Abstract
The dynein activator dynactin is a multiprotein complex with distinct microtubule- and cargo-binding domains. The cargo-binding domain contains a short, actin-like filament of the actin-related protein Arp1, a second actin-related protein, Arp11, and conventional actin. The length of this filament is invariant in dynactin isolated from multiple species and tissues, suggesting that activities that regulate Arp1 polymerization are important for dynactin assembly. Arp11 is present in a protein complex localized at the pointed end of the Arp1 minifilament, whereas actin capping protein (CapZ) is present at the barbed end. Either might cooperate with conventional actin to cap Arp1. We tested the ability of Arp11 to interact with conventional actin and found it could coassemble. Like Arp1, cytosolic Arp11 is found only in dynactin, suggesting that Arp11 and free cytosolic actin do not interact significantly. Recombinant Arp11 and Arp1 were demonstrated to interact by coprecipitation. We developed an in vivo assay for Arp11–Arp1 interaction based on previous observations that Arp1 forms filamentous assemblies when overexpressed in cultured cells. Arp11 significantly decreases the formation of these organized Arp1 assemblies. Finally, this assay was used to confirm the identity of a putative Arp11 homolog in Drosophila melanogaster.
INTRODUCTION
The actin superfamily is present in all eukaryotic organisms whose genome sequences have been determined. Careful analysis of green fluorescent protein (GFP)-tagged actin-related proteins (Arps) in the budding yeast Saccharomyces cerevisiae revealed that they localize to a range of intracellular compartments. Arps can be found in the nucleus (Arp4 and Arp6, Arp7 and Arp9), cytoplasm (Arp1) or both (Arp2 and Arp3) (Andersen et al., 2002; Goodson and Hawse, 2002). Arps seem to function in heterologous pairs (reviewed in Schafer and Schroer, 1999) such as Arp7/Arp9 (Cairns et al., 1999) or Arp2/Arp3 (Kelleher et al., 1995), a fact that remains unexplained to date. Arp functions are as varied as their localization. Nuclear Arps are believed to play a role in chromatin remodeling (Peterson et al., 1998; Boyer and Peterson, 2000; Kato et al., 2001), whereas cytosolic Arps contribute to actin filament assembly and organization (Suetsugu et al., 2001; Weaver et al., 2001) and intracellular vesicular trafficking (Holleran et al., 1998). Arps2 and 3 are important regulators of cytoskeletal dynamics, but other Arps seem to function primarily as structural proteins.
In vertebrate cells, the dynein accessory protein dynactin is required for the processive dynein-based movement of vesicular cargo along microtubules (Gill et al., 1991; Schroer and Sheetz, 1991). Dynactin has also been implicated in cortical anchoring of spindle microtubules (Busson et al., 1998), attachment of interphase microtubules to adhesion plaques (Ligon et al., 2001; Palazzo et al., 2001), and microtubule anchoring at centrosomes (Quintyne et al., 1999). Dynactin contains two different actin-related proteins, Arp1 and Arp11. Arp1 forms a short polymer of uniform size that has proteins bound along its length and at both ends. One end terminates in the conventional actin capping protein CapZ, suggesting an analogy to the Arp1 filament “barbed” end (Schafer et al., 1994). The other end of the Arp1 polymer is associated with a tetrameric protein assembly referred to as the pointed end complex (Eckley et al., 1999). This dynactin subcomplex contains Arp11 plus the dynactin subunits p62, p27, and p25.
Careful analysis of the composition of highly purified dynactin reveals one monomer of conventional β-actin in each dynactin molecule (Schafer et al., 1994; Bingham and Schroer, 1999), although some dynactin preparations are reported to lack actin (Holleran et al., 1996). We assume actin coassembles with Arp1, but its exact location in the 37-nm minifilament is unknown. Either CapZ or p62 might interact with actin, because both proteins can bind actin filaments (Cooper and Pollard, 1985; Garces et al., 1999). Because p62 and CapZ are found at opposite ends of the Arp1 minifilament (Schafer et al., 1994), a rigorous assignment of actin's location within dynactin has not been possible. In vitro translated Arp1 has been reported to bind to, and cycle with, actin (Melki et al., 1993), suggesting actin may be randomly incorporated in the Arp1 minifilament. However, if actin's position in the Arp1 minifilament is fixed, it might provide insight into the assembly pathway of dynactin.
The predicted structure of Arp11 suggests it has the potential to bind conventional actin or Arp1 and possibly contribute to filament assembly. The barbed end face of Arp11 is relatively conserved (Eckley et al., 1999), suggesting it can interact with filament pointed ends. Arp11 might function like Arp2/3 complex to nucleate assembly of either actin or Arp1 filaments. Actin assembly is well known to be governed in this way, but Arp1 has a vanishingly low critical concentration for polymerization (<1 nM) and assembles without a lag phase, suggesting nucleation is not required (Bingham and Schroer, 1999). Interestingly, the filaments formed by pure, isolated Arp1 are variable in length, unlike the Arp1 filaments in dynactin, suggesting that other dynactin components provide a ruler activity that governs Arp1 assembly. Arp1 filaments are also able to undergo end-to-end annealing (Bingham and Schroer, 1999), a behavior never seen for dynactin. CapZ and subunits of the pointed end complex are believed to cap the ends of the Arp1 filament and prevent annealing. Of the four pointed-end complex subunits, Arp11 and p62 are most tightly associated with dynactin and are therefore the dynactin subunits most likely to be bound directly to Arp1.
In the present study, we explore the actin-binding properties of Arp11 in copelleting and cycling assays. Fractionation and immunoblot analysis of cultured cells to characterize the subcellular distribution of cytosolic Arp11 reveal that Arp11 is associated exclusively with dynactin. In vitro translated Arp11 and Arp1 can be coprecipitated, suggesting they bind each other. The absence of a free Arp11 pool in cells allowed us to develop an assay for Arp1/Arp11 interactions in vivo. Coexpression of Arp11 and Arp1 decreases the number of Arp1 cables that form when Arp1 is overexpressed in cultured cells. Finally, we showed that a putative fruit fly homolog of Arp11 acts similarly to vertebrate Arp11, verifying its identity as Arp11 and highlighting the significance of the Arp11/Arp1 interaction.
MATERIALS AND METHODS
T7 Expression Constructs
T7-luciferase (T7 control plasmid; T7 TnT kit) was from Promega (Madison, WI). T7-β-actin was kindly provided by S. Lewis and N. Cowan (Melki et al., 1993) Human Arp1 was cloned in pET3d as described previously (Lees-Miller et al., 1992). To make Arp11, a HindIII fragment from the full-length Arp11 clone (Eckley et al., 1999) was cloned into the pRSET-B (Invitrogen, San Diego, CA) to generate T7-Arp11(23–417). The T7-Arp11 (23–417) construct was digested with NcoI. The smaller fragment was cloned into the pR-SET-B NcoI site to generate T7-Arp11 (23–137) and the larger NcoI fragment was religated to generate T7-Arp11 (138–417). To make a full-length Arp11 construct, we removed upstream, noncoding sequences from the open reading frame by polymerase chain reaction. The upstream primer GCGGATCCATGCATCATCATCATCATCATATGCCGCTCTACGAG appends a 6-histidine tag (His tag) to the N terminus of Arp11. Addition of the downstream primer CCACAATCCAGGACCATGGCCG allowed amplification of a 450-base pair fragment of Arp11 cDNA, with a His tag. The fragment was cloned into T7-Arp11 (138–417) by using BamHI and NcoI to generate T7-His-Arp11 (out of frame). T7-His-Arp11(1–417) was made by digesting T7-His-Arp11(out of frame) with BamHI, filling in overhangs with Klenow fragment (New England BioLabs, Beverly, MA) and religating. T7-His-GFP-Arp11 was made by cloning the appropriate NcoI fragment from GFP-Arp11 (1–417) into the NcoI site of T7-Arp11 (138–417).
In Vitro Transcription/Translation
Coupled transcription/translation reactions were carried out as directed by the manufacturer (TnT; Promega). Plasmid DNA and [35S]methionine were added to TnT mix and the reaction was incubated at 30°C for 90 min. Test proteins were prepared by 20-fold dilution of each TnT reaction with G-buffer (2 mM Tris-Cl, pH 8, 0.2 mM ATP, 0.2 mM CaCl2, 0.5 mM dithiothreitol) followed by a clearing spin to remove aggregates (186k g-hour: 55 krpm, 1 h, 20°C; TLA-55 rotor). In some experiments, 100-μl aliquots were snap frozen in liquid nitrogen and stored at –80°C until use.
Actin Copelleting/Cycling
Skeletal muscle actin or cytoplasmic actin (1-mg aliquots) were purchased from Cytoskeleton (Denver, CO). Actin was diluted in fresh G-buffer to a concentration of 200 μM and cleared of aggregates (186k g, 1 h) before use. Binding and cycling experiments were performed using [35S]d-methionine (PerkinElmer Life Sciences, New Bedford, MA) labeled test proteins that were freshly prepared or frozen in aliquots (see above). Actin was added to the precleared test proteins at a concentration of 5 μM and the sample was split equally into two tubes. Then 10× KMEI (500 mM KCl, 10 mM MgCl2, 10 mM EGTA, 100 mM imidazole, pH 7.0) was carefully added to final concentration of 1× in one tube to start actin polymerization; G-buffer was added to the other. After 4 h at 20°C, actin filaments were pelleted by a 186k g-hour spin. The supernatants were removed and the pellet surfaces gently washed with G-buffer. Next, the pellets were resuspended in G-buffer and incubated on ice overnight to depolymerize the actin filaments. The samples were centrifuged for 186k g-hour to remove insoluble protein aggregates. Then 10× KMEI was added to the supernatant to induce a second round of polymerization. Finally, actin filaments were pelleted at 186k g-hour. Aliquots of the supernatants and pellets from all preceding steps were analyzed by SDS-PAGE and autoradiography. Pellet resuspension buffer (30 mM Tris-HCl, pH 6.8, 5% glycerol, 2.5% β-mercaptoethanol, 0.5 mM EDTA, 6 M urea) was used to carefully resuspend each pellet (after a wash with G-buffer) in the exact volume of the starting supernatant. For actin-binding assays, aliquots of the pellet and supernatant fractions were analyzed by SDS-PAGE. Gels were fixed and stained in 0.5 M sodium salicylate in 30% methanol containing 1 mg/ml Coomassie Blue R-250 for 30 min at room temperature, and then destained in two changes of 0.5 M sodium salicylate in 30% methanol. Gels were dried, scanned (Scanmaker III; Microtek, Redondo Beach, CA) to document the behavior of actin, and exposed to x-ray film for 4–48 h at –80°C with an intensifying screen.
Bead Precipitation
In vitro-translated proteins were prepared as described above. Test protein (untagged Arp1) was diluted 1:10 in buffer and precleared with Talon beads (BD Biosciences Clontech, Palo Alto, CA). The supernatant was used immediately for precipitation studies. His-Arp11 or His-GFP-Arp11 were mixed with fresh Talon beads on ice. Arp1 was added to beads alone, or with beads plus tagged Arp11 and allowed to bind for 15 min on ice. The beads were pelleted at 200 × g for 1 min, the supernatant was removed, and the beads were washed four times before resuspending in buffer at the original volume of binding mixture. Then 50% of each pellet sample was boiled in sample buffer and loaded on a 10% polyacrylamide gel. The gel was processed for autoradiography and exposed to film, as described above.
Cell Fractionation
Cells were harvested by trypsinzation or scraping with a rubber spatula and resuspended in cell lysis buffer [10 mM HEPES, pH 7.4, 150 mM NaCl, 1 μg/ml each leupeptin, pepstatin, Nα-p-tosyl-L-arginine methyl ester, Nα-benzoyl-L-arginine methyl ester, N-tosyl-l-phenylalanine chloromethyl ketone, N-tosyl-l-lysine chloromethyl ketone, and 4-(2-aminoethyl)benzenesulfonyl fluoride]. Cells were pelleted in a Dynac clinical centrifuge at setting 30% for 5 min. The pellet volume was determined and an equal volume of buffer was added to resuspend the cells. Cells were homogenized using a ball bearing homogenizer of inner diameter 8.020 mm containing a ball bearing of 8.014 mm. A nuclear pellet (P1) was prepared by a 9300-rpm spin in a swinging bucket rotor (Eppendorf 5417; 12-place rotor), for 10 min at 4°C. The supernatant was cleared of all membranes by ultracentrifugation at 55 krpm for 1 h at 4°C in a Beckman TLA55 fixed angle rotor. The high-speed cytosol (S2) was saved for further analysis. The pellets (P1 and P2) were resuspended in cell lysis buffer to the same volume as the original cell pellet.
Sucrose Gradient Fractionation
Sucrose gradients (5–20% in G-buffer) were prepared using a 15-ml Hoefer gradient maker and drill-propelled rotary stirrer and pumped into 14 × 89 mm ultraclear tubes. The resulting 11.6-ml gradients were overlaid with up to 500 μl of sample (e.g., diluted in vitro transcription/translation mix or S2), and subjected to ultracentrifugation at 34 krpm for 16.5 h at 4°C in an SW41 rotor. A gradient containing sedimentation standard proteins (thyroglobulin, 18.2S; catalase, 11.6S; β-amylase, 9S; alcohol dehydrogenase, 7.5S; and serum albumin, 4.2S) was included in every run. Fractions (1 ml) were collected with a pump from the bottom of each gradient and stored on ice before SDS-PAGE analysis. Pellets were resuspended in 1 ml of pellet resuspension buffer (see above).
Immunoblotting and Antibodies
Samples were analyzed by SDS-PAGE and transferred to polyvinylidene difluoride membranes (Immobilon-P; Millipore, Bedford, MA) overnight at 20 V in Towbin transfer buffer (Towbin et al., 1979). Transfer was verified by the appearance of prestained Marker proteins (Bio-Rad, Hercules, CA), Pyronin Y tracking dye (Sigma-Aldrich, St. Louis, MO) and Ponceau S (Sigma-Aldrich) stain for total protein. Proteins of interest were detected using monoclonal or polyclonal antibodies followed by alkaline phosphatase-conjugated secondary antibodies and chemiluminescence according to the manufacturer's instructions (Tropix, New Bedford, MA). Dynactin subunits p150Glued and Arp1 were detected with monoclonal antibodies 150B and 45A (Schafer et al., 1994). Monoclonal anti-GFP and antiactin (C4; Lessard, 1988) antibodies were purchased from Babco (Berkeley, CA). A polyclonal serum recognizing Arp11 (Michael Way, Imperial Cancer Research Fund, London, United Kingdom) was generated by immunizing rabbits with synthesized peptides corresponding to the Arp11 N terminus or C termini.
GFP Fusion Protein Expression
An N-terminal GFP fusion with Arp11 (AA 23–417) was created by cloning the HindIII fragment from a full-length Arp11 clone (Eckley et al., 1999) into the HindIII site of pEGFP-C2 (BD Biosciences Clontech). An N-terminal fusion of GFP to the complete Arp11 open reading frame (residues 1–417) was generated in a two-step process. First, a SalI/BstXI fragment from expressed sequence tag (EST) AA250667 was ligated into the above-mentioned construct digested with XhoI and BstXI. Second, a BamHI/BglII fragment (containing the complete Arp11 open reading frame) was cloned into pEGFP-C1 (BglII digested) to make GFP-Arp11 (1–417). The Drosophila melanogaster Arp11 open reading frame (EST LD36140) was fused downstream of GFP by cloning an EcoRI/XhoI fragment into pEGFP-C1 digested with EcoRI and SalI.
Electroporation and Immunostaining
Fifteen micrograms of each DNA construct was mixed with 2 × 106 cells in a volume of 400 μl just before electroporation. HeLa or COS-7 cells were electroporated at 280V, 1600 μF (BTX ECM600) in 4-mm gap cuvettes. Cells were diluted in Opti-MEM (1.5 ml/cuvette) and incubated at room temperature for 20 min before plating onto coverslips in complete medium containing serum. Cell cultures were grown overnight at 37°C under 5% CO2 atmosphere. At 18–24 h postplating, coverslips were fixed for 5 min in –20°C methanol. Fixed cells were rehydrated by three changes of blocking buffer (phosphate-buffered saline + 1% bovine serum albumin). Primary antibodies were diluted into blocking buffer and coverslips were incubated for 30 min at room temperature. After three washes with blocking buffer, the coverslips were incubated with secondary antibodies for 30 min at room temperature. Final washes were performed in the presence of 4,6-diamidino-2-phenylindole (10 ng/ml) to visualize chromatin. Coverslips were mounted in MOWIOL containing 1 mg/ml N-propyl gallate. Cells were viewed through an Axiovert microscope (Carl Zeiss, Jena, Germany) equipped with a 100× Plan-Neofluor objective, numerical aperture 1.3, and GFP/fluorescein and Texas Red filters (Chroma Technologies, New Bedford, MA).
Genomic Analysis
Putative dynactin subunit genes were identified by BLAST search of the predicted gene product database, at the Biotechnology and Biological Sciences Research Council, Institute for Animal Health Chicken EST repository Web site, http://www.chick.umist.ac.uk/, the Berkeley Drosophila Genome project Web site, http://www.fruitfly.org/, the Welcome Trust Sanger Institute Web site, http://www.sanger.ac.uk/Projects/C_elegans/blast_server.shtml, or the Neurospora crassa database (Munich information centre for protein Neurospora crassa database), http://mips.gsf.de/proj/neurospora/. Direct alignments between imported amino acid sequences representing the mouse and putative chicken, fly, worm, or fungus dynactin subunits were performed at the National Center for Biotechnology Information BLAST 2 sequences Web site, http://www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html.
Arp1, CapZ β, CapZα, p25, p27, Arp11, p62, p150Glued, p50 (dynamitin), and p24 were found at the following loci, respectively: D. melanogaster, CG6174, CG17158, CG10540, CG10846, CG17347, CG12235, CG12042, CG9206, CG8269, and CG9893; and Caenorhabditis elegans, Y53F4, M106.5, D2024.6, Y71F9AL, Y54E10A, C49H3.9, C26B2.2, ZK593, C28H8.8, and W02A2.2. Neurospora crassa Arp1 is encoded by Ropy 4, p25 by Ropy-12, Arp11 by Ropy-7, p62 by Ropy-2, and p150Glued by Ropy-3. Chicken (Gallus gallus) sequences were identified in the EST repository except p150Glued and dynamitin whose sequences are published (Gill et al., 1991; Quintyne et al., 1999; Valetti et al., 1999).
RESULTS
Binding and Cycling of Arp11 and Actin
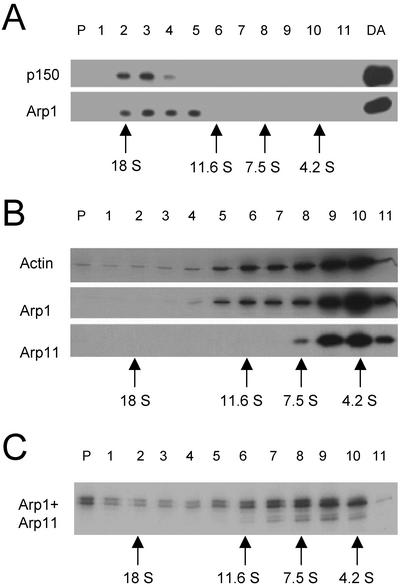
To pursue the question of whether Arp11 can bind and/or copolymerize with conventional actin, we expressed recombinant mouse Arp11 by in vitro translation by using a T7-based expression system (Studier, 1991). Sucrose gradient fractionation of the reticulocyte lysate without added Arp11 cDNA revealed endogenous dynactin which sedimented at about 18S (Figure 1A). Before assaying the interactions of Arp11 with actin, we used velocity sedimentation to determine how much in vitro translated Arp11 was present in a “free pool” and how much had been incorporated into reticulocyte dynactin (Figure 1B). In vitro-translated Arp1, Arp11, and actin were all found predominately near the top of the sucrose gradient, suggesting that they were not incorporated into reticulocyte dynactin during the 90 min required for expression.
Figure 1.
Characterization of endogenous and recombinant dynactin subunits in reticulocyte extracts. (A) Immunoblot analysis of in vitro translation mixture subjected to velocity sedimentation into a 5–20% sucrose gradient. P, gradient pellet; DA, purified bovine brain dynactin. Blots were probed with antibodies to p150Glued and Arp1, and then processed for chemiluminescence. (B) Velocity sedimentation (5–20% sucrose) of in vitro translation mixtures containing in vitro translated β-actin, Arp1, or Arp11. P, gradient pellet. (C) Velocity sedimentation (5–20% sucrose) of an in vitro translation mixture containing cotranslated Arp1 and Arp11. B and C are autoradiograms of the dried gels.
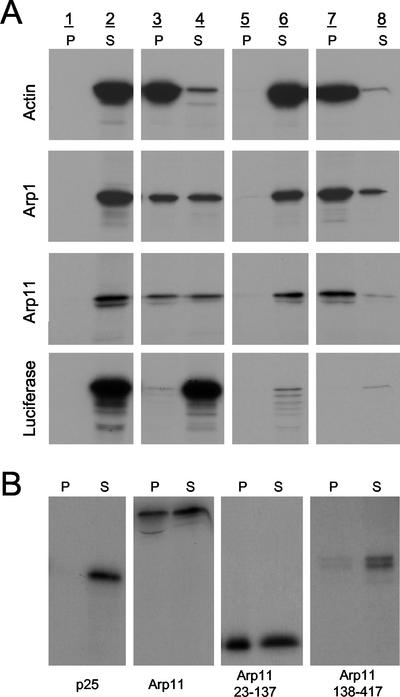
Next, we tested the ability of in vitro translated Arp11 to coassemble with purified actin. The behaviors of in vitro-translated Arp1, human β-actin, and luciferase were analyzed as controls. Proteins were mixed with purified rabbit skeletal muscle actin and subjected to two cycles of actin filament assembly and disassembly (Figure 2A). That none of the test proteins pelleted in G-buffer (lane 1) indicates they all were soluble. To verify that the proteins that copelleted with F-actin (lane 3) were truly coassembled, the resulting F-actin pellet was depolymerized, cleared of aggregates (lane 5), and the supernatant (lane 6) subjected to a second round of polymerization. The final pellets (lane 7) contain proteins that are able to cycle with F-actin. As reported previously (Melki et al., 1993), Arp1 cycled with purified actin. Arp11 was also found to cycle with skeletal muscle actin. A small amount of luciferase was trapped in the initial F-actin pellet but did not cycle. Similar results were obtained when purified platelet cytoplasmic actin was used in place of skeletal muscle actin (our unpublished data).
Figure 2.
Actin coassembly and binding assays. (A) In vitro-translated proteins were mixed with actin and subjected to two cycles of polymerization as described in the text. Equal proportions of pellet and supernatant (P and S) were separated on 11% polyacrylamide gels and subjected to autoradiography. Lanes 1 and 2, G-buffer; lanes 3 and 4, F-buffer; and lanes 5 and 6: The pellet from lane 4 resuspended in G-buffer. Lanes 7 and 8, lane 6 sample plus F-buffer. (B) Arp11 subdomains (23–137 and 138–417) were compared with an Arp11 N-terminal truncation (23–417, as used in A) and p25 (a control dynactin subunit). The P and S lanes are the equivalent of lanes 3 and 4 in A.
We then mapped the Arp11 domain required for actin binding (Figure 2B). An N-terminal fragment (AA 23–137) could copellet with actin, whereas a nonoverlapping C-terminal fragment (AA 138–417) could not. Thus, a minimal actin-binding domain of Arp11 is present in amino acids 23–137.
Arp11 Is Present in Membrane-associated and Cytosolic Pools
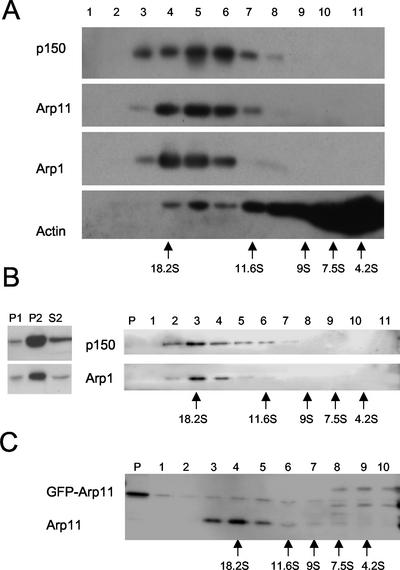
Recombinant Arp1 can bind and coassemble with actin (Figure 2; Melki et al., 1993) but Arp1 is not believed to contribute to actin dynamics in vivo because a free Arp1 pool does not exist in any cell type examined to date (Paschal et al., 1993; Echeverri et al., 1996; Quintyne et al., 1999; Valetti et al., 1999). Arp11 can also bind and coassemble with actin, raising the possibility that it might play a role outside dynactin, perhaps to regulate the behavior of the actin cytoskeleton. To explore this possibility, we first assayed cells for a free pool of Arp11. Cytosol prepared from cultured COS-7 cells was subjected to velocity sedimentation into a 5–20% sucrose gradient and the resulting gradient fractions were analyzed by immunoblotting (Figure 3A). Most cytoplasmic actin sedimented in the low-density fractions; notably, a small but detectable pool was found at about 18S. As expected, the dynactin subunits Arp1 and p150Glued sedimented in a peak at about 18S, consistent with assembly into a large complex. Arp11 behaved indistinguishably from Arp1 and p150Glued, suggesting that it, too, is present only in dynactin. Similar results were obtained using HeLa cytosol (our unpublished data).
Figure 3.
Sucrose gradient analysis of cytosolic and membrane proteins. (A) High-speed cytosol from COS-7 cells was sedimented into a 5–20% sucrose gradient and fractions were analyzed by immunoblotting. Sedimentation standards (analyzed on a separate gradient) are indicated. (B) Subcellular fractionation of HeLa cells. P1, nuclear fraction; P2, high-speed membrane pellet; S2, cytosol Ten percent of the P1, 10% of the P2, and 1% of the S2 were loaded on the gel. The high-speed membrane pellet (P2) was extracted with 0.1% NP40 and the supernatant sedimented into a 5–20% sucrose gradient. Fractions were assayed by immunoblot. (C) Sucrose gradient fractionation of COS-7 cytosol isolated from cells overexpressing GFP-Arp11. Gradient fractions were blotted and probed with anti-Arp11 antibody. The positions of endogenous Arp11 and GFP-Arp11 are indicated. The bands flanking GFP-Arp11 in lanes 8, 9, and 10 are nonspecific immunoreactive bands.
This and all previously published work on dynactin structure and composition have focused on the cytosolic pool of dynactin. When we surveyed different subcellular fractions we found considerable dynactin (i.e., p150Glued, Arp1, and Arp11 subunits) in the high-speed membrane pellet (Figure 3B, P2; Bingham et al., 1998). To determine how similar this pool was to cytosolic dynactin, we solubilized the membrane pellet with nonionic detergent (NP40). NP40 caused complete release of the dynactin subunits p150Glued and Arp1 from the residual membrane pellet (our unpublished data). We then sedimented the resulting NP40-soluble supernatant into a sucrose gradient (Figure 3B). Arp1 and p150Glued both sedimented at approximately 18S, similar to their behavior in NP40 lysates of whole cells (Echeverri et al., 1996; Quintyne et al., 1999; Valetti et al., 1999). This indicates that NP40 does not significantly affect the integrity of membrane-associated dynactin and further reveals that membrane-associated and cytosolic dynactins are similar.
Arp11 Can Bind Arp1 in Solution
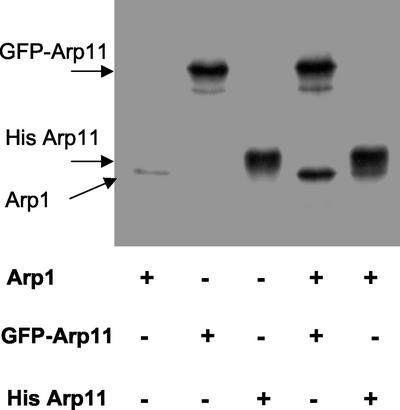
The data in Figure 2 indicate clearly that Arp11 can bind and coassemble with conventional actin. Unfortunately, we could not perform a similar analysis of Arp11/Arp1 binding owing to the difficulty in obtaining purified Arp1 and the fact that Arp1 polymers do not cycle (Bingham, Ph.D. thesis). In the course of the actin cocycling experiments, we noted that in vitro cotranslation of Arp1 and Arp11 yields proteins that sediment further into sucrose gradients than the individual species (Figure 1, compare C and B). This suggested that Arp1 and Arp11 might form complexes in solution. We explored this possibility by examining the ability of the two in vitro translated proteins to be coprecipitated. Arp1 and Arp11 normally migrate very closely on SDS-PAGE, so to ensure they would be resolved we engineered two hexahistidine-tagged forms of Arp11 (His-Arp11 and His-GFP-Arp11). Both were assayed for their ability to coprecipitate untagged Arp1 (Figure 4). Little, if any, Arp1 was precipitated by beads alone, but both His-tagged forms of Arp11 coprecipitated Arp1.
Figure 4.
Coprecipitation of in vitro-translated Arp1 and Arp11. Talon beads were mixed with untagged Arp1, GFP-His-Arp11, His-Arp11, or mixtures of two proteins as indicated below the gel. The beads were then washed and analyzed by SDS-PAGE and autoradiography.
Arp11 Can Associate with Arp1 in Cultured Cells
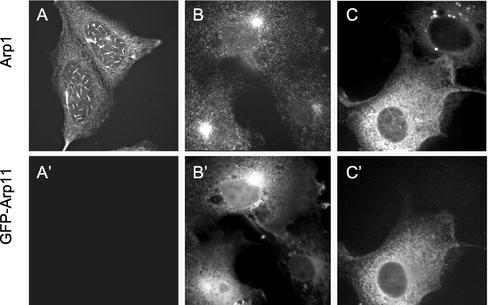
We extended our in vitro analysis of Arp1/Arp11 binding by examining the ability of the two proteins to interact within cells. To do this, we took advantage of the fact that Arp1 forms organized aggregates when overexpressed in cultured PtK cells (Holleran et al., 1996). Arp1 overexpression in HeLa cells yields a variety of localization patterns (Figure 5). The most prevalent is a punctate distribution with an accumulation at centrosomes that is similar to untransfected controls (Figure 5B). However, a subset of cells (30–40%; Table 1) exhibit a striking phenotype in which Arp1 “cables” accumulate on or near nuclei (Figure 5, A and C; Holleran et al., 1996). Some cells contained large Arp1 aggregates (our unpublished data), but these were excluded from further analysis. Arp1-overexpressing cells exhibited variable numbers of cables and the cables varied in size. Overexpression of GFP-tagged Arp11, in contrast, yielded a punctate, diffuse pattern (Figure 5, B′ and C′) in all cells examined. We confirmed that GFP-Arp11 is functional by evaluating its incorporation into dynactin. Sedimentation of a high-speed cytosol into a 5–20% sucrose gradient revealed a sizeable population of GFP-Arp11 that sedimented at 20S (Figure 3C), suggesting it can be assembled into dynactin.
Figure 5.
Phenotypes of Arp1 and GFP-Arp11 transfected HeLa cells. (A–C) Arp1 immunostaining pattern in transfected cells showing (A) Arp1 cables, (B) punctate Arp1, and (C) Arp1 cables and punctae. The cables in C are slightly out of focus but can be seen within the nuclear perimeter. (A′–C′) GFP-Arp11 in the same cells.
Table 1.
Arp11 overexpression suppresses formation of Arp1 cables
| Overexpressed protein(s) | Arp1 cables | Arp1 punctae |
|---|---|---|
| Arp1 alone | 32.5 | 67.5 |
| Arp1 + p25 (control) | 41.0 | 59.0 |
| Arp1 + Arp11 | 20.0 | 80.0 |
| Arp1 + fly Arp11 | 16.0 | 84.0 |
Cells were transfected or cotransfected with the constructs shown, and the Arp1 staining pattern was analyzed. The percentage of cells exhibiting the different phenotypes is indicated herein. Cells containing large aggregates (approximately 20% of the Arp1-expressing population) were not considered in this analysis. p25 is another pointed end complex subunit that was used as a control. The data represent the mean of multiple experiments. More than 100 expressing cells were analyzed in each experiment.
Our coprecipitation analysis (Figure 4) indicate that Arp1 and Arp11 can interact in a reticulocyte lysate. If overexpressed Arp11 is able to form complexes with overexpressed Arp1 in HeLa cells, this might prevent formation of large Arp1 aggregates. We therefore determined the prevalence of the Arp1 overexpression phenotypes (i.e., punctate staining versus cables) in cells coexpressing both proteins (Table 1). The major consequence of cooverexpression of GFP-Arp11 with Arp1 was the reduction of the percentage of cells exhibiting Arp1 cables and a corresponding increase in the percentage of cells showing a diffuse punctate distribution. In cells that still contained cables, they ranged in both number and length, similar to what we saw in cells overexpressing Arp1 alone. Our results suggest that GFP-Arp11 can interact with overexpressed Arp1 to prevent the formation of organized cables. Preliminary analysis did not reveal a readily discernible increase in slowly sedimenting pool of Arp1 (our unpublished data), so we could not characterize this effect further.
The fly genome contains a possible ortholog of Arp11 (Figure 6), the gene for which (CG12235; Bourbon et al., 2002) is essential, as expected for a dynactin subunit (Spradling et al., 1999). However, the degree of relatedness to vertebrate Arp11 is low. CG12235 shares only 40% identity with vertebrate Arp11, compared with the 80% or higher identity seen for fly versus vertebrate Arp1 or β-actin (Table 2). To verify that CG12235 encodes a true Arp11 homolog we tested its ability to interact with Arp1 by using the cooverexpression assay described above. Fly Arp11 caused a decrease in the percentage of cells that contained Arp1 cables (Table 1), indicating that the fly and mouse Arp11 proteins are functionally related.
Figure 6.
Alignment of mouse, human and D. melanogaster (fly) Arp11 polypeptide sequences. Black indicates residues that are identical; open boxes indicate conserved residues with similar chemical character.
Table 2.
Comparison of dynactin subunit sequences
| Identity to mouse (%) | ||||
|---|---|---|---|---|
| Dynactin subunit | G. gallus | D. melanogaster | C. elegans | N. crassa |
| β-actin | 98 | 93 | 93 | 86 |
| Arp1 | 97 | 79 | 70 | 63 |
| CapZ beta | 91 | 78 | 66 | 53 |
| CapZ alpha | 96 | 62 | 51 | 34 |
| p25 | 97 | 58 | 41 | 39 |
| p27 | 90 | 48 | 31 | 22 |
| Arp11 | 87 | 40 | 24 | 20 |
| p62 | 89 | 36 | 26 | 23 |
| p150Glued | 80 | 32 | 27 | 25 |
| p50 (dynamitin) | 68 | 33 | 21 | undetermined |
| p24 | 68 | 25 | 12 | undetermined |
Putative dynactin subunit genes were identified and aligned as described in MATERIALS AND METHODS. Genes are arranged in approximately descending order of homology. Genes encoding potential N. crassa orthologs of p50 or p24 could not be identified.
DISCUSSION
Arp11 is a novel member of the actin superfamily that is present only in dynactin. We have explored possible binding interactions of Arp11 within the dynactin molecule and find that Arp11 can interact with both Arp1 and conventional actin. Although Drosophila Arp11 is less well conserved than either Arp1 or actin, it has effects similar to vertebrate Arp11 when expressed in cultured cells, suggesting the fly and vertebrate proteins are functionally homologous.
Sequence alignments of Arp11 with conventional actin reveal a conserved actin fold (Kabsch and Holmes, 1995) that is punctuated with insertions. Purified native Arp11 contains bound ATP (J. Bingham, unpublished observations) suggesting it indeed folds into an actin-like structure. As in many actin-related proteins (Schafer and Schroer, 1999), the insertions in the Arp11 sequence are found in the middle and at both ends of the protein. All map to sites predicted to be on the surface. We identified a minimal actin-binding fragment as amino acids 23–137, which corresponds roughly to subdomains 1 and 2 in conventional actin (residues 17–153; see Eckley et al., 1999; Holmes et al., 1990; Kabsch et al., 1990). If these subdomains of Arp11 are engaged in contacts with actin or Arp1 it would leave the rest of the protein to bind other proteins such as the pointed end complex subunits p62, p27, p25, and/or cargo components.
Like Arp1, Arp11 is incorporated into a large, rapidly sedimenting complex, most likely dynactin. Biochemical analysis of dynactin subcomplexes allowed Arp11 to be mapped to the pointed end of the dynactin Arp1 minifilament (Eckley et al., 1999). At present, we do not know whether Arp11 binds directly to Arp1 or actin. The pointed end complex protein, p62, may play a role in the association of Arp11 with the Arp1 minifilament. Like Arp11, p62 has been shown to bind to actin (Garces et al., 1999) and Arp1 (Karki et al., 2000) in vitro. If p62 and/or Arp11 is bound to actin, this would place actin at the pointed end of the Arp1 minifilament instead of in association with CapZ as we suggested previously (Schafer et al., 1994). However, preliminary chemical cross-linking experiments (Eckley, unpublished observations) suggest that Arp11 directly contacts p62 and Arp1 but not actin. At present, we favor a model in which Arp11 binds Arp1 directly at the filament pointed end to dock the pointed end complex to the Arp1 filament. Rigorous proof of this will await further study.
Arp11 might play multiple roles in dynactin. Its predicted structure suggests it caps the Arp1 filament to disallow further subunit addition or filament annealing. A potential Arp1 capping activity could not be assayed directly because purified Arp1 is so difficult to obtain. Native Arp 1 can be isolated from purified dynactin (Bingham and Schroer, 1999), but the yield is very low and the resulting protein is so labile as to preclude detailed analysis. Heterologous expression of recombinant Arp1 yields insoluble aggregates even in insect cells (Bingham, Ph.D. thesis). A detailed analysis of the role of Arp11 in Arp1 assembly will require development of a new purification strategy for recombinant Arp1.
Orthologs of Arp11 can be found in most eukaryotes; however, this branch of the actin superfamily is considerably more divergent than actin or Arp1 (Table 2). Neurospora crassa Arp11 (Ropy-7) is barely discernible as an Arp11 ortholog, sharing only 20% identity with its mouse counterpart. A possible Arp11 ortholog has also been identified as the Arp10 gene in the budding yeast S. cerevisiae (Goodson and Hawse, 2002). S. cerevisiae Arp10 was found to interact with Arp1 in a two-hybrid screen (Uetz et al., 2000), but Arp10 deletion mutants do not show the nuclear partitioning defect typical of dynein/dynactin pathway mutants (Eckley, Hoyt, and Schroer, unpublished observations). This suggests that S. cerevisiae Arp10 provides a slightly different function than Arp11 in other species. Both S. cerevisiae and Schizosaccharomyces pombe seem to lack the three other subunits of the pointed end complex (p62, p27, and p25), indicating that this entire structural element may not be required for yeast dynactin function. S. cerevisiae dynactin sediments with a smaller S value than bovine dynactin (15.5S versus 20S; Kahana et al., 1998), consistent with a more streamlined structure.
Fungal and metazoan cells share a number of important differences, a major one being the absence or presence of an open mitosis. Nuclear envelope breakdown, the defining feature of an open mitosis, is facilitated by dynein and dynactin. Binding of these proteins to the envelope (Salina et al., 2002) allows tension to be exerted on the membrane, which contributes to rupture. Overexpression of the dynactin subunit, p62, blocks dynein recruitment to nuclei (Salina et al., 2002), suggesting that this protein, possibly acting in conjunction with other pointed end complex subunits (Quintyne, Eckley, and Schroer, unpublished data), mediates dynactin binding. That budding and fission yeast lack most pointed end complex proteins is consistent with the fact that these cells do not accumulate dynactin on their nuclear envelopes (McMillan and Tatchell, 1994; Kahana et al., 1998; Harata et al., 2000). It is possible that the Arp11 orthologs found in yeasts provide no function other than to cap the Arp1 filament.
In addition to their supporting role in nuclear envelope breakdown, dynein and dynactin have been proposed to participate in the long-range movements of nuclei in cells such as zygotes, neurons, and filamentous fungi (reviewed in Reinsch and Gonczy, 1998; Morris, 2000; Morris, 2003). Mutations in dynein or dynactin genes in the fungi N. crassa and Aspergillus nidulans yield the conspicuous “ropy” or “nud” (nuclear distribution) phenotypes in which nuclear transport into growing hyphae is blocked (Plamann et al., 1994; Xiang et al., 1994; Tinsley et al., 1996; Xiang et al., 1999; Lee et al., 2001). However, the microtubules in these cells are oriented with their plus ends toward the hyphal tip, the wrong orientation to support a simple dynein-based transport process. Although N. crassa contains orthologs of p62 (Ropy-2) and p25 (Ropy-12), Ropy-12 mutants show apparently normal nuclear migration (Lee et al., 2001), leaving the function of these pointed end complex components in filamentous fungi obscure.
Acknowledgments
We thank Drs. Sally Lewis and Nick Cowan (New York University, New York, NY) for the mouse β-actin clone, Drs. Jim Lees-Miller and David Helfman (Cold Spring Harbor Laboratories, Cold Spring Harbor, NY) for the human Arp1 clone, and Dr. M. Way (Imperial Cancer Research Fund, London) for Arp11 antisera. This work was supported by National Institutes of Health grant GM-44589 (to T.A.S.).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–01–0049. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03-01-0049.
References
- Andersen, J.S., Lyon, C.E., Fox, A.H., Leung, A.K., Lam, Y.W., Steen, H., Mann, M., and Lamond, A.I. (2002). Directed proteomic analysis of the human nucleolus. Curr. Biol. 12, 1–11. [DOI] [PubMed] [Google Scholar]
- Bingham, J.B. (1999). Purification and characterization of the actin-related protein, Arpl. Ph.D. thesis, The Johns Hopkins University, Baltimore, MD.
- Bingham, J.B., King, S.J., and Schroer, T.A. (1998). Purification of dynactin and dynein from brain tissue. Methods Enzymol. 298, 171–184. [DOI] [PubMed] [Google Scholar]
- Bingham, J.B., and Schroer, T.A. (1999). Self-regulated polymerization of the actin-related protein Arp1. Curr. Biol. 9, 223–226. [DOI] [PubMed] [Google Scholar]
- Bourbon, H.M., et al. (2002). A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech. Dev. 110, 71–83. [DOI] [PubMed] [Google Scholar]
- Boyer, L.A., and Peterson, C.L. (2000). Actin-related proteins (Arps): conformational switches for chromatin-remodeling machines? Bioessays 22, 666–672. [DOI] [PubMed] [Google Scholar]
- Busson, S., Dujardin, D., Moreau, A., Dompierre, J., and De Mey, J.R. (1998). Dynein and dynactin are localized to astral microtubules and at cortical sites in mitotic epithelial cells. Curr. Biol. 8, 541–544. [DOI] [PubMed] [Google Scholar]
- Cairns, B.R., Schlichter, A., Erdjument-Bromage, H., Tempst, P., Kornberg, R.D., and Winston, F. (1999). Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol. Cell 4, 715–723. [DOI] [PubMed] [Google Scholar]
- Cooper, J.A., and Pollard, T.D. (1985). Effect of capping protein on the kinetics of actin polymerization. Biochemistry 24, 793–799. [DOI] [PubMed] [Google Scholar]
- Echeverri, C.J., Paschal, B.M., Vaughan, K.T., and Vallee, R.B. (1996). Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J. Cell Biol. 132, 617–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckley, D.M., Gill, S.R., Melkonian, K.A., Bingham, J.B., Goodson, H.V., Heuser, J.E., and Schroer, T.A. (1999). Analysis of dynactin subcomplexes reveals a novel actin-related protein associated with the arp1 minifilament pointed end. J. Cell Biol. 147, 307–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces, J.A., Clark, I.B., Meyer, D.I., and Vallee, R.B. (1999). Interaction of the p62 subunit of dynactin with Arp1 and the cortical actin cytoskeleton. Curr. Biol. 9, 1497–1500. [DOI] [PubMed] [Google Scholar]
- Gill, S.R., Schroer, T.A., Szilak, I., Steuer, E.R., Sheetz, M.P., and Cleveland, D.W. (1991). Dynactin, a conserved, ubiquitously expressed component of an activator of vesicle motility mediated by cytoplasmic dynein. J. Cell Biol. 115, 1639–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodson, H.V., and Hawse, W.F. (2002). Molecular evolution of the actin family. J. Cell Sci. 115, 2619–2622. [DOI] [PubMed] [Google Scholar]
- Harata, M., Oma, Y., Tabuchi, T., Zhang, Y., Stillman, D.J., and Mizuno, S. (2000). Multiple actin-related proteins of Saccharomyces cerevisiae are present in the nucleus. J. Biochem. 128, 665–671. [DOI] [PubMed] [Google Scholar]
- Holleran, E.A., Karki, S., and Holzbaur, E.L. (1998). The role of the dynactin complex in intracellular motility. Int. Rev. Cytol. 182, 69–109. [DOI] [PubMed] [Google Scholar]
- Holleran, E.A., Tokito, M.K., Karki, S., and Holzbaur, E.L. (1996). Centractin (ARP1) associates with spectrin revealing a potential mechanism to link dynactin to intracellular organelles. J. Cell Biol. 135, 1815–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, K.C., Popp, D., Gebhard, W., and Kabsch, W. (1990). Atomic model of the actin filament. Nature 347, 44–49. [DOI] [PubMed] [Google Scholar]
- Kabsch, W., and Holmes, K.C. (1995). The actin fold. FASEB J. 9, 167–174. [DOI] [PubMed] [Google Scholar]
- Kabsch, W., Mannherz, H.G., Suck, D., Pai, E.F., and Holmes, K.C. (1990). Atomic structure of the actin:DNase I complex. Nature 347, 37–44. [DOI] [PubMed] [Google Scholar]
- Kahana, J.A., Schlenstedt, G., Evanchuk, D.M., Geiser, J.R., Hoyt, M.A., and Silver, P.A. (1998). The yeast dynactin complex is involved in partitioning the mitotic spindle between mother and daughter cells during anaphase B. Mol. Biol. Cell 9, 1741–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karki, S., Tokito, M.K., and Holzbaur, E.L. (2000). A dynactin subunit with a highly conserved cysteine-rich motif interacts directly with Arp1. J. Biol. Chem. 275, 4834–4839. [DOI] [PubMed] [Google Scholar]
- Kato, M., Sasaki, M., Mizuno, S., and Harata, M. (2001). Novel actin-related proteins in vertebrates: similarities of structure and expression pattern to Arp6 localized on Drosophila heterochromatin. Gene 268, 133–140. [DOI] [PubMed] [Google Scholar]
- Kelleher, J.F., Atkinson, S.J., and Pollard, T.D. (1995). Sequences, structural models, and cellular localization of the actin-related proteins Arp2 and Arp3 from Acanthamoeba. J. Cell Biol. 131, 385–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, I.H., Kumar, S., and Plamann, M. (2001). Null mutants of the neurospora actin-related protein 1 pointed-end complex show distinct phenotypes. Mol. Biol. Cell 12, 2195–2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees-Miller, J.P., Helfman, D.M., and Schroer, T.A. (1992). A vertebrate actin-related protein is a component of a multisubunit complex involved in microtubule-based vesicle motility. Nature 359, 244–246. [DOI] [PubMed] [Google Scholar]
- Lessard, J.L. (1988). Two monoclonal antibodies to actin: one muscle selective and one generally reactive. Cell Motil. Cytoskeleton 10, 349–362. [DOI] [PubMed] [Google Scholar]
- Ligon, L.A., Karki, S., Tokito, M., and Holzbaur, E.L. (2001). Dynein binds to beta-catenin and may tether microtubules at adherens junctions. Nat. Cell Biol 3, 913–917. [DOI] [PubMed] [Google Scholar]
- McMillan, J.N., and Tatchell, K. (1994). The JNM1 gene in the yeast Saccharomyces cerevisiae is required for nuclear migration and spindle orientation during the mitotic cell cycle. J. Cell Biol. 125, 143–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melki, R., Vainberg, I.E., Chow, R.L., and Cowan, N.J. (1993). Chaperonin-mediated folding of vertebrate actin-related protein and gamma-tubulin. J. Cell Biol. 122, 1301–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, N.R. (2000). Nuclear migration. From fungi to the mammalian brain. J. Cell Biol. 148, 1097–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, N.R. (2003). Nuclear positioning: the means is at the ends. Curr. Opin. Cell Biol. 15, 54–59. [DOI] [PubMed] [Google Scholar]
- Palazzo, A.F., Joseph, H.L., Chen, Y.J., Dujardin, D.L., Alberts, A.S., Pfister, K.K., Vallee, R.B., and Gundersen, G.G. (2001). Cdc42, dynein, and dynactin regulate MTOC reorientation independent of Rho-regulated microtubule stabilization. Curr. Biol. 11, 1536–1541. [DOI] [PubMed] [Google Scholar]
- Paschal, B.M., Holzbaur, E.L., Pfister, K.K., Clark, S., Meyer, D.I., and Vallee, R.B. (1993). Characterization of a 50-kDa polypeptide in cytoplasmic dynein preparations reveals a complex with p150GLUED and a novel actin. J. Biol. Chem. 268, 15318–15323. [PubMed] [Google Scholar]
- Peterson, C.L., Zhao, Y., and Chait, B.T. (1998). Subunits of the yeast SWI/SNF complex are members of the actin-related protein (ARP) family. J. Biol. Chem. 273, 23641–23644. [DOI] [PubMed] [Google Scholar]
- Plamann, M., Minke, P.F., Tinsley, J.H., and Bruno, K.S. (1994). Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J. Cell Biol. 127, 139–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyne, N.J., Gill, S.R., Eckley, D.M., Crego, C.L., Compton, D.A., and Schroer, T.A. (1999). Dynactin is required for microtubule anchoring at centrosomes. J. Cell Biol. 147, 321–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinsch, S., and Gonczy, P. (1998). Mechanisms of nuclear positioning. J. Cell Sci. 111, 2283–2295. [DOI] [PubMed] [Google Scholar]
- Salina, D., Bodoor, K., Eckley, D.M., Schroer, T.A., Rattner, J.B., and Burke, B. (2002). Cytoplasmic dynein as a facilitator of nuclear envelope breakdown. Cell 108, 97–107. [DOI] [PubMed] [Google Scholar]
- Schafer, D.A., Gill, S.R., Cooper, J.A., Heuser, J.E., and Schroer, T.A. (1994). Ultrastructural analysis of the dynactin complex: an actin-related protein is a component of a filament that resembles F-actin. J. Cell Biol. 126, 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer, D.A., and Schroer, T.A. (1999). Actin-related proteins. Annu. Rev. Cell Dev. Biol. 15, 341–63. [DOI] [PubMed] [Google Scholar]
- Schroer, T.A., and Sheetz, M.P. (1991). Two activators of microtubule-based vesicle transport. J. Cell Biol. 115, 1309–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling, A.C., Stern, D., Beaton, A., Rhem, E.J., Laverty, T., Mozden, N., Misra, S., and Rubin, G.M. (1999). The Berkeley Drosophila Genome Project gene disruption project: single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153, 135–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier, F.W. (1991). Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J. Mol. Biol. 219, 37–44. [DOI] [PubMed] [Google Scholar]
- Suetsugu, S., Miki, H., Yamaguchi, H., Obinata, T., and Takenawa, T. (2001). Enhancement of branching efficiency by the actin filament-binding activity of N-WASP/WAVE2. J. Cell Sci. 114, 4533–4542. [DOI] [PubMed] [Google Scholar]
- Tinsley, J.H., Minke, P.F., Bruno, K.S., and Plamann, M. (1996). p150Glued, the largest subunit of the dynactin complex, is nonessential in Neurospora but required for nuclear distribution. Mol. Biol. Cell 7, 731–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin, H., Staehelin, T., and Gordon, J. (1979). Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. USA 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz, P., et al. (2000). A. comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627. [DOI] [PubMed] [Google Scholar]
- Valetti, C., Wetzel, D.M., Schrader, M., Hasbani, M.J., Gill, S.R., Kreis, T.E., and Schroer, T.A. (1999). Role of dynactin in endocytic traffic: effects of dynamitin overexpression and colocalization with CLIP-170. Mol. Biol. Cell 10, 4107–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver, A.M., Karginov, A.V., Kinley, A.W., Weed, S.A., Li, Y., Parsons, J.T., and Cooper, J.A. (2001). Cortactin promotes and stabilizes Arp2/3-induced actin filament network formation. Curr. Biol. 11, 370–374. [DOI] [PubMed] [Google Scholar]
- Xiang, X., Beckwith, S.M., and Morris, N.R. (1994). Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc. Natl. Acad. Sci. USA 91, 2100–2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang, X., Zuo, W., Efimov, V.P., and Morris, N.R. (1999). Isolation of a new set of Aspergillus nidulans mutants defective in nuclear migration. Curr. Genet. 35, 626–630. [DOI] [PubMed] [Google Scholar]