Abstract
Calponins and transgelins are members of a conserved family of actin-associated proteins widely expressed from yeast to humans. Although a role for calponin in muscle cells has been described, the biochemical activities and in vivo functions of nonmuscle calponins and transgelins are largely unknown. Herein, we have used genetic and biochemical analyses to characterize the budding yeast member of this family, Scp1, which most closely resembles transgelin and contains one calponin homology (CH) domain. We show that Scp1 is a novel component of yeast cortical actin patches and shares in vivo functions and biochemical activities with Sac6/fimbrin, the one other actin patch component that contains CH domains. Purified Scp1 binds directly to filamentous actin, cross-links actin filaments, and stabilizes filaments against disassembly. Sequences in Scp1 sufficient for actin binding and cross-linking reside in its carboxy terminus, outside the CH domain. Overexpression of SCP1 suppresses sac6Δ defects, and deletion of SCP1 enhances sac6Δ defects. Together, these data show that Scp1 and Sac6/fimbrin cooperate to stabilize and organize the yeast actin cytoskeleton.
INTRODUCTION
Actin filament assembly and organization are regulated by a large number of actin-associated proteins that use a limited set of structural modules to achieve great diversity of activities (Matsudaira, 1991). One such protein module, the calponin homology (CH) domain, is found in actin-associated proteins that cross-link actin filaments (e.g., spectrin, filamin, and fimbrin), link actin to other cytoskeletal systems (e.g., fimbrin and plectin), and form signaling scaffolds (e.g., IQGAP, Vav; reviewed in Gimona et al., 2002). It is well established that a pair of CH domains forms a classic actin binding domain (e.g., α-actinin and fimbrin; reviewed in Matsudaira, 1991). In contrast, calponin family members contain only a single CH domain, and it remains controversial whether this domain can bind to actin filaments (Gimona and Winder, 1998; Fu et al., 2000; Winder, 2003).
The calponin protein family, which includes calponins and transgelins, is characterized by a single CH domain located at the amino terminus and either one or more calponin-like repeats (CLR) located at the carboxy terminus (Prinjha et al., 1994). Both mammalian transgelin and Saccharomyces cerevisiae calponin (Scp1) have a single CLR, whereas mammalian calponin contains three CLRs (Figure 1). The calponin family is highly conserved from yeast to humans. Fungal genomes (S. cerevisiae, Schizosaccharomyces pombe, and Neurospora crassa) contain a single transgelin-like gene, whereas higher eukaryotic genomes have multiple transgelins and calponins. This evolutionary conservation of calponin family proteins suggests that they may have highly conserved functions in vivo, yet our understanding of these functions is limited. Calponin is a regulator of smooth muscle contraction, but the functions of nonmuscle calponins are not as well understood (reviewed in Morgan and Gangopadhyay, 2001).
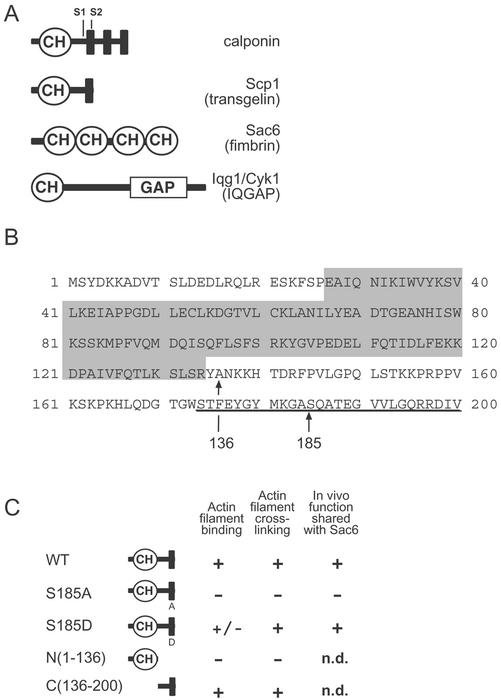
Figure 1.
Domain organization of Scp1 and related proteins. (A) Domain organization of calponin and the three CH domain-containing proteins in budding yeast: Scp1, Sac6, and Iqg1/Cyk1. CH domains are circled, CLRs are indicated as solid rectangles, the two putative actin binding sequences in calponin are labeled (S1 and S2), and the GTPase activating protein (GAP) domain in Iqg1/Cyk1 is boxed. (B) Primary sequence of Scp1. The CH domain is shaded, the CLR is underlined, and the sites of mutations in scp1 are indicated by arrows. (C) Diagram of the mutant constructs generated in this study and summary of their activities and in vivo functions.
Transgelin (also called SM22 and WS3-10; Lees-Miller et al., 1987a; Lawson et al., 1997) was named for its in vitro gelation activity on actin filaments (Shapland et al., 1993), but this activity has been questioned because it does not occur at physiological salt concentrations (see DISCUSSION). In vivo, transgelin localizes to actin structures such as stress fibers (Fu et al., 2000), yet the ability of transgelin to bind directly to actin filaments in vitro also has been disputed (Gimona and Mital, 1998; Morgan and Gangopadhyay, 2001). Increased levels of transgelin expression have been correlated with cell differentiation and senescence, but the function, if any, of transgelins in these processes has not been demonstrated (Thweatt et al., 1992; Liu et al., 1994; Grigoriev et al., 1996). Thus, little is known about the in vitro or in vivo functions of transgelins.
The S. cerevisiae genome contains a single open reading frame with homology to the calponin protein family, and this gene was annotated as S. cerevisiae calponin homolog, Scp1 (Epp and Chant, 1997). However, the domain organization of Scp1 more closely resembles transgelin than calponin. As shown in Figure 1B, Scp1 contains a single CH domain (residues 28–139; shaded) and one calponin-like repeat (residues 174–200; underlined). The yeast genome encodes two other proteins with readily apparent CH domains, Sac6 (fimbrin) and Iqg1/Cyk1 (IQGAP), shown schematically in Figure 1A. IQGAP has a single CH domain, localizes to the bud neck, and functions in cytokinesis, but little is known about its interactions with actin (Epp and Chant, 1997; Lippincott and Li, 1998). Sac6/fimbrin binds to and bundles actin filaments through a tandem pair of actin binding domains, each comprised of two CH domains. Sac6 localizes to cortical actin patches and actin cables and is important for actin organization, endocytosis, and cell polarity in vivo (Drubin et al., 1988; Adams et al., 1991; Kubler and Riezman, 1993).
Herein, we show that the S. cerevisiae transgelin homolog Scp1 is a novel component of the cortical actin cytoskeleton and a bona fide actin filament binding and cross-linking protein. The sequences in Scp1 critical for actin filament binding and cross-linking reside outside of the CH domain. Genetic interactions between SCP1 and SAC6/fimbrin and similar biochemical activities suggest that these two CH domain-containing proteins cooperate in vivo to regulate the stability and organization of the cortical actin cytoskeleton.
MATERIALS AND METHODS
Strains and Growth Conditions
The yeast strains used in this study are listed in Table 1. Standard methods were used for growing and manipulating yeast (Guthrie and Fink, 1991). To generate AGY189, the coding regions of SAC6 and SCP1 in AGY20 were replaced with LEU2 and HIS3, respectively. AGY189 was sporulated and resulting haploid strains of opposite mating type were crossed to generate AGY490, AGY491, AGY492, and AGY493. For growth assays, homozygous diploid strains carrying vectors (pRS313, pRS314, pRS315, and pRS316) and/or plasmids (Table 2) were grown to log phase and then serially diluted fivefold, spotted on plates, and grown for an additional 2–3 d. Latrunculin A sensitivity of cells was measured by halo assays as described previously (Ayscough et al., 1997).
Table 1.
Strains used in this study
| Name | Genotype | Source |
|---|---|---|
| AAY1918 | MATa, ura3, trp1, leu2, his3, prb1, can1, sac6::LEU2, pep4::HIS3, pGal10-SAC6/CEN/URA | Sandrock et al., 1997 |
| AGY20 | MATa/MATα, his3 200/his3 200, leu2-3, 112/leu2-3,112, ura3-52/ura3-52, trp1::HisG/trp1::HisG | Fink laboratory |
| AGY189 | MATa/MATα, his3 200/his3 200, leu2-3, 112/leu2-3,112, ura3-52/ura3-52, trp1::HisG/trp1::HisG, SAC6/sac6::LEU2, SCP1/scp1::HIS3 | This study |
| AGY490* | MATa/MATα, SAC6/SAC6, SCP1/SCP1 | This study |
| AGY491* | MATa/MATα, sac6::LEU2/sac6::LEU2, SCP1/SCP1 | This study |
| AGY492* | MATa/MATα, SAC6/SAC6, scp1::HIS3/scp1::HIS3 | This study |
| AGY493* | MATa/MATα, sac6::LEU2/sac6::LEU2, scp1::HIS3/scp1::HIS3 | This study |
| BJ2168 | MATa, pep4-3, prb1-1122, prc1-407, trp1, ura3-52, leu2, gal2 | Jones, 2002 |
Strains have the same genotype as AGY189, except at SAC6 and SCP1 loci.
Table 2.
Plasmids generated in this study
| Name | Insert | Vector |
|---|---|---|
| pAG9 | GFP-scp1 | pRS316 |
| pAG16 | scp1S185A | pRS316 |
| pAG17 | scp1S185D | pRS316 |
| pAG20 | SCP1 | pRS316 |
| pAG22 | SCP1 | pTrcHisA (Invitrogen) |
| pAG37 | scp1S185D | pTrcHisA (Invitrogen) |
| pAG50 | scp1S185A | pTrcHisA (Invitrogen) |
| pAG179 | SCP1 | pRS426Gal1 |
| pAG202 | scp1N(1-136) | pProEX™HTa (Invitrogen) |
| pAG203 | scp1C(136-200) | pBAT4 |
Plasmid Construction
The coding region of the SCP1 gene (YOR367w), plus 397 bases of sequence upstream of the translation start site, was amplified by polymerase chain reaction (PCR) from wild-type yeast genomic DNA. The PCR product was cloned into the ClaI and SmaI sites of pRS316 (Sikorski and Hieter, 1989), generating pAG20. For additional SCP1 constructs, we introduced by site-directed mutagenesis (QuikChange kit; Stratagene, La Jolla, CA), a BglII site at the start codon of SCP1 in pAG20, generating pAG3. To construct an amino terminal green fluorescent protein (GFP)-SCP1 fusion plasmid (pAG9), we cloned GFP as a BamHI fragment from plasmid B3355 (Fink laboratory collection) into the BglII site of pAG3. To generate an Escherichia coli expression amino terminal hexahistidine-SCP1 fusion construct (pAG22), SCP1 was excised from pAG3 as a BglII-XhoI fragment and cloned into the BamHI and XhoI sites of pTrcHisA (Invitrogen, Carlsbad, CA). To generate SCP1 Gal-overexpression plasmids (pAG179), the BglII-XhoI SCP1 fragment was cloned into the BamHI and XhoI sites of pRS426Gal1 (Christianson et al., 1992). Point mutations in SCP1 (S185A and S185D) were generated by site-directed mutagenesis as described above. To generate N136 and 136C constructs, sequences coding for the amino terminus and carboxy terminus of Scp1 were amplified by PCR and cloned into NcoI and HindIII sites of pProET™HTa (Invitrogen) and pBAT4 (Peranen et al., 1996). All mutant scp1 constructs were sequenced to verify that no additional mutations had been introduced.
Protein Purification
Yeast actin was purified as described previously (Goode et al., 1999). His6-tagged Scp1 proteins were expressed in BL21/DE3 E. coli and purified on nickel resin as per manufacturer's instructions (QIA-GEN, Valencia, CA). Peak fractions eluted from the nickel column were pooled and fractionated on a monoQ (5/5) column by using an AKTA FPLC (Amersham Biosciences, Piscataway, NJ). Peak fractions were pooled, concentrated in a Centricon 10 device (Millipore, Bedford, MA), and exchanged into HEKG5 buffer (20 mM HEPES, pH 7.5, 1 mM EDTA, 50 mM KCl, 5% glycerol). The proteins were aliquoted, frozen in liquid nitrogen, and stored at –80°C. Untagged carboxyl-terminal fragment of Scp1 was expressed in E. coli. Cells were lysed in HEKG5 buffer by using french press, and the lysate was clarified by centrifugation at 313,000 × g at 4°C for 30 min (high-speed supernatant; HSS). HSS was fractionated on a 1-ml HiTrap SP column (Amersham Biosciences). Peak fractions were pooled, diluted in low salt buffer, and fractionated on a Mono S column. Peak fractions were concentrated and fractionated on a Superdex 75 (5/30) column (Amersham Biosciences) equilibrated in HEKG5 buffer. Peak fractions were pooled concentrated, aliquoted, frozen in liquid nitrogen, and stored at –80°C. Untagged full-length Scp1 was purified from yeast overexpressing SCP1 (BJ2168 carrying pAG179). One liter of cells was grown to mid-log phase in SC-His medium with 2% raffinose. Then, 2% galactose was added to the medium, and cells were grown for an additional 12 h and harvested by centrifugation. The cell pellet was resuspended in 0.3 volume of water and frozen in droplets in liquid nitrogen. Next, the frozen yeast cells were lysed in a coffee grinder by using liquid nitrogen, described under “Lab Protocols” on the Goode Laboratory Web site at www.bio.brandeis.edu/goodelab. An HSS was generated in HEKG5 buffer supplemented with 0.5 mM dithiothreitol (DTT) and protease inhibitors as described previously (Goode et al., 1999). The HSS was fractionated on a 1-ml HiTrap SP column (Amersham Biosciences), and proteins were eluted with a linear salt gradient (50–500 mM KCl). Scp1 eluted at approximately 200 mM KCl. Peak fractions were pooled and concentrated to 3 ml in a Centricon 10 device and then fractionated on a Superdex 75 (26/60) column (Amersham Biosciences) equilibrated in HEKG5 buffer. Peak fractions were pooled, concentrated as described above, aliquoted, frozen in liquid nitrogen, and stored at –80°C. Sac6 was purified as described above for untagged Scp1 with the following exceptions: AAY1918 strain was used for galactose induction; after the HSS was fractionated on a HiTrap Q column, the Sac6-containing fractions were pooled, desalted, and fractionated on a monoQ (5/5) column. Peak fractions were pooled, concentrated in Centricon 10 devices, and fractionated on a Superose12 (5/30) gel filtration column (Amersham Biosciences) equilibrated in HEKG5 buffer. Sac6 peak fractions were pooled, concentrated, aliquoted, frozen in liquid nitrogen, and stored at –80°C. Tpm1 was purified as described previously (Liu and Bretscher, 1989).
Actin Filament Binding and Cross-Linking Assays
Yeast actin was assembled as follows. Actin (50 μM) in G-buffer (10 mM Tris, pH 7.5, 0.2 mM CaCl2, 0.2 mM DTT, 0.2 mM ATP) was thawed on ice, precleared by centrifugation, and 20× initiation mix (10 mM ATP, 40 mM MgCl2, 1 M KCl) was added to induce polymerization. Reactions were incubated for 1 h at 25°C. Then, actin filaments were diluted in F-buffer (10 mM Tris, pH 7.5, 0.2 mM CaCl2, 0.2 mM DTT, 0.7 mM ATP, 2 mM MgCl2, and 50 mM KCl), purified proteins (Scp1, Sac6, and Tpm1), and/or HEKG5 buffer was added, and the reactions were incubated at room temperature for 1 h. For low-speed pelleting assays, reactions were centrifuged for 10 min at 10,000 × g, 4°C. For high-speed pelleting assays, actin filaments were pelleted by centrifugation for 30 min at 313,000 × g in a TLA100 rotor (Beckman Coulter, Fullerton, CA). In both assays, supernatants and pellets were fractionated on SDS-PAGE gels, stained with Coomassie, and bands were quantified by densitometry with NIH Image (version 1.61, available at sippy.nimh.nih.gov). The binding constant (Kd) of Scp1 for actin filaments was defined as the concentration of Scp1 at which half-maximal Scp1 binding occurred. For light scattering assays, yeast actin was thawed on ice, diluted in G-buffer, and mixed with 20× initiation mix plus Scp1 and/or HEKG5 buffer. Light scattering was monitored overtime at 360 nm in a fluorescence spectrophotometer (Photon Technology International, Lawrenceville, NJ) held at a constant temperature of 25°C. Apparent viscometry of actin solutions was measured by the falling ball assay, performed as described previously (Pollard and Cooper, 1982). Rabbit muscle G-actin (Cytoskeleton, Denver, CO) was clarified by centrifugation for 30 min at 313,000 × g 4°C in a TLA100 rotor. Capillary tubes were loaded with actin (4.2 μM), initiation salts, and varying concentrations of His6-Scp1 (0, 0.23, 0.46, 0.93, 1.4, 1.8, and 3.7 μM) and incubated at room temperature for 1 h before falling ball measurements. For electron microscopy, 2 μl of reactions were spotted onto freshly ionized carbon-coated grids, stained with 1% uranyl acetate, and visualized using a Phillips EM410 transmission electron microscope.
Actin Filament Disassembly Kinetics
Yeast actin (with 1% pyrene labeled rabbit skeletal muscle actin; Cytoskeleton) was assembled at 35 μM as described above. Actin filaments (7 μl) in F-buffer was mixed with 52.5 μl of F-buffer and 7 μl of Scp1 and/or HEKG5 buffer and incubated for 10 min at room temperature. Actin filaments were agitated by vortexing for 10 s, and then mixed with 3.5 μl of latrunculin A (400 μM) in a cuvette. The final reactions contained 3.5 μM actin, 20 μM latrunculin A, and variable concentrations of Scp1 (0–2 μM). The depolymerization kinetics of pyrene-labeled actin filaments was monitored by excitation at 365 nm and emission at 407 nm in a fluorescence spectrophotometer held at a constant temperature of 25°C.
Fluorescence Light Microscopy
Images of cells were acquired using a Nikon TE300 inverted fluorescence microscope equipped with a Hamamatsu Orca charge-coupled device camera controlled by Openlab software (Improvision, Lexington, MA). The localization pattern of GFP-Scp1 fusion protein was examined in live yeast cells grown to log phase. To disrupt the actin cytoskeleton, cultures were treated with 200 μM latrunculin A for 5 min before imaging (Figure 2A). Colocalization of GFP-Scp1 and actin (Figure 2B) was performed essentially as described previously (Warren et al., 2002). Briefly, 1 ml of exponentially growing cells was fixed with 70% ethanol on ice for 10 min, and cells were pelleted by centrifugation at 3000 × g and resuspended in 100 μl of phosphate-buffered saline buffer plus 1 mg/ml bovine serum albumin and 10 μl of rhodamine-phalloidin (300 U in 1.5 ml of methanol; Molecular Probes, Eugene, OR). After incubation on ice for 5 min, cells were washed three times in phosphate-buffered saline buffer and mounted on a slide for imaging.
Figure 2.
Localization of GFP-Scp1 to cortical actin patches. (A) Localization of GFP-Scp1 in live yeast cells untreated, treated with dimethyl sulfoxide, or treated with 200 μM latrunculin A in dimethyl sulfoxide. (B) Colocalization of GFP-Scp1 and rhodamine phalloidin actin staining in fixed cells. Bar, 5 μm.
RESULTS
Scp1 Localizes to Cortical Actin Patches In Vivo
To investigate the in vivo function of Scp1, we first examined localization of Scp1 in yeast cells. We were unable to localize endogenous Scp1 by using anti-Scp1 antibodies or hemagglutinin (HA)-tagged Scp1 by using anti-HA antibodies, likely because Scp1 is expressed at low levels (see below). Therefore, we examined the localization of GFP-Scp1 expressed under the control of the SCP1 promoter from a low copy plasmid. As shown in Figure 2A, GFP-Scp1 localized to motile cortical patches, largely polarized in the bud, but also present in the mother cell. The GFP-Scp1 patches disappeared rapidly after cells were treated briefly with 200 μM latrunculin A, an actin monomer-sequestering agent (Figure 2A). Thus, filamentous actin is required for GFP-Scp1 localization. In fixed cells, GFP-Scp1 patches colocalized with rhodamine-phalloidin stained actin patches, demonstrating that GFP-Scp1 localizes to actin patches (Figure 2B). Although all GFP-Scp1 patches overlapped with actin patches, ∼16% of actin patches (n = 89) did not have a corresponding GFP signal. This leaves open the possibility that some actin patches do not contain Scp1.
Deletion of SCP1 Enhances sac6Δ Phenotypes
To further study SCP1 in vivo function, we generated a complete deletion of the SCP1 gene. This mutation alone had no salient phenotype in haploid or diploid cells, but did show specific genetic interactions with sac6Δ. Among the many phenotypes tested for the scp1Δ single mutants were growth at a full range of temperatures, growth under various stresses (e.g., NaCl, caffeine, and benomyl), cell morphology, bipolar budding pattern, actin cytoskeleton organization, and endocytosis (assayed by lucifer yellow, FM4-64 uptake and Ste6 internalization). The only detectable phenotype of scp1Δ was a modest but reproducible sensitivity to latrunculin A (Table 3). Given the high degree of functional redundancy among components of cortical actin patches (reviewed in Pruyne and Bretscher, 2000; Goode and Rodal, 2001), we tested for synthetic genetic interactions between SCP1 and other genes that regulate actin function. The scp1Δ mutants showed synthetic defects only with sac6Δ, but not with abp1Δ, aip1Δ, arp2-1, cap2Δ, cof1-22, crn1Δ, end3Δ, las17Δ, pan1-4, rvs167Δ, sla1Δ, sla2Δ, srv2Δ, tpm1Δ, or tpm2Δ. Deletion of SCP1 enhanced many phenotypes of sac6Δ, including temperature and caffeine sensitivity (Figure 3A), salt sensitivity (our unpublished observation), and latrunculin A sensitivity (Table 3). Deletion of SCP1 did not further enhance the actin organization or endocytosis phenotypes of sac6Δ cells, which already have depolarized actin cytoskeleton and fail to accumulate lucifer yellow dye in the vacuole (our unpublished observations).
Table 3.
Synthetic latrunculin A sensitivities of scp1 and sac6 mutations
| Relevant genotype | Relative sensitivity to latrunculin A |
|---|---|
| Wild-type | 1.0 |
| sac6Δ | 2.2 |
| scp1Δ | 1.4 |
| sac6Δ scp1Δ | 3.5 |
| sac6Δ scp1Δ, pSCP1/CEN | 2.0 |
The latrunculin A sensitivities of cells were measured by halo assays as described previously (Ayscough et al., 1997). Relative sensitivity was defined as the ratio of latrunculin A concentrations required to produce halos of the same diameter for wild-type and mutant strains.
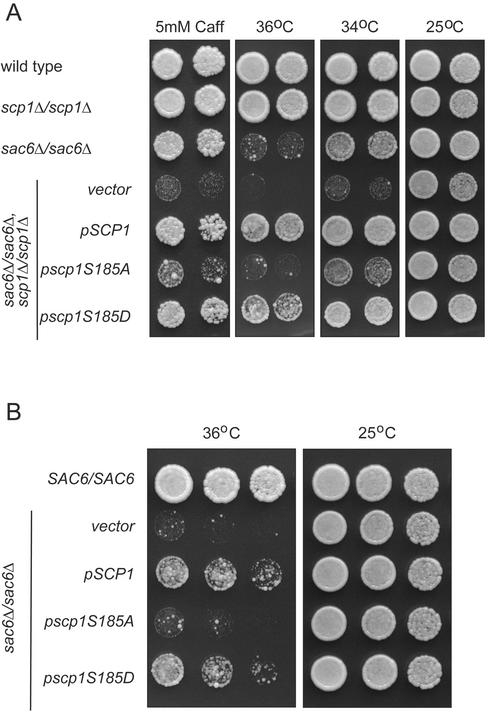
Figure 3.
Genetic interactions between SCP1 and SAC6. (A) Synthetic genetic interactions of scp1 with a sac6Δ null mutation. Homozyogous diploid yeast (AGY490, AGY491, AGY492, or AGY 493) carrying vector alone (pRS316) or SCP1 plasmids (pAG20, pAG16, and pAG17) were serially diluted and grown on YPD at different temperatures in the presence or absence of 5 mM caffeine. (B) Suppression of sac6Δ null mutant phenotypes by additional copies of SCP1. Serial dilutions of wild-type (AGY490) and sac6Δ/sac6Δ (AGY491) homozygous diploid yeast cells carrying vector (pRS316) or low copy SCP1 plasmids (pAG20, pAG16, and pAG17) grown at different temperatures.
The scp1Δ sac6Δ double mutant cells provided a genetic background that permits direct testing of the Scp1 function in vivo. Mutation of a conserved serine residue in the CLR of mammalian calponin (Ser175) and transgelin (Ser184) disrupts actin binding in vitro (Tang et al., 1996; Fu et al., 2000). To test whether the analogous residue in Scp1 (S185; Figure 1B) is important for in vivo function, we generated two substitutions (S185A and S185D). scp1Δ sac6Δ cells transformed with wild-type or mutant SCP1 constructs were analyzed for growth phenotypes. Both wild-type SCP1 and scp1S185D suppressed the growth defects of scp1Δ sac6Δ cells, indicating that these constructs restore SCP1 function. In contrast, cells expressing scp1S185A grew only slightly better than control cells carrying an empty vector (Figure 3A). Stable expression of the mutant proteins was verified by immunoblotting (our unpublished observations). These data indicate that the conserved serine residue (located in the CLR) is critical for Scp1 function in vivo.
Additional Copies of SCP1 Partially Suppress the sac6Δ Growth Phenotype
SCP1 expressed from a low copy plasmid suppressed the temperature sensitivity of sac6Δscp1Δ double mutant cells (Figure 3A). To investigate the basis of this effect, we quantified the expression levels of actin, Sac6, and Scp1 in cells, comparing cell extracts to standard curves of purified proteins (actin, Sac6, and Scp1) by immunoblotting. The level of Scp1 (∼0.01 ng/μg total cellular protein) was considerably lower than that of Sac6 (∼0.15 ng/μg) and actin (∼1 ng/μg). From these values, we calculated that the molar ratio of actin, Sac6, and Scp1 in cells is ∼65:6:1. The expression level of Scp1 in sac6Δscp1Δ cells carrying a low copy SCP1 plasmid was two- to threefold higher than endogenous Scp1 levels in wild-type cells (our unpublished observations). This suggested that extra copies of SCP1 can suppress sac6Δ cell growth defects. To test this hypothesis directly, we transformed sac6Δ cells with low copy plasmids expressing wild-type and mutant Scp1 proteins. As shown in Figure 3B, a wild-type SCP1 plasmid partially suppressed the temperature sensitivity of the sac6Δ mutant. scp1S185D also partially suppressed the sac6Δ phenotype, whereas scp1S185A showed no suppression. Thus, low-level overexpression of SCP1 partially suppresses the temperature-sensitive growth phenotype of the sac6Δ mutant, and a specific mutation in SCP1 abolishes this suppression.
Scp1 Binds to and Cross-Links Actin Filaments In Vitro
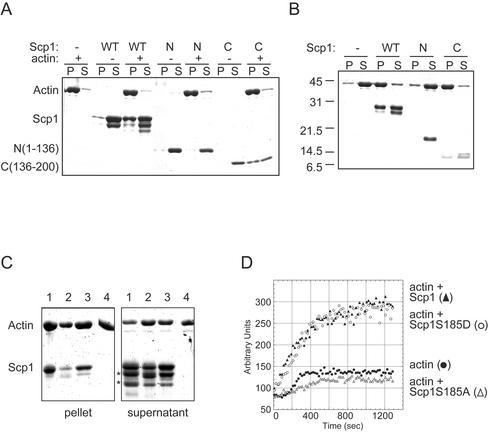
To test whether Scp1 interacts directly with actin filaments in vitro, we overexpressed Scp1 in yeast by using a galactose-inducible promoter, purified the protein, and measured its ability to bind actin filaments in a high-speed cosedimentation assay. As shown in Figure 4, A and B, Scp1 bound to yeast actin filaments in a concentration-dependent manner with micromolar binding affinity (Kd = 0.7 μM) and a molar saturation stoichiometry of 1:2 Scp1 to actin. Hexahistidine (His6)-tagged Scp1 expressed and purified from E. coli bound to actin filaments with a similar affinity (Figure 4C).
Figure 4.
Binding of Scp1 to actin filaments. (A) Cosedimentation assay using 2.5 μM yeast actin filaments and varying concentrations of Scp1. The samples are labeled below the pellet (P) and supernatant (S) lanes: A, 0.5 μM; B, 1 μM; C, 2 μM; D, 3 μM; and E, 4 μM. Half of the supernatant was loaded in each lane compared with pellet. (B) Resulting binding curve. The amount of Scp1 bound (micromolar) in each reaction was calculated from densitometry measurements of the gel in A and plotted versus the total concentration of Scp1 in the reaction. (C) Cosedimentation assay using 5 μM yeast actin filaments and/or 1 μM His6-Scp1 or untagged Scp1. Equivalent amounts of pellet and supernatant were loaded in each lane. The samples are labeled below the pellet and supernatant lanes: 1, actin alone; 2, Scp1; 3, actin and Scp1; 4, His6-Scp1; and 5, actin and His6-Scp1. Note that in A, a contaminant that does not pellet with actin is marked with an asterisk; and in C, a proteolytic fragment of His6-Scp1 is visible below the full-length protein.
Mammalian calponin family members have been shown to cross-link actin filaments (Shapland et al., 1993; Kolakowski et al., 1995; Tang et al., 1997). Using several complementary approaches, we found that Scp1 has a similar activity. First, His6-Scp1 increased light scattering of yeast actin filaments in a concentration-dependent manner (Figure 5A), suggesting that Scp1 organized filaments into larger structures (e.g., bundles or networks). Second, we analyzed the reactions from the light scattering experiment in a low-speed pelleting assay. In the absence of Scp1, most actin remained in the supernatant as expected, but with increasing amounts of Scp1, actin shifted to the pellet (Figure 5B). This concentration-dependent increase in actin pelleting correlated with the observed increase in light scattering (Figure 5A). To ensure that actin cross-linking by His6-Scp1 was not due to the tag, we tested untagged Scp1 in the low-speed actin-pelleting assay. Figure 5C shows that the effects of 1 μM untagged Scp1 are nearly identical to the effects of 1 μM His6-Scp1. These results demonstrate that Scp1 cross-links actin filaments in vitro.
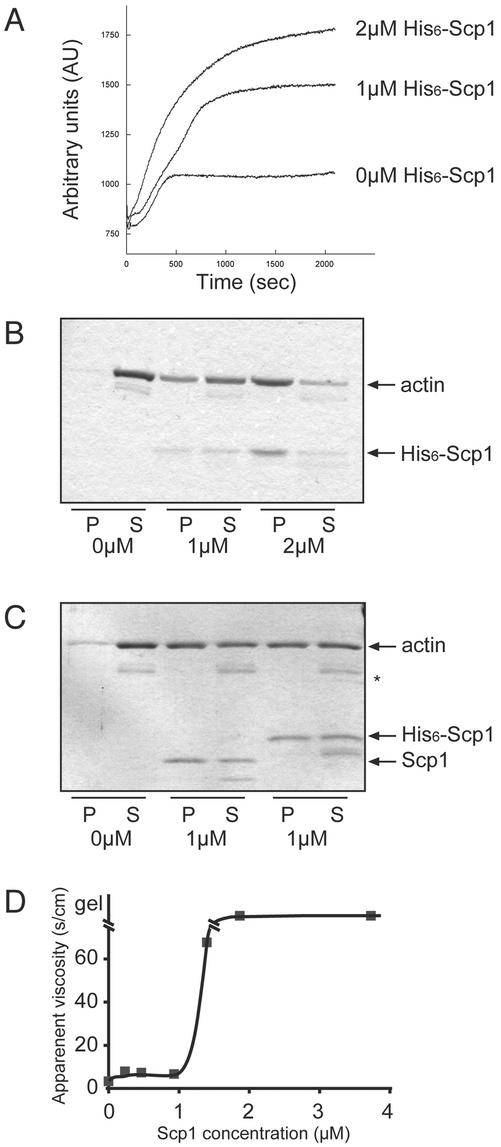
Figure 5.
Actin filament cross-linking by Scp1. (A) Actin filament assembly and organization monitored by light scattering. Monomeric yeast actin (4 μM) was polymerized in the absence or presence of His6-Scp1 (1 or 2 μM) and monitored for change in light scattering (360 nm) overtime. (B) Low-speed pelleting assay for actin filament cross-linking. The reactions shown in A were centrifuged at low speed (10,000 × g) to precipitate cross-linked actin filament structures. Pellets and supernatants were analyzed by SDS-PAGE and Coomassie staining. (C) Low-speed pelleting assay of 2.5 μM actin filaments incubated with and without untagged Scp1 (middle two lanes) or His6-Scp1 (last two lanes). Note that a proteolytic fragment of actin is marked by a single asterisk. (D) Effect of His6-Scp1 on viscosity of actin filaments in solution. Actin (4 μM) was polymerized in capillary tubes in the presence of varying concentrations of His6-Scp1 (0, 0.23, 0.46, 0.93, 1.4, 1.8, and 3.7 μM). Viscosity was measured after 1-h incubation. Viscosity is inversely proportional to the velocity of the ball moving through the sample (Pollard and Cooper, 1982). Gelation (as indicated by the ball-bearing remaining stationary beneath the meniscus) was observed at Scp1 concentrations above 1.4 μM.
To characterize the actin cross-linking activity of Scp1 further, we used the falling ball assay (Pollard and Cooper, 1982) to measure the apparent viscosity of the actin filament solution in the presence and in the absence of His6-Scp1. The apparent viscosity of a 4 μM actin filament solution increased markedly when Scp1 concentration exceeded 1 μM (Figure 5D). We also examined by electron microscopy negatively stained actin filaments in the presence and in the absence of His6-Scp1. In the absence of Scp1, actin filaments were distributed evenly throughout the grid (Figure 6A). When actin was polymerized in the presence of Scp1, actin filaments formed loose bundles tangled into networks (Figure 6, B and C). Scp1 cross-linked bundles seemed wavy, and the spacing between filaments in a bundle was not uniform. In contrast, Sac6/fimbrin bundles were straight with uniform spacing between filaments (Figure 6D).
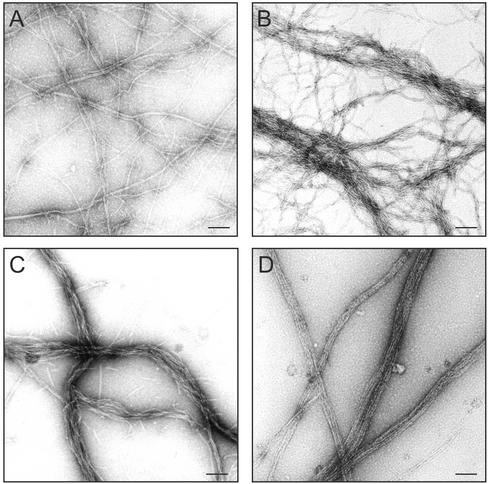
Figure 6.
Electron micrographs of negatively stained actin filaments cross-linked by Scp1 or Sac6. Actin filaments (15 μM) were polymerized in the presence of control buffer (A), 7.5 μM His6-Scp1 (B and C), or 7.5 μM Sac6 (D), negatively stained with uranyl acetate, and photographed at 21,000× magnification. Bar, 100 nm.
The Carboxy Terminus of Scp1 Alone Can Cross-Link Actin Filaments
To better understand the molecular mechanism of actin binding and cross-linking by Scp1, we expressed and purified amino-terminal N(1–136) and carboxyl-terminal C(136–200) fragments of Scp1 (Figure 1C). Although we attempted to generate both His6-tagged and untagged constructs in E. coli, we were able to isolate only the His6-tagged N(1–136) and untagged C(136–200). We tested the purified proteins for actin binding in the high-speed pelleting assays. In contrast to the full-length Scp1, the N(1–136) fragment did not copellet with actin filaments (Figure 7A). The C(136–200) fragment, on the other hand, copelleted with actin filaments, indicating that at least one actin-binding site resides in the carboxy terminus of Scp1. We also tested the ability of the truncated proteins to cross-link actin filaments. In a low-speed pelleting assay, actin remained in the supernatant in the absence of Scp1 and in the presence of N(1–136) (Figure 7B). However, actin was found mostly in the pellet in the presence of the carboxyl-terminal fragment or the full-length Scp1. Therefore, untagged carboxy terminus of Scp1 is sufficient to cross-link actin filaments.
Figure 7.
Effects of Scp1 mutations on actin filament binding and cross-linking. (A) High-speed actin filament cosedimentation assay. Reactions contained 0 or 5 μM yeast actin filaments and 5 μM Scp1: full-length His6-Scp1, amino terminus His6-Scp1N(1–136), or carboxy terminus Scp1C(136–200) (designated WT, N, and C, respectively). Actin filaments were pelleted by high-speed centrifugation and equivalent amounts of the pellet and supernatant were analyzed by SDS-PAGE and Coomassie staining. (B) Low-speed pelleting assay for actin filament cross-linking. Actin filaments (5 μM) were mixed with 5 μM Scp1 [His6-Scp1, His6-Scp1N(1–136), Scp1C(136–200)] or control buffer and centrifuged for 10 min at 10,000 × g. The pellets and supernatants were analyzed as described above. (C) High-speed actin filament cosedimentation assay comparing wild-type and mutant Scp1 proteins. Yeast actin filaments (10 μM) were mixed with 6 μM His6-Scp1 (wild-type, S185A, or S185D mutant) or buffer alone. Actin filaments were pelleted by high-speed centrifugation and pellets and supernatants were analyzed by SDS-PAGE and Coomassie staining. Lanes: 1, actin and Scp1; 2, actin and Scp1 S185A; 3, actin and Scp1 S185D; and 4: actin alone. Note that proteolytic fragments of Scp1 are marked by asterisks. (D) Actin filament assembly and organization monitored by light scattering. Monomeric yeast actin (10 μM) was polymerized in the absence or presence of 5 μM wild-type or mutant His6-Scp1 and monitored for change in light scattering (360 nm) over time.
In addition, we addressed whether a specific residue in the carboxy terminus (S185) critical for in vivo function of Scp1 (see above), was also important for actin filament binding. We purified the His6-tagged S185A and S185D mutant Scp1 proteins and compared their ability to bind and cross-link actin filaments with the wild-type Scp1. Scp1S185D bound to actin filaments in a high-speed actin pelleting assay (Figure 7C), cross-linked actin filaments in the low-speed actin pelleting assay (our unpublished observations), and increased light scattering of the actin filaments similar to wild-type Scp1 (Figure 7D). In contrast, Scp1S185A had greatly diminished actin binding and cross-linking activities (Figure 7, C and D; our unpublished observations). These results suggest that Scp1S185D retains much of the wild-type Scp1 interaction with actin. However, Scp1S185D showed reduced actin binding affinity compared with wild-type Scp1 at higher salt concentrations (150 mM KCl; our unpublished observations). Therefore, the Scp1S185D interaction with actin may be weakened, but not nearly to the extent of Scp1S185A.
Scp1, Like Fimbrin, Decreases the Rate of Actin Filament Disassembly
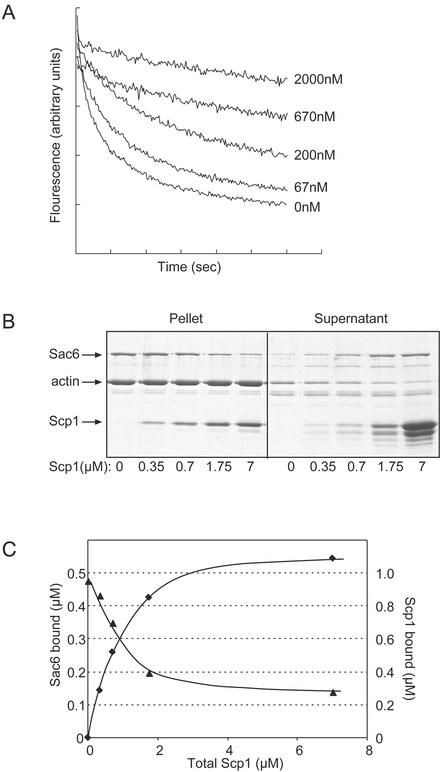
Both Sac6/fimbrin (Bretscher, 1981; Adams et al., 1991) and Scp1 (Figures 4 and 5) bind to and cross-link actin filaments; in addition, Sac6 stabilizes actin filaments against disassembly (see Figure 5 in Goode et al., 1999). To address whether Scp1 similarly can stabilize actin filaments, we compared pyrene-actin filament disassembly kinetics in the presence and absence of His6-Scp1. In the absence of Scp1, actin filaments disassembled rapidly, and pyrene fluorescence reached steady state by 600 s (Figure 8A). His6-Scp1 reduced the rate of actin filament disassembly in a concentration-dependent manner. Similar effects were observed using untagged Scp1 (our unpublished observations). Thus, Scp1, like Sac6/fimbrin, stabilizes actin filaments.
Figure 8.
Activities of Scp1 on actin filaments similar to Sac6/fimbrin. (A) Effects of His6-Scp1 on the rate of actin filament depolymerization. Actin filament disassembly was initiated by the addition of 20 μM latrunculin A to 2 μM preformed actin filaments (1% pyrene labeled) in the presence of 0, 67, 200, 670, and 2000 nM His6-Scp1, and change in pyrene-actin fluorescence was monitored overtime. (B) Competition of Scp1 with Sac6 for binding to actin filaments. Cosedimentation assay using 2 μM yeast actin filaments 0.5 μM Sac6, and variable concentrations of His6-Scp1 (0, 0.35, 0.7, 1.75, and 7 μM). The pellets and supernatants were analyzed by SDS-PAGE and Coomassie staining. (C) The concentrations of Sac6 (▴) and Scp1 (⋄) bound to actin filaments were calculated from gel densitometry and plotted versus the total concentration of His6-Scp1 in the reactions.
Scp1 and Sac6 Compete for Actin Filament Binding In Vitro
Given that Scp1 and Sac6/fimbrin show genetic interactions and have similar biochemical activities on actin, we investigated whether they also have overlapping binding sites on actin filaments. We tested the ability of Scp1 to compete with Sac6 for binding to actin filaments in a high-speed cosedimentation assay, by using constant concentrations of Sac6 (0.5 μM) and actin filaments (2 μM) and variable concentrations of His6-Scp1 (0–7 μM). In the absence of Scp1, nearly 90% of the Sac6 bound to actin filaments (Figure 8, B and C). The percentage of Sac6 bound to actin filaments decreased proportionally with increasing concentrations of Scp1. To test the specificity of the competition, we assayed Sac6 binding to actin in the presence of another actin binding protein, tropomyosin (Tpm1). A range of Tpm1 concentrations (1–10 μM) had no effect on Sac6 binding to actin filaments (our unpublished observations). Thus, Scp1 specifically competes with Sac6 for binding to actin.
DISCUSSION
Calponins and transgelins comprise one of the most widely conserved actin-associated protein families, yet their in vivo function in nonmuscle cells has remained elusive. Furthermore, controversy has surrounded the issue of whether transgelins even bind to actin filaments and/or localize to actin structures in vivo (reviewed in Small and Gimona, 1998; Morgan and Gangopadhyay, 2001). Herein, we have shown unambiguously that the yeast transgelin homolog Scp1 binds directly to actin filaments, cross-links and stabilizes actin filaments in vitro, and localizes to actin filament structures in vivo. Furthermore, we established that the sites on Scp1 necessary and sufficient for actin cross-linking reside in the carboxy terminus, outside the CH domain. We also provide the first in vivo evidence for transgelin cellular function, showing that Scp1 cooperates with Sac6 (fimbrin) to organize and stabilize the actin cytoskeleton. Finally, our mutant analysis revealed a correlation between in vivo phenotypes and in vitro activities on actin, demonstrating that actin binding is required for Scp1 in vivo functions.
Activities of Scp1 on Actin Filaments In Vitro
Scp1 binds directly to actin filaments with an affinity (Kd of ∼0.7 μM) similar to that reported for other members of the calponin family: 1 μM for calponin (Lu et al., 1995; Tang et al., 1996) and 1.3 and 1.4 μM for transgelin (Shapland et al., 1993; Kobayashi et al., 1994). Scp1 also cross-links actin filaments. Previous studies have reported actin filament bundling for calponin (Kolakowski et al., 1995) and actin filament gelation for transgelin (Shapland et al., 1993). However, the ability of transgelin to cross-link actin filaments has been questioned (Morgan and Gangopadhyay, 2001), in part because the gelation activity occurs specifically in low ionic strength buffer and is blocked by the addition of 10 mM KCl (Shapland et al., 1993). Using four independent assays, we showed that His6-Scp1 and untagged Scp1 each cross-link actin filaments in buffer containing 50 mM KCl. One possible explanation for the discrepancy between previous results and ours is that previous experiments tested transgelin and actin from different species, whereas our experiments used transgelin (Scp1) and actin from the same organism. This raises the possibility that other transgelins besides Scp1 also cross-link actin filaments.
By what mechanism does Scp1 cross-link actin filaments? Cross-linking requires either the presence of two actin binding sites within a single polypeptide chain or dimerization of an actin binding protein. All of the data available for calponin family members suggest that they do not dimerize, because they behave as monomers in sedimentation velocity, sedimentation equilibrium, and gel filtration experiments (Lees-Miller et al., 1987b; Stafford et al., 1995). Similarly, we found no evidence for Scp1 dimerization by using several methods: gel filtration, yeast two-hybrid assay, and coimmunoprecipitation of HA-tagged Scp1 with untagged Scp1 (our unpublished observations). We cannot rule out the possibility that Scp1 dimerizes (e.g., it may dimerize specifically when bound to actin). However, based on the available data, we speculate that Scp1 (and possibly other calponins) cross-link actin filaments via two distinct actin binding sites.
The location of the two sites required for actin filament cross-linking was revealed by the analysis of the mutant proteins. One actin binding site probably resides in the CLR (Figure 1B), because specific mutation of a single residue in the CLR (S185A) abolished actin filament cross-linking and greatly reduced actin binding affinity of Scp1. These results are in agreement with the previous studies that identified the analogous serine residue to be critical for actin binding of other calponin family members (Winder et al., 1993; Tang et al., 1996; Fu et al., 2000). These data also suggest that the mechanism of actin binding by calponins is highly conserved.
Our analysis of the truncated Scp1 constructs revealed the location of a second site required for actin filament cross-linking. The amino-terminal Scp1 fragment His6-N(1–136) containing CH domain did not bind to actin filaments, whereas the carboxy-terminal fragment C(136–200) not only bound to actin filaments, but also cross-linked them. It is formally possible that the hexa-histidine tag interfered with the actin binding of the amino-terminal fragment, yet it seems unlikely, given that the full-length his-tagged Scp1 bound to actin filaments. In addition, single CH domains of mammalian calponin and transgelin are also not sufficient for in vitro binding to actin filaments (Gimona and Mital, 1998; Fu et al., 2000). The second actin binding site or dimerization site of Scp1 must reside in the sequences between the CH domain and the CLR. This site may be analogous to the actin binding site S1 of calponin (Mezgueldi et al., 1995; Mino et al., 1998; Figure 1A). Although the sequence of S1 is not conserved among calponin isoforms or in transgelins (Gimona and Mital, 1998), this region is enriched in positively charged amino acids in all calponin family members. Mutations of the positively charged amino acids in this region decrease actin binding affinity of calponin and transgelin (Gong et al., 1993; Fu et al., 2000). Further mutational analysis of Scp1 will be required for precise identification of the residues required for cross-linking of the actin filaments.
Does the CH domain of Scp1 contribute to actin binding? Our data show clearly that the CH domain is neither sufficient nor necessary for actin filament binding by Scp1, yet Scp1 competes for actin binding with Sac6/fimbrin, which binds actin via two tandem pairs of CH domains. This apparent discrepancy may be explained by a recently proposed model. Based on comparing cryo-electron microscopy reconstructions of calponin and fimbrin decorated actin filaments, it was suggested that the CH domain of calponin may serve as a “locator” domain, helping to position the true actin binding motifs in calponin (reviewed in Winder, 2003). The CH domain of Scp1 may act similarly.
Scp1 Functions with Sac6 to Regulate the Actin Cytoskeleton In Vivo
Our genetic and biochemical data, as well as subcellular localization, reveal a functional relationship between Scp1 and Sac6/fimbrin. Both GFP-Scp1 (this work) and Sac6 (Drubin et al., 1988) localize to cortical actin patches. Sac6 was also reported to colocalize faintly with actin cables by immunofluorescence; however, this was not observed with GFP-Sac6 (Doyle and Botstein, 1996). Therefore, it is possible that Scp1 localizes in vivo to both actin patches and cables, but that we have only been able to detect patch localization with GFP-Scp1. Biochemical analyses show that Scp1 and Sac6 have similar activities on actin. Like Sac6/fimbrin, Scp1 cross-links actin filaments in vitro. Sac6 cross-links actin filaments into tight bundles, and Scp1 cross-links actin into loose bundles and networks. Scp1, like Sac6/fimbrin, not only organizes actin filaments but also decreases the rate of actin filament disassembly (filament stabilization). The shared role of SAC6 and SCP1 in stabilizing the actin cytoskeleton is supported further by the latrunculin A sensitivities of sac6Δ and scp1Δ mutant cells. Together, these in vitro and in vivo observations suggest that Scp1 and Sac6 cooperate in organizing and stabilizing the actin cytoskeleton.
The overlapping genetic functions of SCP1 and SAC6 may be related to their relative abundance in cells and their ability to compete for actin binding. Using quantitative immunoblotting, we defined the in vivo molar ratios of actin, Sac6, and Scp1 to be ∼65:6:1 (actin to Sac6 to Scp1). The higher levels of Sac6 compared with Scp1 suggest that Sac6 may provide the more “dominant” activity on actin. This idea is supported by the relative strengths of their respective null phenotypes. Furthermore, this could explain why as little as two- to threefold higher expression of Scp1 partially suppresses defects in sac6Δ cells.
To demonstrate the importance of the Scp1–actin interaction for in vivo functions, we have used a mutant of Scp1 that has a weak affinity for actin filaments in vitro. scp1S185A failed to suppress loss of SAC6 or loss of SCP1 function in a sac6Δ background. On the other hand, scp1S185D mutant, which retained actin filament binding in vitro, suppressed the phenotypes associated with the loss of SCP1 and SAC6 in vivo. These results provide strong evidence that Scp1–actin interactions are required for in vivo functions of Scp1 shared with Sac6.
A functional relationship between calponins and fimbrin may be conserved in other organisms. Both protein families are widely expressed in different vertebrate tissues and have overlapping subcellular locations. In fibroblasts, fimbrin and calponin are both found on stress fibers (Shapland et al., 1988; Messier et al., 1993; Babb et al., 1997; Jiang et al., 1997;), where they might function together to regulate actin cross-linking and stabilization. In addition, fimbrin and calponin may play a role in adhesive actin structures, linking the actin cytoskeleton to the cell membrane. Fimbrin is found at focal adhesions and podosomes (Messier et al., 1993; Babb et al., 1997), and calponin is found in dense plaques, a type of adherence junction similar to podosomes and focal adhesions (North et al., 1994). Finally, it has been proposed that fimbrin may link the actin cytoskeleton to the vimentin intermediate filament cytoskeleton, and a vimentin-binding site has been mapped to the first CH domain of fimbrin (Correia et al., 1999). A similar function for calponin has been suggested by in vitro binding studies and overlapping in vivo localization of desmin and calponin (North et al., 1994; Mabuchi et al., 1996; Wang and Gusev, 1996). Thus, calponin and fimbrin may have shared in vivo functions that are conserved across a wide range of organisms.
Supplementary Material
Acknowledgments
We thank Brian Cali for generous help during the initiation of this project, Nicki Watson for assistance with microscopy, and Heath Balcer and Avital Rodal for helpful advice and critical reading of the manuscript. The microscopy was conducted in the W.M. Keck Foundation Biological Imaging Facility (Whitehead Institute, Cambridge, MA). A.G. was supported by a predoctoral fellowship from the Howard Hughes Medical Institute. B.G. was supported by a Pew Scholars award, a Basil O'Conner award, and a grant from the National Institutes of Health (GM-63691). P.M. was supported by a grant from the National Institutes of Health (GM-/AR57418). G.F. was supported by a grant from the National Institutes of Health (GM-35010).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–01–0028. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-01-0028.
Abbreviations used: CH calponin homology, CLR calponin-like repeat. Supplementary video: Changes in GFP-Scp1 localization over time (consecutive frames taken at 3-s intervals).
Online version of this article contains video material for some figures. Online version is available at www.molbiolcell.org.
References
- Adams, A.E., Botstein, D., and Drubin, D.G. (1991). Requirement of yeast fimbrin for actin organization and morphogenesis in vivo. Nature 354, 404–408. [DOI] [PubMed] [Google Scholar]
- Ayscough, K.R., Stryker, J., Pokala, N., Sanders, M., Crews, P., and Drubin, D.G. (1997). High rates of actin filament turnover in budding yeast and roles for actin in establishment and maintenance of cell polarity revealed using the actin inhibitor latrunculin-A. J. Cell Biol. 137, 399–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babb, S.G., Matsudaira, P., Sato, M., Correia, I., and Lim, S.S. (1997). Fimbrin in podosomes of monocyte-derived osteoclasts. Cell Motil. Cytoskeleton 37, 308–325. [DOI] [PubMed] [Google Scholar]
- Bretscher, A. (1981). Fimbrin is a cytoskeletal protein that crosslinks F-actin in vitro. Proc. Natl. Acad. Sci. USA 78, 6849–6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson, T.W., Sikorski, R.S., Dante, M., Shero, J.H., and Hieter, P. (1992). Multifunctional yeast high-copy-number shuttle vectors. Gene 110, 119–122. [DOI] [PubMed] [Google Scholar]
- Correia, I., Chu, D., Chou, Y.H., Goldman, R.D., and Matsudaira, P. (1999). Integrating the actin and vimentin cytoskeletons. Adhesion-dependent formation of fimbrin-vimentin complexes in macrophages. J. Cell Biol. 146, 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle, T., and Botstein, D. (1996). Movement of yeast cortical actin cytoskeleton visualized in vivo. Proc. Natl. Acad. Sci. USA 93, 3886–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin, D.G., Miller, K.G., and Botstein, D. (1988). Yeast actin-binding proteins: evidence for a role in morphogenesis. J. Cell Biol. 107, 2551–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp, J.A., and Chant, J. (1997). An IQGAP-related protein controls actin-ring formation and cytokinesis in yeast. Curr. Biol. 7, 921–929. [DOI] [PubMed] [Google Scholar]
- Fu, Y., Liu, H., Forsythe, S., Kogut, P., McConville, J., Halayko, A., Camoretti-Mercado, B., and Solway, J. (2000). Mutagenesis analysis of human SM22: characterization of actin binding. J. Appl. Physiol. 89, 1985–1990. [DOI] [PubMed] [Google Scholar]
- Gimona, M., Djinovic-Carugo, K., Kranewitter, W.J., and Winder, S.J. (2002). Functional plasticity of CH domains. FEBS Lett. 513, 98–106. [DOI] [PubMed] [Google Scholar]
- Gimona, M., and Mital, R. (1998). The single CH domain of calponin is neither sufficient nor necessary for F-actin binding. J. Cell Sci. 111, 1813–1821. [DOI] [PubMed] [Google Scholar]
- Gimona, M., and Winder, S. (1998). Single calponin homology domains are not actin-binding domains. Curr. Biol. 8, R674–R675. [DOI] [PubMed] [Google Scholar]
- Gong, B.J., Mabuchi, K., Takahashi, K., Nadal-Ginard, B., and Tao, T. (1993). Characterization of wild type and mutant chicken gizzard alpha calponin expressed in E. coli. J. Biochem. 114, 453–456. [DOI] [PubMed] [Google Scholar]
- Goode, B.L., Wong, J.J., Butty, A.C., Peter, M., McCormack, A.L., Yates, J.R., Drubin, D.G., and Barnes, G. (1999). Coronin promotes the rapid assembly and cross-linking of actin filaments and may link the actin and microtubule cytoskeletons in yeast. J. Cell Biol. 144, 83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goode, B.L., and Rodal, A.A. (2001). Modular complexes that regulate actin assembly in budding yeast. Curr. Opin. Microbiol. 4, 703–712. [DOI] [PubMed] [Google Scholar]
- Grigoriev, V.G., Thweatt, R., Moerman, E.J., and Goldstein, S. (1996). Expression of senescence-induced protein WS3–10 in vivo and in vitro. Exp. Gerontol. 31, 145–157. [DOI] [PubMed] [Google Scholar]
- Guthrie, C., and Fink, R. (1991). Guide to yeast genetics and molecular biology. Methods Enzymol. 194, 1–933. [PubMed] [Google Scholar]
- Jiang, Z., Grange, R.W., Walsh, M.P., and Kamm, K.E. (1997). Adenovirus-mediated transfer of the smooth muscle cell calponin gene inhibits proliferation of smooth muscle cells and fibroblasts. FEBS Lett. 413, 441–445. [DOI] [PubMed] [Google Scholar]
- Jones, E. (2002). Vacuolar proteases and proteolytic artifacts in Saccharomyces cerevisiae. Methods Enzymol. 351, 127–150. [DOI] [PubMed] [Google Scholar]
- Kobayashi, R., Kubota, T., and Hidaka, H. (1994). Purification, characterization, and partial sequence analysis of a new 25-kDa actin-binding protein from bovine aorta: a SM22 homolog. Biochem. Biophys. Res. Commun. 198, 1275–1280. [DOI] [PubMed] [Google Scholar]
- Kolakowski, J., Makuch, R., Stepkowski, D., and Dabrowska, R. (1995). Interaction of calponin with actin and its functional implications. Biochem. J. 306, 199–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubler, E., and Riezman, H. (1993). Actin and fimbrin are required for the internalization step of endocytosis in yeast. EMBO J. 12, 2855–2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson, D., Harrison, M., and Shapland, C. (1997). Fibroblast transgelin and smooth muscle SM22alpha are the same protein, the expression of which is down-regulated in many cell lines. Cell Motil. Cytoskeleton 38, 250–257. [DOI] [PubMed] [Google Scholar]
- Lees-Miller, J.P., Heeley, D.H., and Smillie, L.B. (1987a). An abundant and novel protein of 22 kDa (SM22) is widely distributed in smooth muscles. Purification from bovine aorta. Biochem. J. 244, 705–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees-Miller, J.P., Heeley, D.H., Smillie, L.B., and Kay, C.M. (1987b). Isolation and characterization of an abundant and novel 22-kDa protein (SM22) from chicken gizzard smooth muscle. J. Biol. Chem. 262, 2988–2993. [PubMed] [Google Scholar]
- Lippincott, J., and Li, R. (1998). Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J. Cell Biol. 140, 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., and Bretscher, A. (1989). Purification of tropomyosin from Saccaromyces cerevisiae and identification of related proteins in Schizosaccharomyces and physarum. Proc. Natl. Acad. Sci. USA 86, 90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S., Thweatt, R., Lumpkin, Jr., C.K., and Goldstein, S. (1994). Suppression of calcium-dependent membrane currents in human fibroblasts by replicative senescence and forced expression of a gene sequence encoding a putative calcium-binding protein. Proc. Natl. Acad. Sci. USA 91, 2186–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, F.W., Freedman, M.V., and Chalovich, J.M. (1995). Characterization of calponin binding to actin. Biochemistry 34, 11864–11871. [DOI] [PubMed] [Google Scholar]
- Mabuchi, K., Li, Y., Tao, T., and Wang, C.L. (1996). Immunocytochemical localization of caldesmon and calponin in chicken gizzard smooth muscle. J. Muscle Res. Cell Motil. 17, 243–260. [DOI] [PubMed] [Google Scholar]
- Matsudaira, P. (1991). Modular organization of actin cross-linking proteins. Trends Biochem. Sci. 16, 87–92. [DOI] [PubMed] [Google Scholar]
- Messier, J.M., Shaw, L.M., Chafel, M., Matsudaira, P., and Mercurio, A.M. (1993). Fimbrin localized to an insoluble cytoskeletal fraction is constitutively phosphorylated on its headpiece domain in adherent macrophages. Cell Motil. Cytoskeleton 25, 223–233. [DOI] [PubMed] [Google Scholar]
- Mezgueldi, M., Mendre, C., Calas, B., Kassab, R., and Fattoum, A. (1995). Characterization of the regulatory domain of gizzard calponin. Interactions of the 145–163 region with F-actin, calcium-binding proteins, and tropomyosin. J. Biol. Chem. 270, 8867–8876. [DOI] [PubMed] [Google Scholar]
- Mino, T., Yuasa, U., Nakamura, F., Naka, M., and Tanaka, T. (1998). Two distinct actin-binding sites of smooth muscle calponin. Eur. J. Biochem. 251, 262–268. [DOI] [PubMed] [Google Scholar]
- Morgan, K., and Gangopadhyay, S. (2001). Invited review: cross-bridge regulation by thin filament-associated proteins. J. Appl. Physiol. 91, 953–962. [DOI] [PubMed] [Google Scholar]
- North, A.J., Gimona, M., Cross, R.A., and Small, J.V. (1994). Calponin is localised in both the contractile apparatus and the cytoskeleton of smooth muscle cells. J. Cell Sci. 107, 437–444. [DOI] [PubMed] [Google Scholar]
- Peranen, J., Rikkonen, M., Hyvonen, M., and Kaariainen, L. (1996). T7 vectors with modified T7lac promoter for expression of proteins in Escherichia coli. Anal. Biochem. 236, 371–373. [DOI] [PubMed] [Google Scholar]
- Pollard, T.D., and Cooper, J.A. (1982). Methods to characterize actin filament networks. Methods Enzymol. 85, 211–233. [DOI] [PubMed] [Google Scholar]
- Prinjha, R.K., Shapland, C.E., Hsuan, J.J., Totty, N.F., Mason, I.J., and Lawson, D. (1994). Cloning and sequencing of cDNAs encoding the actin cross-linking protein transgelin defines a new family of actin-associated proteins [published erratum in Cell Motil. Cytoskeleton 1994;29(4):383]. Cell Motil. Cytoskeleton 28, 243–255. [DOI] [PubMed] [Google Scholar]
- Pruyne, D., and Bretscher, A. (2000). Polarization of cell growth in yeast. J. Cell Sci. 113, 571–585. [DOI] [PubMed] [Google Scholar]
- Sandrock, T.M., O'Dell, J.L., and Adams, A.E. (1997). Allele-specific suppression by formation of new protein-protein interactions in yeast. Genetics 147, 1635–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapland, C., Hsuan, J.J., Totty, N.F., and Lawson, D. (1993). Purification and properties of transgelin: a transformation and shape change sensitive actin-gelling protein. J. Cell Biol. 121, 1065–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapland, C., Lowings, P., and Lawson, D. (1988). Identification of new actin-associated polypeptides that are modified by viral transformation and changes in cell shape. J. Cell Biol. 107, 153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small, J.V., and Gimona, M. (1998). The cytoskeleton of the vertebrate smooth muscle cell. Acta Physiol. Scand. 164, 341–348. [DOI] [PubMed] [Google Scholar]
- Stafford, W.F., 3rd, Mabuchi, K. Takahashi, K., and Tao, T. (1995). Physical characterization of calponin. A circular dichroism, analytical ultracentrifuge, and electron microscopy study. J. Biol. Chem. 270, 10576–10579. [DOI] [PubMed] [Google Scholar]
- Tang, D.C., Kang, H.M., Jin, J.P., Fraser, E.D., and Walsh, M.P. (1996). Structure-function relations of smooth muscle calponin. The critical role of serine 175. J. Biol. Chem. 271, 8605–8611. [DOI] [PubMed] [Google Scholar]
- Tang, J.X., Szymanski, P.T., Janmey, P.A., and Tao, T. (1997). Electrostatic effects of smooth muscle calponin on actin assembly. Eur. J. Biochem. 247, 432–440. [DOI] [PubMed] [Google Scholar]
- Thweatt, R., Lumpkin, Jr., C.K., and Goldstein, S. (1992). A novel gene encoding a smooth muscle protein is overexpressed in senescent human fibroblasts. Biochem. Biophys. Res. Commun. 187, 1–7. [DOI] [PubMed] [Google Scholar]
- Wang, P., and Gusev, N.B. (1996). Interaction of smooth muscle calponin and desmin. FEBS Lett. 392, 255–258. [DOI] [PubMed] [Google Scholar]
- Warren, D.T., Andrews, P.D., Gourlay, C.W., and Ayscough, K.R. (2002). Sla1p couples the yeast endocytic machinery to proteins regulating actin dynamics. J. Cell Sci. 115, 1703–1715. [DOI] [PubMed] [Google Scholar]
- Winder, S.J. (2003). Structural insights into actin binding, branching, and bundling proteins. Curr. Opin. Cell Biol. 15, 14–22. [DOI] [PubMed] [Google Scholar]
- Winder, S.J., Allen, B.G., Fraser, E.D., Kang, H.M., Kargacin, G.J., and Walsh, M.P. (1993). Calponin phosphorylation in vitro and in intact muscle. Biochem. J. 296, 827–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.