Abstract
The accumulation of aberrantly folded proteins can lead to cell dysfunction and death. Currently, the mechanisms of toxicity and cellular defenses against their effects remain incompletely understood. In the endoplasmic reticulum (ER), stress caused by misfolded proteins activates the unfolded protein response (UPR). The UPR is an ER-to-nucleus signal transduction pathway that regulates a wide variety of target genes to maintain cellular homeostasis. We studied the effects of ER stress in budding yeast through expression of the well-characterized misfolded protein, CPY*. By challenging cells within their physiological limits to resist stress, we show that the UPR is required to maintain essential functions including protein translocation, glycosylation, degradation, and transport. Under stress, the ER-associated degradation (ERAD) pathway for misfolded proteins is saturable. To maintain homeostasis, an “overflow” pathway dependent on the UPR transports excess substrate to the vacuole for turnover. The importance of this pathway was revealed through mutant strains compromised in the vesicular trafficking of excess CPY*. Expression of CPY* at levels tolerated by wild-type cells was toxic to these strains despite retaining the ability to activate the UPR.
INTRODUCTION
During biosynthesis, most polypeptides fold and assemble into their native conformations assisted by chaperones and folding catalysts. In the secretory pathway, nascent polypeptides are first translocated across the endoplasmic reticulum (ER) membrane via the Sec61 translocon pore (Walter and Johnson, 1994). They enter the lumen in an unfolded state and their subsequent maturation can involve considerable complexity. In addition to folding, these proteins may integrate into membranes and/or be subjected to covalent modifications including glycosylation, disulfide bond formation, and lipid linkage. In the ER, and in other parts of the cell, errors in any biosynthetic step or arising by mutation can result in the synthesis of aberrant proteins. Left unchecked, these proteins may have detrimental consequences as underscored by the numerous human diseases including Alzheimer's, Huntington's, and cystic fibrosis (reviewed in Carrell and Gooptu, 1998; Kim and Arvan, 1998; Kopito and Ron, 2000).
To assure that only properly folded proteins are transported to their sites of function, a mechanism termed “ER quality control” monitors the folding state of newly synthesized proteins (reviewed in Brodsky and McCracken, 1999; Ellgaard and Helenius, 2001). Through this mechanism, immature proteins are kept in the ER until they are fully folded. In mammals, the ER lectins calnexin and calreticulin play an important role in the retention of incompletely folded proteins (Zhang et al., 1997). ER quality control also targets proteins that cannot fold for destruction via the ER-associated protein degradation pathway (ERAD; Finger et al., 1993). Here, misfolded proteins are either retained statically in the ER or transported to the Golgi and retrieved (Hammond and Helenius, 1994; Vashist et al., 2001; Yamamoto et al., 2001). Next, the misfolded proteins are translocated back to the cytosol, probably through the same translocon pore used for import (Pilon et al., 1997; Plemper et al., 1997; Zhou and Schekman, 1999). On the cytosolic face of the ER membrane, substrates are ubiquitinated and degraded by the 26S proteasome (Ward et al., 1995; Hiller et al., 1996; Bays et al., 2001).
Recently, a physiological link was established between ER quality control and a stress-inducible pathway known as the unfolded protein response (UPR) (Casagrande et al., 2000; Friedlander et al., 2000; Ng et al., 2000; Travers et al., 2000). The UPR is a conserved signal transduction pathway that mediates communication between the ER and nucleus (reviewed in Patil and Walter, 2001; Spear and Ng, 2001). The connection was intriguing since the UPR was known to be essential in resisting ER stress caused by pharmacological agents (Cox et al., 1993; Mori et al., 1993). Genome-wide expression analysis revealed ERAD-related genes among the wide array of UPR targets (Travers et al., 2000). Together, these studies suggested an important aspect of UPR-mediated homeostasis is to rid aberrant proteins via ERAD. Indeed, modest defects were observed in UPR-deficient strains' ability to degrade ERAD substrates (Casagrande et al., 2000; Ng et al., 2000; Travers et al., 2000). However, the surprising breadth of the UPR transcriptional program suggests that its role in ER stress tolerance might require other functions in addition to ERAD.
The cytotoxic effects of misfolded proteins are well documented (Kim and Arvan, 1998; Plemper and Wolf, 1999; Kopito and Sitia, 2000). In the secretory pathway, the hypersensitivity of UPR mutants to agents that disrupt ER protein folding (tunicamycin and DTT) suggested that the unfolded protein response might play a protective role against their effects (Cox et al., 1993; Mori et al., 1993). Other studies have demonstrated that genetic methodologies can provide important insight into ER stress tolerance. The overexpression of the heterologous protein Δpro (a mutant version of an aspartic proteinase from Rhizopus niveus) was shown to be harmful to UPR-deficient strains but tolerated in wild-type cells (Umebayashi et al., 1999). This is a more favorable approach because it eliminates potential indirect effects associated with the use of pharmacological agents. However, the basis of its toxicity is unclear because Δpro is not a substrate of the ERAD pathway (Umebayashi et al., 2001).
In this study, we examined the role of the UPR in the stress tolerance of misfolded proteins. To study the effects of ER stress, we challenged cells with a well-characterized misfolded version of carboxypeptidase Y called CPY* (Finger et al., 1993). To be within the physiological range of stress resistance, we calibrated expression of the protein to be well tolerated in wild-type cells whereas lethal to UPR-deficient cells. In the mutant cells, CPY* led to severe defects in ER function including protein translocation, glycosylation, ER- to-Golgi transport, and degradation—functions that are normal in wild-type cells when identically challenged. Surprisingly, overexpression of CPY* was without detrimental effect to the growth of several ERAD mutants. In these strains, the protein is degraded at a rate similar to wild-type cells. This observation led to the discovery of an alternative degradative pathway for excess misfolded protein. In UPR mutants, degradation of overexpressed CPY* was severely impaired, suggesting that both pathways are dependent on the UPR regulation. These studies reveal new roles of the UPR in alleviating ER stress and provide an expanded physiological basis for the UPR transcriptional program.
MATERIALS AND METHODS
Plasmids Used in This Study
Plasmids were constructed using standard cloning protocols (Sambrook et al., 1989). HA epitope-tagged CPY* contained in plasmid pDN436 was described previously (Ng et al., 2000). GFP-ALP expression vector was a gift of S. Emr (University of California, San Diego, CA; Cowles et al., 1997).
A galactose-inducible version of prc1-1 was constructed by digesting pDN436 (carrying HA-tagged CPY*) with AccI, treated with T4 DNA polymerase, and subsequently digested with SphI. The release insert was inserted into pTS210 (YCp50 with the GAL1/10 promoter) to yield pES28. An integration version of GAL-CPY* was constructed by releasing the gene as a SalI and EcoRI (site filled and destroyed by T4 DNA polymerase) fragment and ligated into the SalI/SmaI sites of pRS305 (Sikorski and Hieter, 1989) creating pES26. pES26 is cleaved with EcoRI before integration. pES67 is the GAL-CPY* construct cloned into pRS315 (Sikorski and Hieter, 1989).
Strains and Antibodies
Yeast strains used in this study are described in Table 1. Anti-HA mAb (HA.11) was purchased from Covance Research Products (Richmond, CA). Anti-Kar2p antibody was provided by Peter Walter (University of California, San Francisco, CA). Anti-ALP and anti-CPS antisera were gifts from Chris Burd (University of Pennsylvania) and Scott Emr (University of California, San Diego, CA). Anti-PrA was a gift from Tom Stevens (University of Oregon, Eugene, OR). Anti-α-1,6 mannose polyclonal antiserum provided by Howard Riezman (Biozentrum, University of Basel, Switzerland). Affinity-purified anti-GFP antibody was provided by Sarah Rice and Ron Vale (University of California, San Francisco, CA). Anti-Gas1p antiserum was raised against a GST-fusion protein containing the amino-proximal amino acids 40–289 of Gas1p. Covance Research Products performed antiserum production.
Table 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| W303a | MATa,leu2-3, 112, his3-11, trp1-1, ura3-1, can1-100, ade2-1 | P. Walter (UCSF) |
| ESY248 | MATaire1::TRP1, W303 background | This study |
| DNY419 | MATa,ire1::TRP1, leu2-3, 112, his3-11::HIS3-UPRE LacZ, trp1-1, ura3-1, can1-100, ade2-1 | Ng et al. (2000) |
| ESY39 | MATa,leu2-3, 112, his3-11::HIS3-UPRE LacZ, trp1-1, ura3-1, can1-100, ade2-1 | This study |
| ESY233 | MATa,leu2-3, 112::LEU2-GAL-CPYHA,* his3-11, trp1-1, ura3-1, can1-100, ade2-1 | This study |
| ESY234 | MATa,cue1::TRP1, leu2-3, 112::LEU2-GAL-CPYHA,* his3-11, trp1-1, ura3-1, can1-100, ade2-1 | This study |
| ESY235 | MATa,der1::KANMX, leu2-3, 112::LEU2-GAL-CPYHA,* his3-11, trp1-1, ura3-1, can1-100, ade2-1 | This study |
| ESY236 | MATa,hrd1::KANMX, leu2-3, 112::LEU2-GAL-CPYHA,* his3-11, trp1-1, ura3-1, can1-100, ade2-1 | This study |
| ESY237 | MATa,ire1::TRP1, leu2-3, 112::LEU2-GAL-CPYHA,* his3-11, trp1-1, ura3-1, can1-100, ade2-1 | This study |
| ESY258 | MATa, pDN436, W303 background | This study |
| ESY259 | MATa,cue1::TRP1, pDN436, W303 background | This study |
| ESY260 | MATa,der1::KANMX, pDN436, W303 background | This study |
| ESY261 | MATa,hrd1::KANMX, pDN436, W303 background | This study |
| ESY342 | MATa, pES67, W303 background | This study |
| ESY343 | MATa,cue1::TRP1, pES67, W303 background | This study |
| ESY344 | MATa,der1::KANMX, pES67, W303 background | This study |
| ESY345 | MATa,hrd1::KANMX, pES67, W303 background | This study |
| ESY347 | MATa,per17-1, pES67 W303 background | This study |
| ESY349 | MATa,pep4::HIS3, pES67, W303 background | This study |
| ESY350 | MATa, pES67, pGFP-ALP, W303 background | This study |
| ESY357 | MATa,pep4::HIS3, pES67, pGFP-ALP, W303 background | This study |
| ESY386 | MATa,erv29::KANMX, pES67, W303 background | This study |
Cell Labeling, Immunoprecipitation, and Cycloheximide Chase Analysis
GAL-CPY* Overexpression Cells containing the GAL-CPY* gene were grown at 30°C in synthetic media containing the appropriate amino acids and 2% galactose to early to midlog phase before processing. For experiments involving Δire1 cells, all strains were grown in synthetic media containing 3% raffinose and 50 μg/ml myo-inositol. To initiate induction, galactose was added to 2%. Cells were then grown for 6 h before processing.
Metabolic pulse-chase Analysis Cell labeling and immunoprecipitation were carried out as described previously (Vashist et al., 2001). Cell labeling was performed in the presence of 0.75 mg/ml bovine serum albumin (BSA). Where indicated, N-linked carbohydrates were removed by treatment with 300 U endoglycosidase H (New England Biolabs, Inc., Beverly, MA) according to the manufacturer's protocol.
Cycloheximide-chase Analysis Cells were grown as described above. Cessation of protein synthesis was initiated with the addition of 100 μg/ml cycloheximide. Equal cell numbers were collected, and samples prepared as described (Kushnirov, 2000); 0.2 OD600 cell equivalents were resolved by SDS-PAGE, transferred to nitrocellulose, and probed using HA.11 (1:10,000 dilution). and HRP-conjugated secondary antibody. Proteins were visualized by enhanced chemiluminescence (Pierce, Rockford, IL).
Measurement of General Translation Cells were grown to log phase in synthetic complete media (SC) lacking cysteine and methionine. Tunicamycin was added to 2.5 μg/ml for incubation with aeration at 30°C. At specific time points after addition of the drug, equal cell numbers were collected and pulse-labeled for 10 min with [35S]methionine/cysteine. TCA was added to 10% to terminate labeling. Equal volumes (3 μl) of detergent lysates were resolved by SDS-PAGE and visualized by autoradiography.
Indirect Immunofluorescence Microscopy
Immunofluorescence was performed using a modified protocol from Vashist et al. (2001) and Guthrie and Fink (1991). Yeast strains were grown in SC media containing the appropriate amino acids and 2% galactose to log phase. Formaldehyde (EM grade; Polysciences, Inc., Warrington, PA) was added to 3.7% at 30°C for 1 h. After fixation, cells were washed with 0.1 M potassium phosphate buffer (pH 7.5). Cells were spheroplasted by incubation in spheroplasting buffer (1.0 mg/ml zymolyase 20T [ICN Biomedicals, Aurora, OH], 0.1 M potassium phosphate, pH 7.5, 0.1% 2-mercaptoethanol, 1.2 M sorbitol) for 30–45 min at 30°C. The cells were then washed once with PBS, 1.2 M sorbitol. For strains not expressing GFP-ALP (Figure 6A), 30 μl of cell suspension was applied to poly-l-lysine–coated slides for 1 min and washed with PBS. Slides were immersed in acetone for 5 min at –20°C and allowed to dry. Strains expressing GFP-ALP (see Figure 6B) cells were resuspended in 2% SDS, 1.2 M sorbitol for 2 min, washed extensively with PBS, 1.2 M sorbitol, and applied to slides. PBS-block, 30 μl (3% BSA in PBS), was added to each well for 30 min. Primary antibodies were incubated for 1 h, whereas secondary antibodies were incubated for 45 min, with three to five PBS-block washes after each application. Primary antibodies α-HA, α-Kar2p, or α-GFP were diluted to 1:1000, 1:5000, and 1:1000, respectively. Secondary antibodies AlexaFluor 488 goat α-rabbit and AlexaFluor 546 goat α-mouse (Molecular Probes, Inc., Eugene, OR) were diluted 1:1000 for working concentrations. Images were captured using a Spot 2 cooled CCD camera (Diagnostic Instruments, Inc., Sterling Heights, MI) mounted to a Zeiss Axioplan epifluorescence microscope (Carl Zeiss, Inc., Thornwood, NY).
Figure 6.
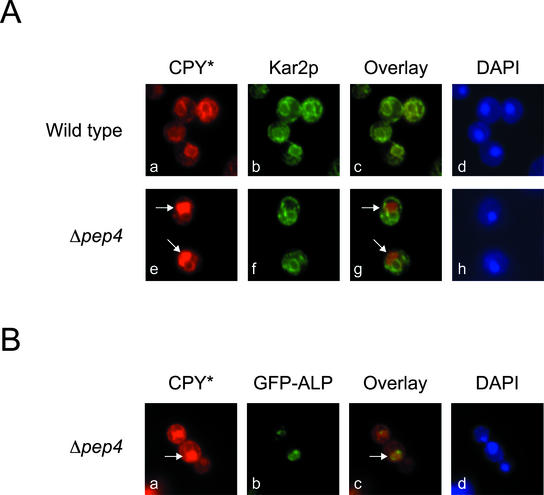
On saturation of ERAD, excess CPY* is transported to the vacuole for degradation. (A) Wild-type (ESY342) and Δpep4 (ESY349) were grown at 30°C in media containing 2% galactose to induce CPY* expression. Cells fixed and permeabilized on glass slides were decorated with mouse anti-HA mAb (CPY*) and anti-Kar2p antiserum (Kar2p). Antibody complexes were bound with Alexa Fluor 546 goat α-mouse and Alexa Fluor 488 goat α-rabbit antibodies for visualization (CPY* in the red channel; Kar2p in the green channel). (B) ESY357 cells (Δpep4, pGFP-ALP, pES67) grown as in A were probed using anti-HA (CPY*) and anti-GFP (GFP-ALP) followed by Alexa Fluor 546 goat α-mouse and Alexa Fluor 488 goat α-rabbit secondary antibodies (CPY* in the red channel; GFP-ALP in the green channel). Staining with DAPI (Ad, Ah, and Bd) indicates the positions of nuclei.
RESULTS
The UPR Abrogates the Toxicity of CPY* Overexpression
An HA-tagged version of CPY* tightly regulated by the strong GAL1 promoter (GAL-CPY*) was constructed and integrated into the genomes of wild-type and UPR-deficient strains of the budding yeast Saccharomyces cerevisiae. As shown in Figure 1A, wild-type cells induced for CPY* overexpression (Galactose) grew nearly as well as control cells not carrying the construct. By contrast, induction of CPY* expression was lethal to Δire1 cells (Figure 1A, Galactose). The difference was not caused by the shift in carbon source because strains lacking GAL-CPY*, but otherwise identical, grew equally well on the same media (Figure 1A, Galactose, compare upper sectors). Previous studies on the expression of the heterologous protein Δpro under similar conditions had only a modest effect on Δire1 cells, suggesting CPY* might be intrinsically more toxic by comparison (Umebayashi et al., 1999). The expression level (greater than tenfold higher from the GAL1 promoter than endogenous) is an important consideration for our analysis because the TDH3 promoter of similar strength had the same effect whereas CPY* expressed from the more moderate PRC1 and CUP1 promoters were better tolerated by UPR-deficient strains (Ng et al., 2000 and unpublished results).
Figure 1.
Activation of the unfolded protein response is required for the tolerance of overexpressed CPY*. (A) Wild-type and Δire1 cells containing GAL1-driven CPY* were streaked onto plates containing YP glucose (expression off) or YP galactose (expression on) and incubated for 2 and 3 d at 30°C, respectively. (B) Wild-type and Δire1 cells containing a single copy of the UPRE-lacZ reporter were assayed for β-galactosidase activity (Ng et al., 2000) after induction of CPY* for 6 h. For comparison, activity was measured from wild-type and Δire1 cells not expressing CPY* and treated with tunicamycin to monitor rate of induction under conditions of extreme stress (2 μg/ml for 6 h). The data reflect three independent experiments with the SE of the mean indicated.
As IRE1 encodes a key regulator of the UPR, the data suggest that the pathway provides a protective function against CPY* toxicity. Consistent with this view, we observed a strong UPR induction in wild-type cells overexpressing CPY* that was absent in Δire1 cells (Figure 1B). We previously showed that the UPR is modulated according to the physiological needs of the cell (Ng et al., 2000). Thus, the observed level of induction reflects the extent needed for stress resistance. For comparison, control cells treated with the glycosylation inhibitor tunicamycin exhibit a higher response. These data show that UPR activation to CPY* overexpression has not reached its maximum level and provide additional evidence that our conditions fall within the functional range of the UPR.
Translational Repression Is Not an Aspect of the Yeast UPR
Tolerance of ER stress by a translational repression mechanism is an important part of the mammalian UPR (Harding et al., 1999, 2000). Although a key component of the mechanism, PERK, is absent in yeast, transcriptional repression of ribosomal genes has been observed after UPR induction by tunicamycin (Nierras and Warner, 1999). Thus, it seemed conceivable that a similar strategy is used in yeast. We tested this possibility by analyzing the overall protein synthesis in wild-type cells after UPR induction with tunicamycin. As shown in Figure 2, except for some variation of a small number of proteins, overall protein synthesis remained uniform during the time course. These data show that translational repression is not part of the immediate UPR response in yeast. Furthermore, translational repression was not observed in wild-type cells constitutively expressing levels of misfolded proteins lethal to UPR mutant strains (unpublished results). Taken together, these data show that the budding yeast UPR program does not include a translational repression mechanism to tolerate ER stress.
Figure 2.
Translational repression is not an aspect of the S. cerevisiae UPR. Wild-type cells were treated with the glycosylation inhibitor tunicamycin for the indicated times, followed by a 10-min pulse-label with [35S]methionine/cysteine. Labeled proteins were resolved by electrophoresis through a 10% SDS polyacrylamide gel (top panel). To monitor the efficacy of tunicamycin treatment, endogenous CPY was immunoprecipitated from lysates of each time point and resolved by SDS-PAGE (bottom panel). An asterisk denotes the position of nonglycosylated pro-CPY. The positions of ER pro-CPY (p1) and Golgi pro-CPY(p2) are indicated.
The UPR Is Required to Maintain a Variety of Cellular Functions during ER Stress
Although the genomic transcriptional program of the UPR is known, it was unclear how it alleviates the stress caused by misfolded proteins (Travers et al., 2000). Guided by the genomic data, we analyzed specific functions in wild-type and UPR mutants challenged by CPY* synthesis. We first analyzed its turnover because several ERAD genes are UPR targets and moderately expressed substrates are less efficiently degraded in Δire1 cells (Casagrande et al., 2000; Ng et al., 2000; Travers et al., 2000). Because CPY* overexpression is lethal to UPR mutants, experiments were performed after induction by galactose. Metabolic pulse-chase experiments show that CPY* is degraded efficiently in wild-type cells but is more stable in Δire1 cells (Figure 3, A and B). Quantification of the data was performed after digestion by endoglycosidase H (Endo H), an enzyme that specifically cleaves N-linked carbohydrates. This was necessary because we noticed a diffuse trail of CPY* radioactivity when overexpressed in wild-type cells but not in Δire1 cells (Figure 3A, hyperglycosylated CPY*). After digestion, the pattern collapsed to a single species, suggesting extensive modification of core carbohydrates in the Golgi as previously observed for CPY* stabilized in a strain deleted of the DER1 gene (Knop et al., 1996a). The extent of stabilization correlates well with CPY* toxicity. CPY* expressed from its weaker endogenous promoter is not lethal to UPR-deficient cells. Under that condition, CPY* degradation was decreased only modestly (Ng et al., 2000; Travers et al., 2000).
Figure 3.
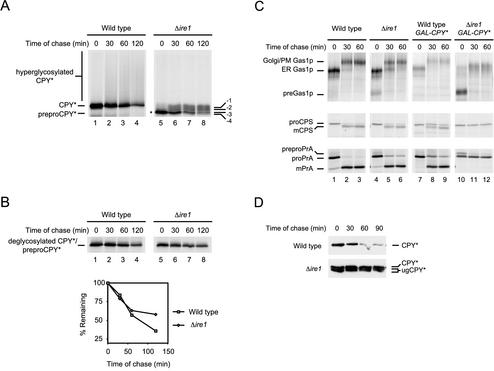
CPY* overexpression causes the loss of multiple functions in Δire1 cells. (A) Wild-type and Δire1 cells overexpressing CPY* were pulse-labeled for 10 min with [35S]methionine/cysteine followed by a cold chase as indicated. CPY* was immunoprecipitated from detergent lysates and resolved by SDS-PAGE. The positions of untranslocated CPY* (preproCPY* and asterisk), underglycosylated CPY* (-1, -2, -3, -4), CPY*, and hyperglycoslyated CPY* are indicated. (B) CPY* was deglycosylated using Endo H but otherwise prepared and analyzed as described in A. Quantification of CPY* decay was performed by phosphorimager analysis (bottom panel). (C) Wild-type and Δire1 cells nonexpressing or overexpressing CPY* were pulse-labeled for 10 min with [35S]methionine/cysteine followed by a cold chase at the indicated times. Endogenous Gas1p, PrA, and CPS were immunoprecipitated from detergent lysates, separated by SDS-PAGE, and visualized by autoradiography. For CPS and PrA, carbohydrates were removed using Endo H to unambiguously identify the relevant species. The underglycosylation defect of Δire1 cells increased the complexity of some forms. Positions of untranslocated (pre-), ER, Golgi/plasma membrane (Golgi/PM), and mature forms (mCPS and mPrA) of each protein are indicated. (D) CPY* stability was monitored using cycloheximide chase analysis. Equal cell numbers of wild-type or Δire1 strains overexpressing CPY* were collected at the indicated times after the addition of cycloheximide. Proteins from cell lysates were separated by SDS-PAGE and analyzed by immunoblotting. Positions of CPY* and underglycosylated CPY* are indicated (CPY* and ugCPY*).
Further defects were observed in the processing of CPY* in Δire1 cells. We previously observed impairment of ER protein translocation in Δire1 cells caused by CPY* expression and limiting translocation factors (Ng et al., 2000). The defect is even more severe with CPY* overexpression (Figure 3A, lane 5, preproCPY*). The defect is not restricted to CPY* as we also observed impaired translocation of the endogenous proteins Gas1p and PrA (Figure 3C, right panels). The kinetics of CPY* degradation was also unusual. After the pulse, a fraction was degraded rapidly in Δire1 cells. After 60 min of chase, the remainder was highly stable (Figure 3, A and B, right panels). The bimodal behavior could be explained by distinct CPY* populations, one in the lumen of the ER and the other, the untranslocated cytosolic precursor (Figure 3A, preproCPY*). Because a portion of newly synthesized CPY* mislocalizes to the cytosol, we hypothesized that this population is degraded, whereas the fraction entering the lumen remains stable. We tested this notion by measuring the turnover of CPY* using the “cycloheximide chase” method (Gardner et al., 1998). By contrast to a metabolic pulse-chase, this experiment analyzes the fate of total CPY* after a block in synthesis. Under these conditions, preproCPY* would represent only a minor fraction of the total because its appearance is transient during the pulse-chase experiment (Figure 3A, lanes 5–8). As shown in Figure 3D, total CPY* is turned over in control cells but highly stable in Δire1 cells. This experiment confirms that the luminal form of CPY* (with a substantial fraction underglycosylated; see below) accumulates stably in Δire1 cells.
In addition to the precursor, we also noticed other fast migrating forms of CPY* from Δire1 cells not present in wild-type (Figure 3A, labeled -1, -2, -3, -4). These species were not observed previously when CPY* was expressed moderately in this strain (Ng et al., 2000). This is reminiscent of the characteristic CPY underglycosylation pattern in mutants defective for N-linked glycosylation (te Heesen et al., 1992). CPY* underglycosylation was confirmed after treatment with Endo H. As shown in Figure 3B, the bands collapsed to a single species equal to the deglycosylated CPY* control. The defect is not confined to CPY* because we also observe underglycosylation of endogenous proteins (Figure 3C, Gas1p). Because glycosylation is normal in wild-type cells under the same conditions (Figure 3A), these data show that the impairment caused by the stress of misfolded proteins is alleviated by the UPR. Indeed, many genes involved in the synthesis (e.g., DPM1, RFT1), and transfer (OST2, OST3) of oligosaccharide moieties to secretory proteins are induced after UPR activation (Ng et al., 2000; Travers et al., 2000).
We recently reported that the degradation of CPY* by the ERAD pathway requires its transport and retrieval from the Golgi (Vashist et al., 2001). Thus, we wondered whether its stabilization is due, in part, to a defect in vesicular trafficking. For this, we examined the transport of the well-characterized cargo proteins Gas1p (plasma membrane), carboxypeptidase S (CPS, vacuole), and proteinase A (PrA, vacuole) (Klionsky et al., 1988; Nuoffer et al., 1991; Spormann et al., 1992). As shown in Figure 3C, overexpression of CPY* in wild-type cells had only a slight effect on the transport of these proteins. In Δire1 cells, however, a severe block in Gas1p transport was observed as indicated by the accumulation of the ER form (Figure 3C, Gas1p panel). This is likely a general transport block since the maturation of both CPS and PrA was also completely defective (Figure 3C, CPS and PrA panels).
The ERAD Pathway Is Not Essential for the Tolerance of CPY* Overexpression
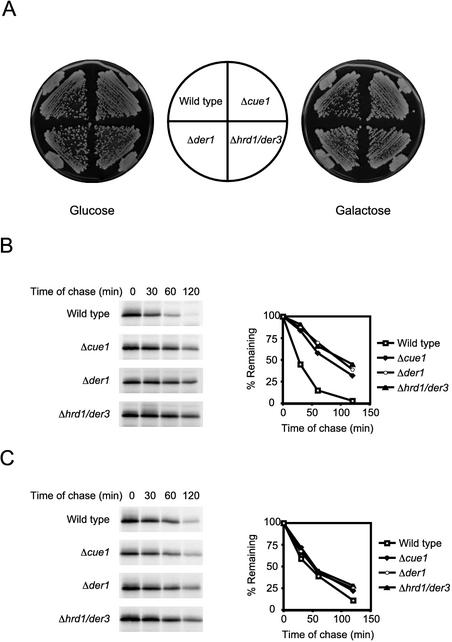
The extent of luminal CPY* stabilization suggested that the ERAD pathway is strongly impaired in Δire1 cells. Consistent with this notion, genes of the ERAD pathway (e.g., DER1, HRD1/DER3, HRD3, UBC7, and SEC61) are upregulated by the UPR upon ER stress (Travers et al., 2000). By extension, we wondered whether the toxicity of CPY* in Δire1 cells is due to the inability to eliminate it through the ERAD pathway. To address the question, we tested cell viability of several ERAD defective strains challenged by CPY* overexpression. Contrary to our expectations, strains deleted of the CUE1, DER1, and HRD1/DER3 genes grew no worse than the wild-type control under these conditions, indicating that the ERAD pathway is not essential for the tolerance of misfolded proteins (Figure 4A).
Figure 4.
ERAD mutants tolerate the stress of CPY* overexpression and efficiently degrades the protein by an alternative pathway. (A) Wild-type, Δcue1, Δder1, and Δhrd1/der3 strains with an integrated GAL-CPY* gene streaked on media containing YP glucose (Glucose) to repress expression or YP galactose to induce expression (Galactose). Plates were incubated at 30°C for 2 and 3 d, respectively. (B) Wild-type and ERAD mutant strains containing plasmid pDN436 (moderate CPY* expression controlled by its native promoter) were analyzed by pulse-chase analysis. CPY* was immunoprecipitated, treated with Endo H to remove N-linked carbohydrates, and analyzed by SDS-PAGE. (C) Wild-type and ERAD mutants transformed with plasmid pES67 (CPY* under the control of the strong GAL1 promoter) were analyzed as described for strains in B. Quantitative analysis was performed using a phosphorimager and plotted to the right of autoradiograms.
The result was surprising because we expected that the inability to clear an ER overloaded of misfolded proteins would be deleterious. It suggested that the accumulation of misfolded proteins is either benign in these strains or alternative pathways exist to remove them. To distinguish between these possibilities, we monitored the degradation of CPY* in wild-type and ERAD mutant strains by metabolic pulse-chase analysis. First, we confirmed the efficacy of the mutations in these strains by observing CPY* stabilization under moderate expression levels (Figure 4B). On overexpression, CPY* is degraded in the mutants nearly as rapidly as wild-type (Figure 4C). Because CPY* is synthesized more than tenfold higher under these conditions (unpublished results), we conclude that a robust alternative pathway is activated to rid misfolded proteins when ERAD is saturated or absent.
An ER-to-Vacuole Pathway Functions to Degrade Excess CPY*
A direct route to the vacuole has been observed for some abnormal proteins not subject to ERAD (Hong et al., 1996; Holkeri and Makarow, 1998). Therefore, we envisioned the possibility that excess CPY* might be diverted to the vacuole (a compartment analogous to metazoan lysosomes) for degradation. This seemed reasonable because genes required for the transport of proteins from the ER to the vacuole are upregulated upon ER stress as well as those encoding several vacuolar proteases (Travers et al., 2000). In addition, GAL1-regulated CPY* is stabilized in the ER-to-Golgi transport mutants sec18-1 and sec12-4 (unpublished results). We tested our hypothesis by measuring the turnover of overexpressed CPY* by metabolic pulse-chase and cycloheximide chase assays with a strain deleted of the PEP4 gene. PEP4 is required for the activation of most vacuolar proteases thereby making Δpep4 vacuoles deficient in proteolytic activity (Ammerer et al., 1986). In this strain, CPY* was stabilized compared with wild-type, suggesting that some degradation requires proteolytic enzymes dependent on PEP4 (Figure 5A). Because the ERAD pathway is fully functional in Δpep4 cells (Knop et al., 1996b), the partial stabilization reflects the fraction of CPY* that cannot be degraded by ERAD. Indeed, the cycloheximide chase experiment shows the inability to clear excess protein when vacuolar function is compromised (Figure 5B). This aspect of ER quality control may have eluded detection previously because PEP4 dependence was only revealed by CPY* expression levels sufficient to saturate the ERAD pathway.
Figure 5.
Vacuolar proteases degrade excess CPY*. (A) Wild-type and Δpep4 cells containing plasmid pES67 (CPY* controlled by the GAL1 promoter) were analyzed by pulse-chase analysis as described for Figure 3. (B) Wild-type and Δpep4 cells overexpressing CPY* (plasmid pES67) were analyzed by cycloheximide chase analysis as described in Figure 3D.
Although the genetic analysis suggested degradation in the vacuole, we sought to determine whether CPY* is transported there using indirect immunofluorescence. In both wild-type and Δpep4 strains, CPY* was found in the ER as shown by colocalization with the ER marker Kar2p (Figure 6, compare Aa and Ae to panels antibody and Af, respectively). In Δpep4 cells, CPY* was also found at sites distinct from the ER and nuclear envelope (Figure 6, Ae and Ag, arrows). We determined these to be vacuoles because the non-ER CPY* colocalizes with the vacuolar marker, GFP-tagged alkaline phosphatase (Figure 6, Ba–Bc). Because little CPY* can be detected in vacuoles of wild-type cells, these data indicate that degradation is rapid after its delivery to the vacuole.
Stress-Tolerance Mutants Are Defective in the ER-to-Golgi Trafficking of CPY*
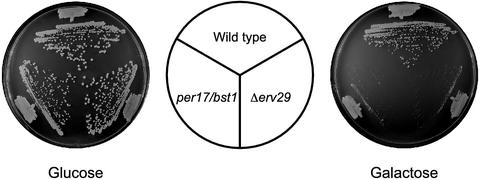
Our data suggest that the trafficking and turnover of misfolded proteins might play important roles in the stress tolerance of overexpressed CPY*. Interestingly, recent reports showed that strains with mutations in the PER17/BST1 and ERV29 genes seem to be defective in the transport and degradation of misfolded proteins (Caldwell et al., 2001; Vashist et al., 2001). Thus, we wondered whether such defects, in turn, would compromise tolerance of misfolded secretory proteins. We addressed the question by challenging per17/bst1 and Δerv29 cells with CPY* overexpression as performed with the UPR and ERAD deficient strains. As shown in Figure 7, both strains grow similarly to wild-type (Glucose) but the mutants grew poorly when CPY* expression was induced (Galactose). We next measured the turnover and transport of CPY* in these strains. Pulse-chase analysis showed that CPY* was partially stabilized in per17/bst1 cells (Figure 8, A and B) and strongly stabilized in Δerv29 cells (Figure 8, C and D). CPY* transport was measured by analyzing the acquisition of α-1,6 mannose, a carbohydrate modification that occurs in the Golgi apparatus (Herscovics and Orlean, 1993). This was performed by using anti-α-1,6 mannose antibodies as a second step in a sequential immunoprecipitation to measure the fraction of modified CPY*. As shown in Figure 8, B and D, modification of CPY* by α-1,6 mannose is sharply reduced in the mutants compared with wild-type. Thus, we conclude that CPY* is stabilized in the per17/bst1 and Δerv29 cells, and its transport from the ER-to-Golgi is also severely compromised. Importantly, the observed tolerance defect is not a consequence of a compromised UPR because these strains can activate the UPR to the same level as wild-type when treated with tunicamycin (unpublished results).
Figure 7.
The PER17/BST1 and ERV29 genes are required for the stress-tolerance of overexpressed CPY*. Wild-type (ESY342), per17/bst1 (ESY347) and Δerv29 (ESY386) cells transformed with pES67 (GAL-CPY*) were streaked onto glucose (expression off) or galactose plates (expression on) and incubated at 30°C for 2 and 3 d, respectively.
Figure 8.
The PER17/BST1 and ERV29 genes are required for the degradation and transport of overexpressed CPY*. (A) and (B) Wild-type (ESY342) and per17/bst1 (ESY347) cells overexpressing CPY* were subjected to pulse-chase analysis. CPY* was immunoprecipitated from detergent lysates and divided into two aliquots. From one CPY* was released from protein A resin and immunoprecipitated using anti-α 1,6-mannose antibodies. Immunoprecipitated proteins were treated with Endo H, and resolved by SDS-PAGE. Quantitative analysis was performed using a phosphorimager and plotted in B. The fraction of CPY* gaining α 1,6-mannose addition was determined by quantifying the ratio of CPY* immunoprecipitated with anti-α 1,6 antibody vs. the CPY* immunoprecipitated using anti-HA antibody. (C) and (D) Wild-type (ESY342) and Δerv29 (ESY386) cells overexpressing CPY* were subjected to pulse-chase analysis as in A and B.
DISCUSSION
Countering the effects of aberrant proteins is an important aspect of cellular homeostasis because as much as 30% of all nascent polypeptides misfold as a normal course of synthesis (Schubert et al., 2000; Turner and Varshavsky, 2000). These proteins can be restored by folding factors or removed altogether by degradative pathways. In the ER, the UPR is a key part of the process. Although recognized nearly a quarter century ago, recent advances have provided a detailed understanding of the pathway. In metazoans, there are at least three ER sensors that comprise the UPR. The first, Ire1p, is conserved among all eukaryotes and functions to splice messages of UPR regulatory factors (Cox and Walter, 1996; Kawahara et al., 1997; Shen et al., 2001; Yoshida et al., 2001; Calfon et al., 2002). The second, an ER-bound UPR-specific transcription factor termed ATF6 (with α and β isoforms) is released by intramembrane proteolysis upon activation (Haze et al., 1999; Ye et al., 2000). A third sensor, PERK, attenuates general translation to reduce the biosynthetic load on the organelle (Shi et al., 1998; Harding et al., 1999). This activity has been shown to play an important role in stress-tolerance in higher eukaryotes (Harding et al., 2000; Shen et al., 2001). Interestingly, BiP seems to play a role in the regulation of all three sensors (Bertolotti et al., 2000; Okamura et al., 2000; Shen et al., 2002). In yeast, the UPR appears to be less complex with only the Ire1p/Rlg1p/Hac1p circuit required. Indeed, we observed that a translational attenuation mechanism is not part of the yeast program (Figure 2). As a model system, yeast offers a tractable means to study this aspect of the pathway in the absence of other UPR outputs.
Previously, because of the small number of known target genes, the UPR was widely viewed as a pathway regulating secretory protein folding capacity by adjusting levels of ER chaperones and folding factors. Genome-wide expression analysis revealed the transcriptional program to be far more complex (Travers et al., 2000). From yeast, at least 381 upregulated genes covering a wide variety of functions were identified under conditions of severe ER stress. Among targets not directly related to folding, genes involved in ERAD are strongly induced. This was appealing because it suggested that the pathway regulates a means of ridding irreversibly damaged proteins. Direct analysis showed that UPR mutants are indeed partially impaired in degrading ERAD substrates (Casagrande et al., 2000; Ng et al., 2000; Travers et al., 2000). Interestingly, many UPR targets function in the secretory pathway beyond the ER. This would seem appropriate when the cell needs to increase its load of normal secretory proteins. However, it was unclear how membrane trafficking activities including vacuolar transport would serve to reduce the cytotoxicity of misfolded proteins in the ER.
Our previous efforts to understand ER quality control mechanisms provided a physiological basis for the necessity to upregulate transport functions to and from the Golgi apparatus. We showed that although some misfolded proteins are retained statically in the ER, others are transported to the Golgi and retrieved for degradation by ERAD (Vashist et al., 2001). Thus, a signaling pathway that monitors the level of misfolded proteins play an important role in ridding them. In the present study, we show that the ERAD pathway is saturable and excess substrate is transported to the vacuole for degradation. This transport does not require the CPY sorting factor Vps10p (Marcusson et al., 1994). Strains deleted of VPS10 degrade overexpressed CPY* as efficiently as wild-type without additional secretion into the media (unpublished results). The importance of removing excess misfolded protein from the ER was demonstrated by the growth sensitivity of the per17/bst1 and erv29 mutants (Figure 7). In these mutants, excess CPY* is poorly degraded while failing to traffic from the ER (Figure 8). Interestingly, the site, rather than the extent, of accumulation is more important for the manifestation of CPY* toxicity. Although pep4 cells accumulated similar amounts of excess CPY* as the per17/bst1 and Δerv29 mutants, they are not sensitive to CPY* overexpression, presumably because the accumulation occurs in the vacuole instead of the ER. Taken together, our data support the notion that stress tolerance of misfolded proteins requires their removal from the ER by any means available.
In previous studies, the expression of the mutant heterologous protein, Δpro, was also observed to be toxic to UPR-compromised cells (Umebayashi et al., 1999). Although some cellular functions were observed to be impaired, some effects of Δpro are different from CPY*. First, much higher levels of Δpro seem to be needed to elicit ER stress. Expression of Δpro from the GAL1 promoter at low copy as we performed with CPY* did not compromise the growth of a UPR mutant (Umebayashi et al., 1999). Even at the further elevated levels of Δpro expression needed to elicit stress, protein glycosylation was unaffected and its effect on ERAD function is not known. In addition, overexpression of the UPR target BiP was sufficient to abrogate Δpro toxicity, suggesting it as the limiting factor. By contrast, we found that BiP overexpression at various levels failed to suppress the lethality of Δire1 cells challenged with CPY* (unpublished results). This is consistent with the view that multiple functions of the UPR program are required to alleviate CPY* toxicity. The differences between these studies could be explained by a recent report showing that Δpro is not a substrate of ERAD (Umebayashi et al., 2001). Therefore, it is likely to cause toxicity through a different mechanism, possibly by exhausting the available pool of BiP.
In a recent report that assessed the fate of increased CPY* expression, excess CPY* was degraded efficiently. Their studies revealed a new role for Rsp5p, an E3 ubiquitin ligase not previously known to be involved in ERAD (Haynes et al., 2002). We consider it likely that a portion of the CPY* expressed in our system utilizes the alternative E3 ligase. In the current study, CPY* was expressed to the extent that the ERAD ubiquitin/proteasomal pathway became saturated. This differs from the previous study where saturation of DER1/HRD1-dependent degradation was observed but saturation of ERAD was not. The difference is likely due to distinct approaches to CPY* overexpression. In the previous study, increased expression was accomplished using a CPY*-bearing multicopy 2-μm plasmid. Under that condition, expression levels vary according to plasmid copy number, which is stochastically distributed within cell populations (Haynes et al., 2002). In addition, expression levels would be limited to the tolerance level of the host, making it difficult to analyze strains sensitive to misfolded proteins. By using the strong regulated GAL1 promoter at low copy, it was possible to overcome these limitations by more uniform overexpression of CPY*. Indeed this approach allowed the observation of the ER-to-vacuole degradative pathway and the stress-sensitivity of IRE1, PER17/BST1, and ERV29 mutant strains.
Using information gleaned from this and other studies on the UPR's role in stress tolerance, we propose the following model. Accumulation of misfolded proteins promotes the widespread loss of ER function—possibly by tying up essential factors. Induction of the UPR alleviates this stress by increasing the synthesis of limiting factors. These would include factors involved in protein translocation, folding, and glycosylation, many of which are known transcriptional targets (Ng et al., 2000; Travers et al., 2000). To rid the cell of the offending proteins, the UPR expands quality control functions including ERAD and vesicle trafficking. Under more severe stress, when ERAD is saturable, an alternative pathway to the vacuole is utilized to degrade excess substrate. Among higher eukaryotes, a similar mechanism has not yet been described. However, for them, the need to upregulate trafficking and lysosomal functions might be less important. The translational repression mechanism might sufficiently limit the load of newly synthesized proteins so the regulation of ER functions alone would suffice to restore homeostasis.
Acknowledgments
We thank Shilpa Vashist and Woong Kim for valuable discussions. We are grateful to Chris Burd, Scott Emr, Sarah Rice, Howard Riezman, Randy Schekman, Tom Stevens, Ron Vale, and Peter Walter for providing strains, plasmids, and antibodies. This work was supported by a grant from the National Institutes of Health to D.T.W.N. (GM059171).
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–11–0717. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-11-0717.
References
- Ammerer, G., Hunter, C.P., Rothman, J.H., Saari, G.C., Valls, L.A., and Stevens, T.H. (1986). PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol. Cell. Biol. 6, 2490–2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays, N.W., Gardner, R.G., Seelig, L.P., Joazeiro, C.A., and Hampton, R.Y. (2001). Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat. Cell Biol 3, 24–29. [DOI] [PubMed] [Google Scholar]
- Bertolotti, A., Zhang, Y., Hendershot, L.M., Harding, H.P., and Ron, D. (2000). Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol 2, 326–332. [DOI] [PubMed] [Google Scholar]
- Brodsky, J.L., and McCracken, A.A. (1999). ER protein quality control and proteasome-mediated protein degradation. Semin. Cell Dev. Biol. 10, 507–513. [DOI] [PubMed] [Google Scholar]
- Caldwell, S.R., Hill, K.J., and Cooper, A.A. (2001). Degradation of endoplasmic reticulum (ER) quality control substrates requires transport between the ER and Golgi. J. Biol. Chem. 276, 23296–23303. [DOI] [PubMed] [Google Scholar]
- Calfon, M., Zeng, H., Urano, F., Till, J.H., Hubbard, S.R., Harding, H.P., Clark, S.G., and Ron, D. (2002). IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96. [DOI] [PubMed] [Google Scholar]
- Carrell, R.W., and Gooptu, B. (1998). Conformational changes and disease—serpins, prions, and Alzheimer's. Curr. Opin. Struct. Biol. 8, 799–809. [DOI] [PubMed] [Google Scholar]
- Casagrande, R., Stern, P., Diehn, M., Shamu, C., Osario, M., Zuniga, M., Brown, P.O., and Ploegh, H. (2000). Degradation of proteins from the ER of S. cerevisiae requires an intact unfolded protein response pathway. Mol. Cell 5, 729–735. [DOI] [PubMed] [Google Scholar]
- Cowles, C.R., Odorizzi, G., Payne, G.S., and Emr, S.D. (1997). The AP-3 adaptor complex is essential for cargo-selective transport to the yeast vacuole. Cell 91, 109–118. [DOI] [PubMed] [Google Scholar]
- Cox, J.S., Shamu, C.E., and Walter, P. (1993). Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73, 1197–1206. [DOI] [PubMed] [Google Scholar]
- Cox, J.S., and Walter, P. (1996). A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87, 391–404. [DOI] [PubMed] [Google Scholar]
- Ellgaard, L., and Helenius, A. (2001). ER quality control: towards an understanding at the molecular level. Curr. Opin. Cell Biol. 13, 431–437. [DOI] [PubMed] [Google Scholar]
- Finger, A., Knop, M., and Wolf, D.H. (1993). Analysis of two mutated vacuolar proteins reveals a degradation pathway in the endoplasmic reticulum or a related compartment of yeast. Eur. J. Biochem. 218, 565–574. [DOI] [PubMed] [Google Scholar]
- Friedlander, R., Jarosch, E., Urban, J., Volkwein, C., and Sommer, T. (2000). A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat. Cell Biol 2, 379–384. [DOI] [PubMed] [Google Scholar]
- Gardner, R., Cronin, S., Leader, B., Rine, J., Hampton, R., and Leder, B. (1998). Sequence determinants for regulated degradation of yeast 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell 9, 2611–2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie, C., and Fink, G. R. (eds.) (1991). Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press, Inc.
- Hammond, C., and Helenius, A. (1994). Quality control in the secretory pathway: retention of a misfolded viral membrane glycoprotein involves cycling between the ER, intermediate compartment, and Golgi apparatus. J. Cell Biol. 126, 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding, H.P., Zhang, Y., Bertolotti, A., Zeng, H., and Ron, D. (2000). Perk is essential for translational regulation and cell survival during the unfolded protein response. Mol Cell 5, 897–904. [DOI] [PubMed] [Google Scholar]
- Harding, H.P., Zhang, Y., and Ron, D. (1999). Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397, 271–274. [DOI] [PubMed] [Google Scholar]
- Haynes, C.M., Caldwell, S., and Cooper, A.A. (2002). An HRD/DER-independent ER quality control mechanism involves Rsp5p-dependent ubiquitination and ER-Golgi transport. J. Cell Biol. 158, 91–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haze, K., Yoshida, H., Yanagi, H., Yura, T., and Mori, K. (1999). Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell 10, 3787–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herscovics, A., and Orlean, P. (1993). Glycoprotein biosynthesis in yeast. FASEB J. 7, 540–550. [DOI] [PubMed] [Google Scholar]
- Hiller, M.M., Finger, A., Schweiger, M., and Wolf, D.H. (1996). ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science 273, 1725–1728. [DOI] [PubMed] [Google Scholar]
- Holkeri, H., and Makarow, M. (1998). Different degradation pathways for heterologous glycoproteins in yeast. FEBS Lett. 429, 162–166. [DOI] [PubMed] [Google Scholar]
- Hong, E., Davidson, A.R., and Kaiser, C.A. (1996). A pathway for targeting soluble misfolded proteins to the yeast vacuole. J. Cell Biol. 135, 623–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara, T., Yanagi, H., Yura, T., and Mori, K. (1997). Endoplasmic reticulum stress-induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol. Biol. Cell 8, 1845–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, P.S., and Arvan, P. (1998). Endocrinopathies in the family of endoplasmic reticulum (ER) storage diseases: disorders of protein trafficking and the role of ER molecular chaperones. Endocr. Rev. 19, 173–202. [DOI] [PubMed] [Google Scholar]
- Klionsky, D.J., Banta, L.M., and Emr, S.D. (1988). Intracellular sorting and processing of a yeast vacuolar hydrolase: proteinase A propeptide contains vacuolar targeting information. Mol. Cell. Biol. 8, 2105–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop, M., Finger, A., Braun, T., Hellmuth, K., and Wolf, D.H. (1996a). Der1, a novel protein specifically required for endoplasmic reticulum degradation in yeast. EMBO J. 15, 753–763. [PMC free article] [PubMed] [Google Scholar]
- Knop, M., Hauser, N., and Wolf, D.H. (1996b). N-Glycosylation affects endoplasmic reticulum degradation of a mutated derivative of carboxypeptidase yscY in yeast. Yeast 12, 1229–1238. [DOI] [PubMed] [Google Scholar]
- Kopito, R.R., and Ron, D. (2000). Conformational disease. Nat. Cell Biol. 2, E207–E209. [DOI] [PubMed] [Google Scholar]
- Kopito, R.R., and Sitia, R. (2000). Aggresomes and Russell bodies. Symptoms of cellular indigestion? EMBO Rep. 1, 225–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov, V.V. (2000). Rapid and reliable protein extraction from yeast. Yeast 16, 857–860. [DOI] [PubMed] [Google Scholar]
- Marcusson, E.G., Horazdovsky, B.F., Cereghino, J.L., Gharakhanian, E., and Emr, S.D. (1994). The sorting receptor for yeast vacuolar carboxypeptidase Y is encoded by the VPS10 gene. Cell 77, 579–586. [DOI] [PubMed] [Google Scholar]
- Mori, K., Ma, W., Gething, M.J., and Sambrook, J. (1993). A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 74, 743–756. [DOI] [PubMed] [Google Scholar]
- Ng, D.T., Spear, E.D., and Walter, P. (2000). The unfolded protein response regulates multiple aspects of secretory and membrane protein biogenesis and endoplasmic reticulum quality control. J. Cell Biol. 150, 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierras, C.R., and Warner, J.R. (1999). Protein kinase C enables the regulatory circuit that connects membrane synthesis to ribosome synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 274, 13235–13241. [DOI] [PubMed] [Google Scholar]
- Nuoffer, C., Jeno, P., Conzelmann, A., and Riezman, H. (1991). Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae GAS1 protein to the plasma membrane. Mol. Cell. Biol. 11, 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura, K., Kimata, Y., Higashio, H., Tsuru, A., and Kohno, K. (2000). Dissociation of Kar2p/BiP from an ER sensory molecule, Ire1p, triggers the unfolded protein response in yeast. Biochem. Biophys. Res. Commun. 279, 445–450. [DOI] [PubMed] [Google Scholar]
- Patil, C., and Walter, P. (2001). Intracellular signaling from the endoplasmic reticulum to the nucleus: the unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13, 349–355. [DOI] [PubMed] [Google Scholar]
- Pilon, M., Schekman, R., and Romisch, K. (1997). Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 16, 4540–4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plemper, R.K., Bohmler, S., Bordallo, J., Sommer, T., and Wolf, D.H. (1997). Mutant analysis links the translocon and BiP to retrograde protein transport for ER degradation. Nature 388, 891–895. [DOI] [PubMed] [Google Scholar]
- Plemper, R.K., and Wolf, D.H. (1999). Retrograde protein translocation: ERADication of secretory proteins in health and disease. Trends Biochem. Sci. 24, 266–270. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.M., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. Plainview: Cold Spring Harbor Laboratory Press.
- Schubert, U., Anton, L.C., Gibbs, J., Norbury, C.C., Yewdell, J.W., and Bennink, J.R. (2000). Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature 404, 770–774. [DOI] [PubMed] [Google Scholar]
- Shen, J., Chen, X., Hendershot, L., and Prywes, R. (2002). ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 3, 99–111. [DOI] [PubMed] [Google Scholar]
- Shen, X. et al. (2001). Complementary signaling pathways regulate the unfolded protein response and are required for C. elegans development. Cell 107, 893–903. [DOI] [PubMed] [Google Scholar]
- Shi, Y., Vattem, K.M., Sood, R., An, J., Liang, J., Stramm, L., and Wek, R.C. (1998). Identification and characterization of pancreatic eukaryotic initiation factor 2 alpha-subunit kinase, PEK, involved in translational control. Mol. Cell. Biol. 18, 7499–7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter, P. (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spear, E., and Ng, D.T. (2001). The unfolded protein response: no longer just a special teams player. Traffic 2, 515–523. [DOI] [PubMed] [Google Scholar]
- Spormann, D.O., Heim, J., and Wolf, D.H. (1992). Biogenesis of the yeast vacuole (lysosome). The precursor forms of the soluble hydrolase carboxypeptidase yscS are associated with the vacuolar membrane. J. Biol. Chem. 267, 8021–8029. [PubMed] [Google Scholar]
- te Heesen, S., Janetzky, B., Lehle, L., and Aebi, M. (1992). The yeast WBP1 is essential for oligosaccharyltransferase activity in vivo and in vitro. EMBO J. 11, 2071–2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers, K.J., Patil, C.K., Wodicka, L., Lockhart, D.J., Weissman, J.S., and Walter, P. (2000). Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101, 249–258. [DOI] [PubMed] [Google Scholar]
- Turner, G.C., and Varshavsky, A. (2000). Detecting and measuring cotranslational protein degradation in vivo. Science 289, 2117–2120. [DOI] [PubMed] [Google Scholar]
- Umebayashi, K., Fukuda, R., Hirata, A., Horiuchi, H., Nakano, A., Ohta, A., and Takagi, M. (2001). Activation of the Ras-cAMP signal transduction pathway inhibits the proteasome-independent degradation of misfolded protein aggregates in the endoplasmic reticulum lumen. J. Biol. Chem. 276, 41444–41454. [DOI] [PubMed] [Google Scholar]
- Umebayashi, K., Hirata, A., Horiuchi, H., Ohta, A., and Takagi, M. (1999). Unfolded protein response-induced BiP/Kar2p production protects cell growth against accumulation of misfolded protein aggregates in the yeast endoplasmic reticulum. Eur J. Cell Biol. 78, 726–738. [DOI] [PubMed] [Google Scholar]
- Vashist, S., Kim, W., Belden, W.J., Spear, E.D., Barlowe, C., and Ng, D.T. (2001). Distinct retrieval and retention mechanisms are required for the quality control of endoplasmic reticulum protein folding. J. Cell Biol. 155, 355–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, P., and Johnson, A.E. (1994). Signal Sequence Recognition and Protein Targeting to the Endoplasmic Reticulum Membrane. Annu. Rev. Cell Biol. 10, 87–119. [DOI] [PubMed] [Google Scholar]
- Ward, C.L., Omura, S., and Kopito, R.R. (1995). Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83, 121–127. [DOI] [PubMed] [Google Scholar]
- Yamamoto, K., Fujii, R., Toyofuku, Y., Saito, T., Koseki, H., Hsu, V.W., and Aoe, T. (2001). The KDEL receptor mediates a retrieval mechanism that contributes to quality control at the endoplasmic reticulum. EMBO J. 20, 3082–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, J., Rawson, R.B., Komuro, R., Chen, X., Dave, U.P., Prywes, R., Brown, M.S., and Goldstein, J.L. (2000). ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6, 1355–1364. [DOI] [PubMed] [Google Scholar]
- Yoshida, H., Matsui, T., Yamamoto, A., Okada, T., and Mori, K. (2001). XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891. [DOI] [PubMed] [Google Scholar]
- Zhang, J.X., Braakman, I., Matlack, K.E., and Helenius, A. (1997). Quality control in the secretory pathway: the role of calreticulin, calnexin, and BiP in the retention of glycoproteins with C-terminal truncations. Mol. Biol. Cell 8, 1943–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, M., and Schekman, R. (1999). The engagement of Sec61p in the ER dislocation process. Mol. Cell 4, 925–934. [DOI] [PubMed] [Google Scholar]