamann et al., 1999; Hobbie et al., 2000). These similarities suggest that AXR6, MP and BDL function in the same pathway during embryogenesis. The molecular characterization of the gene products provides strong support for this view. MP encodes ARF5 while BDL encodes IAA12 (Hardtke and Berleth, 1998; Hamann et al., 2002). These two proteins interact in a yeast two-hybrid test and are co-expressed during early embryogenesis, consistent with a model in which BDL negatively regulates MP function (Hamann et al., 2002). Our demonstration that AXR6 encodes CUL1 strongly suggests that degradation of BDL/IAA12 requires the action of an SCF. A model describing the role of these proteins is presented in Figure 8. We propose that the axr6 mutations affect the levels of functional SCF resulting in abnormal accumulation of BDL and subsequent repression of MP function. It is also possible that an APC containing the HBT protein contributes to BDL/IAA12 degradation (Blilou et al., 2002). However, the axr6 phenotype suggests that one or more SCF complexes are primarily responsible for degradation of these repressors during embryogenesis. This view is also consistent with the phenotype of the recently described axr1 rce1 double mutant (Dharmasiri et al., 2003). These plants also have a seedling phenotype that is similar to axr6, bdl and mp. The axr1 and rce1 mutations affect SCF function indirectly by reducing the levels of RUB-modified CUL1. The APC also has a cullin-related subunit, but this protein is not modified by RUB (Yeh et al., 2000).
Fig. 8. Role of AXR6/CUL1 during embryonic development. In wild-type plants (left side) embryonic development requires auxin-dependent degradation of BDL/IAA12 leading to transcription of MP target genes (Mattsson et al., 2003). MP transcriptional activators can act either as monomers or dimers (Tiwari et al., 2003). Possible target genes include AtHB8 and AtHB20 (Mattsson et al., 2003). In contrast, the axr6 mutations affect the interaction between ASK1 and CUL1 and reduce the number of functional SCF complexes (right side). As a result BDL–MP heterodimers are stabilized. Low levels of MP activity lead to abnormal embryonic development.
The identity of MP-regulated genes is unclear. In a recent study, Mattsson et al. (2003) showed that expression of the HD-ZIP genes AtHB8 and AtHB20 is increased in plants overexpressing MP (Mattsson et al., 2003). AtHB8 has been implicated in vascular development and may be an MP target in provascular cells (Baima et al., 2001; Mattsson et al., 2003). Similarly, the identity of the FPB involved in BDL destruction is unknown. The TIR1 gene is expressed during embryogenesis but the tir1 mutants do not have an embryonic phenotype (Gray et al., 1999). However, genetic studies indicate that TIR1 and several closely related FBPs have overlapping functions leaving open the possibility that SCFTIR1 and/or close relatives are involved in Aux/IAA degradation during embryogenesis (N.Dharmasiri, S.Dharmasiri and M.Estelle, unpublished data).
CUL1 is required for auxin signaling during postembryonic development
The postembryonic phenotype of axr6 plants indicates that CUL1 and SCF complexes are required for normal auxin response throughout development (Hobbie et al., 2000). Heterozygous axr6 plants have fewer lateral roots, decreased root gravitropism and shorter but more numerous inflorescences. Since these plants retain one wild-type CUL1 gene they presumably retain significant CUL1 function. A further reduction in CUL1 activity may result in additional defects. Indeed transgenic lines with reduced CUL1 levels have novel phenotypes not observed in the axr6 or cul1 knockout lines. For example, light-grown T1 seedlings with reduced CUL1 levels had much longer hypocotyls suggesting that a negative regulator of light signaling was accumulating in these seedlings. Defects in root growth and inflorescence elongation were also observed.
The most dramatic defects are in meristem growth and organ initiation. In young CUL1-deficient seedlings, the SAM was enlarged compared with the control. This may be related to the much slower rate of leaf initiation in these plants resulting in accumulation of cells in the meristem. After producing several leaves the SAM arrested in a pin-like structure. At this point axillary meristems produced additional rosette leaves and eventually an inflorescence. However, the inflorescence meristem also failed to initiate lateral organs, and terminated in a pin-like structure similar to that observed in the pin and pid mutants. Unlike these mutants, the pin meristems on CUL1-deficient plants were insensitive to applied auxin indicating a defect in auxin response (Reinhardt et al., 1998) (D.Reinhardt and C.Kuhlemeier, unpublished data).
Based on studies of organ initiation in Arabidopsis and tomato, Reinhardt and co-workers proposed that organ initiation requires the local accumulation of auxin (Reinhardt et al., 2000; Kuhlemeier and Reinhardt, 2001; Reinhardt and Kuhlemeier, 2001). Since auxin induces degradation of Aux/IAA proteins and the frequency of plants with a pin inflorescence increases ∼5-fold in a tir1 background, the failure to initiate lateral organs is probably related to accumulation of Aux/IAA proteins. Hence, our results are consistent with the hypothesis of Rheinhardt and co-workers and further suggest that auxin promotes organ initiation by stimulating degradation of Aux/IAA proteins at the sites of initiation.
RUB conjugation pathway is required for meristem function
The axr1 mutants are deficient in the RUB activating enzyme AXR1-RCE1 and have reduced RUB-CUL1 levels (del Pozo et al., 2002). The consequences of this change are decreased auxin and jasmonate response and diverse defects in growth and development (Lincoln et al., 1990; del Pozo et al., 2002; Tiryaki and Staswick, 2002; Xu et al., 2002). However, gross defects in meristem structure and function have not been observed, leaving open the possibility that the RUB pathway is not required in this tissue. We find that the effects of reduced CUL1 levels were dramatically enhanced in an axr1 background indicating that the pathway is required in the meristem. The lack of meristem defects in the axr1 mutant is probably due to genetic redundancy. The Arabidopsis genome contains a gene that is closely related to AXR1, that we have called AXR1 RELATED 1 (AXL1). Plants that carry mutations in both genes have an embryo lethal phenotype (N.Dharmasiri and M.Estelle, unpublished data).
Genetic screens have led to the recovery of many genes that function in auxin response. So far, these genes fall into three groups: members of the ARF gene family (MP/ARF5, NPH4/ARF7, ETT/ARF3), members of the Aux/IAA gene family [BDL/IAA12, AXR2/IAA7 and many others, see Reed (2001) for a review], and genes that function in SCF-dependent protein degradation (AXR1, AXR6, TIR1). Each of these genes is likely to have specific functions in auxin response except for AXR1 and AXR6, both of which have a general role in SCF function. The fact that mutations in CUL1 result in striking auxin-related phenotypes is indicative of the importance of the SCF for auxin response and the central role of auxin in plant growth and development.
Materials and methods
Plant material
Arabidopsis thaliana plants were grown in soil and in sterile culture at 20–23°C with continuous light. For culture experiments, seeds were surface sterilized for 10 min in 30% (v/v) bleach, 0.01% Triton X-100, washed three times with sterile water and spread on plates using 0.1% agar. Auxin resistance was scored on day 5 of growth on minimal medium containing different concentrations of 2,4-dichlorophenoxyacetic acid. Analyses of segregating populations and identification of auxin resistant plants were done as described in Hobbie et al. (2000).
Generation of 35S::CUL1 Arabidopsis plants
CUL1 overexpression constructs were generated by cloning CUL1 cDNA or mutated CUL1 cDNA carrying a point mutation in the RUB modification site (del Pozo et al., 1999) behind a 35S CaMV promoter into the binary vector pROKII using SacI/KpnI sites. Transgenic Arabidopsis plants were generated using a floral dip protocol according to Clough and Bent (1998).
Mapping
Heterozygous axr6-2 (Columbia) plants were crossed into Landsberg erecta mutants, ga1 and prl. Selection, analysis and DNA isolation from F3 families was performed as described in Hobbie et al. (2000).
RNA expression analyses
Col-0, axr6-1 and axr6-2 seedlings were pre-selected for 5 days on minimal medium containing 40 nM 2,4-D. Auxin-resistant seedlings were then transferred to medium without 2,4-D and grown for 2 additional days. For 2,4-D induction, plants were transferred to liquid minimal medium containing 50 µM 2.4-D for 1 h. RNA extraction, gel electrophoresis and blotting were done using standard techniques.
In vitro translation and RUB modification
CUL1 cDNA was cloned into the SmaI site of pBluescript (±) (Stratagene) and used for introduction of axr6-1 and axr6-2 point mutations by a Stratagene mutagenesis kit with complementary primer pairs (axr6-1mut, 5′- GGTCAGATGGCTATCCCGCGTCTTCTACTA CCTTGACCG; axr6-1complement, 5′-CGGTCAAGGTAGTAGAAGA CGCGGGATAGCCATCTGACC; axr6-2mut, 5′-GGTCAGATGGCTA TCCCGCATCTTCTACTACCTTGACCG; axr6-2complement, 5′-CGG TCAAGGTAGTAGAAGATGCGGGATAGCCATCTGACC). Coupled transcription/translation reactions of CUL1 and mutant cDNAs (1 mg each) were performed in the TNT-T7 coupled system (Promega, Madison, WI) using [35S]trans-labeled methionine (ICN). After a 90 min reaction time, GST–RUB1, 3 mM ATP, 0.1 mM DTT, 5 mM MgCl2 and 10 U/ml inorganic pyrophosphatase were added to the reaction and the mixtures were incubated at 25°C for 15 min. Reactions were stopped by adding 4× SDS/DTT loading buffer and boiling for 10 min. Proteins were resolved on an SDS–PAGE/10% acrylamide gel. Products were detected by autoradiography.
Protein extraction, immunoprecipitations and GST pulldowns
Proteins were extracted from plant tissue using a standard extraction buffer [100 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5% (v/v) Igepal]. Samples were left 10 min on ice before centrifugation for 15 min in a microcentrifuge. Supernatant containing 30 µg protein was used for western blot analysis while up to 3 mg protein was used for immunoprecipitation (IP) or pulldown experiments. For IPs and pulldowns, extracts were incubated at 4°C with primary antibody or GST fusion proteins. Proteins were recovered from the extract by adding protein agarose beads (Sigma, St Louis, MO) or glutathionine beads (Sigma) and incubating for 1 h. Beads were washed four times with extraction buffer before loading on a SDS–PAGE/10% acrylamide. Detection of proteins on nitrocellulose membrane was done using an ECL kit (Amersham, Arlington Heights, IL).
Pulse–chase experiments
axr6/+ seedlings were identified by growth on medium containing 2,4-D. Wild-type and mutant seedlings were incubated for 3.5 h in liquid sugar-free minimal medium supplemented with 200 µCi of trans-labeled [35S]methionine. After washing with water, seedlings were either directly frozen in liquid nitrogen or incubated for 17 min with 1 mM cold methionine/cysteine (Sigma) and cycloheximide (125 µg/ml) before freezing in liquid nitrogen. Protein extraction, immunoprecipitation and detection of AXR2/IAA7 were done as described previously (Gray et al., 2002).
Immunolocalization of CUL1
Paraffin embedded 9-day-old seedlings grown in sterile culture were sectioned (10 µm) and transferred to Superfrost Plus Micro Slides (VWR). Before incubation with the primary antibody, sections were deparaffinized in xylene, rehydrated in an decreasing ethanol series, treated for 20 min with 0.2 M HCl and protease K (1 µg/ml) and were re-fixed in a freshly prepared 4% paraformaldehyde solution. Sections were incubated overnight at 4°C with an affinity purified αCUL1 as primary antibody. Detection of the primary antibody was done using a secondary biotin-conjugated antibody and biotin–avidin detection kit with peroxidase activity (Vector Laboratories, Burkingame, CA).
Exogenous application of auxin and scanning electron microscopy
For local treatments of apices, IAA (Fluka, Buchs, Switzerland) 100 mM stock solutions in DMSO were dissolved in a pre-warmed (50°C) paste consisting of lanolin with 2.5% paraffin (Merck) according to Reinhardt et al. (2000) to give a final concentration of 1 mM IAA. The paste was manually applied directly to Arabidopsis inflorescence apices on the intact plant. For microscopic analysis, apices were viewed with an S-3500N variable pressure scanning electron microscope from Hitachi (Tokyo, Japan), equipped with a cool stage. Lanolin paste in digital images was pseudocolored for clarity.
Acknowledgments
Acknowledgements
We thank John Mendenhall for advice and help on sectioning, Pascal Genschik for the gift of the RBX1 antibody, Patricia Springer for the prl mutant, ABRC for other seeds and clones, and Cereon for access to the Col-0/Ler polymorphism database. L.H. would like to thank the following Adelphi students who contributed to mapping AXR6: Helen Stutz, Andrea Pierro, Nancy Yang Liu, Susanne Sherman, Kimberly Kranz, Stephanie Wasser, Anthony Cucci, Medina Vernon, Yana Gladysheva, Theresa Cesarski and Georgeta Badrajan. This work was supported by grants from the DFG (3224/1-1 to H.H.), the NIH (43644 to M.E.), the DOE (DE-FG03-99ER20327 to M.E.) and NSF (DBI 0115870 to M.E. and IBN 998926 to L.H.). The work in Bern was supported by grant 31-55540.98 of the Swiss National Science Foundation to C.Kuhlemeier and D.R.
References
- Baima S., Possenti,M., Matteucci,A., Wisman,E., Altamura,M.M., Ruberti,I. and Morelli,G. (2001) The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol., 126, 643–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett S.R.M., Alvarez,J., Bossinger,G. and Smyth,D.R. (1995) Morphogenesis in pinoid mutants of Arabidopsis thaliana. Plant J., 8, 505–520. [Google Scholar]
- Berleth T. and Jurgens,G. (1993) The role of the monopteros gene in organising the basal body region of the Arabidopsis embryo. Development, 118, 575–587. [Google Scholar]
- Blilou I. et al. (2002) The Arabidopsis HOBBIT gene encodes a CDC27 homolog that links the plant cell cycle to progression of cell differentiation. Genes Dev., 16, 2566–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J. and Bent,A.F. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J., 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Davies P.J. (1995) The plant hormones: their nature, occurrence and functions. In Davies,P.J. (ed.), Plant Hormones: Physiology, Biochemistry and Molecular Biology. Kluwer Academic, Dordrecht, The Netherlands, pp. 1–12. [Google Scholar]
- del Pozo J.C., Timpte,C., Tan,S., Callis,J. and Estelle,M. (1998) The ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science, 280, 1760–1763. [DOI] [PubMed] [Google Scholar]
- del Pozo J.C., Dharmasiri,S., Hellmann,H., Walker,L., Gray,W.M. and Estelle,M. (2002) AXR1-ECR1-dependent conjugation of RUB1 to the Arabidopsis cullin AtCUL1 is required for auxin response. Plant Cell, 14, 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshaies R.J. (1999) SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol., 15, 435–467. [DOI] [PubMed] [Google Scholar]
- Dharmasiri S., Dharmasiri,N., Hellmann,H. and Estelle,M. (2003) The RUB/Nedd8 conjugation pathway is required for early development in Arabidopsis. EMBO J., 22, 1762–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farras R. et al. (2001) SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J., 20, 2742–2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa M., Zhang,Y., McCarville,J., Ohta,T. and Xiong,Y. (2000) The CUL1 C-terminal sequence and ROC1 are required for efficient nuclear accumulation, NEDD8 modification and ubiquitin ligase activity of CUL1. Mol. Cell. Biol., 20, 8185–8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagne J.M., Downes,B.P., Shiu,S.H., Durski,A.M. and Vierstra,R.D. (2002) The F-box subunit of the SCF E3 complex is encoded by a diverse superfamily of genes in Arabidopsis. Proc. Natl Acad. Sci. USA, 99, 11519–11524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W.M. et al. (1999) Identification of an SCF ubiquitin–ligase complex required for auxin response in Arabidopsis thaliana. Genes Dev., 13, 1678–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray W.M., Kepinski,S., Rouse,D., Leyser,O. and Estelle,M. (2001) Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature, 414, 271–276. [DOI] [PubMed] [Google Scholar]
- Gray W.M., Hellmann,H., Dharmasiri,S. and Estelle,M. (2002) Role of the Arabidopsis RING-H2 protein RBX1 in RUB modification and SCF function. Plant Cell, 14, 2137–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen G. and Guilfoyle,T. (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol., 49, 373–385. [PubMed] [Google Scholar]
- Hamann T., Mayer,U. and Jurgens,G. (1999) The auxin-insensitive bodenlos mutation affects primary root formation and apical-basal patterning in the Arabidopsis embryo. Development, 126, 1387–1395. [DOI] [PubMed] [Google Scholar]
- Hamann T., Benkova,E., Baurle,I., Kientz,M. and Jurgens,G. (2002) The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev., 16, 1610–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke C.S. and Berleth,T. (1998) The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J., 17, 1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann H. and Estelle,M. (2002) Plant development: regulation by protein degradation. Science, 297, 793–797. [DOI] [PubMed] [Google Scholar]
- Hobbie L., McGovern,M., Hurwitz,L.R., Pierro,A., Liu,N.Y., Bandyopadhyay,A. and Estelle,M. (2000) The axr6 mutants of Arabidopsis thaliana define a gene involved in auxin response and early development. Development, 127, 23–32. [DOI] [PubMed] [Google Scholar]
- Kuhlemeier C. and Reinhardt,D. (2001) Auxin and phyllotaxis. Trends Plant Sci., 6, 187–189. [DOI] [PubMed] [Google Scholar]
- Kuroda H., Takahashi,N., Shimada,H., Seki,M., Shinozaki,K. and Matsui,M. (2002) Classification and expression analysis of Arabidopsis F-box-containing protein genes. Plant Cell Physiol., 43, 1073–1085. [DOI] [PubMed] [Google Scholar]
- Lincoln C., Britton,J.H. and Estelle,M. (1990) Growth and development of the axr1 mutants of Arabidopsis. Plant Cell, 2, 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E. and Reed,J.W. (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol., 49, 387–400. [PubMed] [Google Scholar]
- Lyapina S., Cope,G., Shevchenko,A., Serino,G., Tsuge,T., Zhou,C., Wolf,D.A., Wei,N. and Deshaies,R.J. (2001) Promotion of NEDD8-CUL1 conjugate cleavage by COP9 signalosome. Science, 292, 1382–1385. [DOI] [PubMed] [Google Scholar]
- Mattsson J., Ckurshumova,W. and Berleth,T. (2003) Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol., 131, 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Ueda,J., Komaki,M.K., Bell,C.J. and Shimura,Y. (1991) Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell, 3, 677–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart C.M. (2001) Mechanisms underlying ubiquitination. Annu. Rev. Biochem., 70, 503–533. [DOI] [PubMed] [Google Scholar]
- Przemeck G.K., Mattsson,J., Hardtke,C.S., Sung,Z.R. and Berleth,T. (1996) Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta, 200, 229–237. [DOI] [PubMed] [Google Scholar]
- Ramos J.A., Zenser,N., Leyser,H.M. and Callis,J. (2001) Rapid degradation of Aux/IAA proteins requires conserved amino acides of domain II and is proteasome-dependent. Plant Cell, 13, 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed J.W. (2001) Roles and activities of Aux/IAA proteins in Arabidopsis. Trends Plant Sci., 6, 420–425. [DOI] [PubMed] [Google Scholar]
- Reinhardt D. and Kuhlemeier,C. (2001) Phyllotaxis in higher plants. In Veit,B. (ed.), Meristematic Tissues in Plant Growth and Development. Sheffield Academic Press, Sheffield, UK, pp. 172–212. [Google Scholar]
- Reinhardt D., Wittwer,F., Mandel,T. and Kuhlemeier,C. (1998) Localized upregulation of a new expansin gene predicts the site of leaf formation in the tomato meristem. Plant Cell, 10, 1427–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt D., Mandel,T. and Kuhlemeier,C. (2000) Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell, 12, 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruegger M., Dewey,E., Gray,W.M., Hobbie,L., Turner,J. and Estelle,M. (1998) The TIR1 protein of Arabidopsis functions in auxin response and is related to human SKP2 and yeast grr1p. Genes Dev., 12, 198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C., Serino,G., Callis,J., Crosby,W.L., Lyapina,S., Deshaies,R.J., Gray,W.M., Estelle,M. and Deng,X.W. (2001) Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIR1 in mediating auxin response. Science, 292, 1379–1382. [DOI] [PubMed] [Google Scholar]
- Shen W.H. et al. (2002) Null mutation of AtCUL1 causes arrest in early embryogenesis in Arabidopsis. Mol. Biol. Cell, 13, 1916–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer J.D., Gurian-West,M., Clurman,B. and Roberts,J.M. (1999) Cullin-3 targets cyclin E for ubiquitination and controls S phase in mammalian cells. Genes Dev., 13, 2375–2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiryaki I. and Staswick,P.E. (2002) An Arabidopsis mutant defective in jasmonate response is allelic to the auxin-signaling mutant axr1. Plant Physiol., 130, 887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S.B., Wang,X.J., Hagen,G. and Guilfoyle,T.J. (2001) AUX/IAA proteins are active repressors and their stability and activity are modulated by auxin. Plant Cell, 13, 2809–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S.B., Hagen,G. and Guilfoyle,T. (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell, 15, 533–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen V., Wolkenfelt,H., de Vrieze,G., Weisbeek,P. and Scheres,B. (1998) The HOBBIT gene is required for formation of the root meristem in the Arabidopsis embryo. Development, 125, 521–531. [DOI] [PubMed] [Google Scholar]
- Winston J.T., Chu,C. and Harper,J.W. (1999) Culprits in the degradation of cyclin E apprehended. Genes Dev., 13, 2751–2757. [DOI] [PubMed] [Google Scholar]
- Xu L., Liu,F., Lechner,E., Genschik,P., Crosby,W.L., Ma,H., Peng,W., Huang,D. and Xie,D. (2002) The SCF(COI1) ubiquitin–ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell, 14, 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E.T., Gong,L. and Kamitani,T. (2000) Ubiquitin-like proteins: new wines in new bottles. Gene, 248, 1–14. [DOI] [PubMed] [Google Scholar]
- Zenser N., Ellsmore,A., Leasure,C. and Callis,J. (2001) Auxin modulates the degradation rate of Aux/IAA proteins. Proc. Natl Acad. Sci. USA, 98, 11795–11800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng N. et al. (2002) Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature, 416, 703–709. [DOI] [PubMed] [Google Scholar]