Abstract
The transmembrane 9 (TM9) family of proteins contains numerous members in eukaryotes. Although their function remains essentially unknown in higher eukaryotes, the Dictyostelium discoideum Phg1a TM9 protein was recently reported to be essential for cellular adhesion and phagocytosis. Herein, the function of Phg1a and of a new divergent member of the TM9 family called Phg1b was further investigated in D. discoideum. The phenotypes of PHG1a, PHG1b, and PHG1a/PHG1b double knockout cells revealed that Phg1a and Phg1b proteins play a synergistic but not redundant role in cellular adhesion, phagocytosis, growth, and development. Complementation analysis supports a synergistic regulatory function rather than a receptor role for Phg1a and Phg1b proteins. Together, these results suggest that Phg1 proteins act as regulators of cellular adhesion, possibly by controlling the intracellular transport in the endocytic pathway and the composition of the cell surface.
INTRODUCTION
Members of the transmembrane 9 (TM9) family are characterized by the presence of a large variable extracellular N-terminal domain followed by nine putative transmembrane domains in their conserved C termini (see drawing in Figure 6A) (Chluba-de Tapia et al., 1997). Due to this high degree of conservation in sequence and predicted topology (Chluba-de Tapia et al., 1997; Schimmoller et al., 1998), genes encoding TM9 proteins can be unambiguously identified in eukaryotic genomes. The family includes many members in organisms ranging from yeast to plants and human. Although conservation throughout evolution strongly suggests an important biological role, their function(s) remains largely unknown (Chluba-de Tapia et al., 1997).
Figure 6.
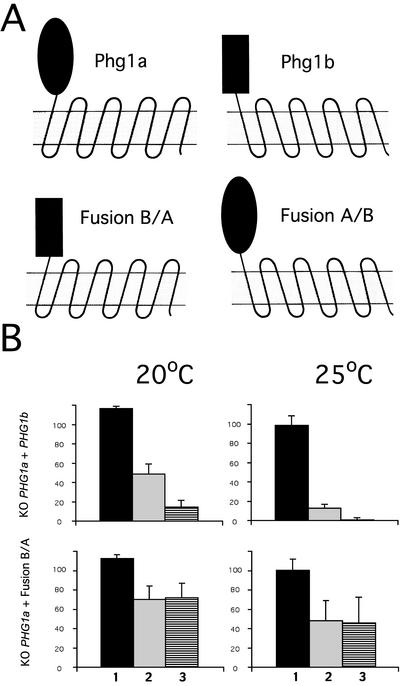
Phg1a and Phg1b proteins are not functionally redundant. (A) Plasmids encoding chimeras composed of the extracellular domain of the Phg1a protein and the membrane portion of Phg1b (A/B fusion) or of the extracellular domain of Phg1b and the membrane portion of Phg1a (B/A fusion) were constructed. (B) Phagocytosis rate of three substrates by PHG1a knockout cells expressing either Phg1b or the B/A fusion protein was tested: 1, plain latex beads in phosphate buffer; 2, E. coli in HL5; and 3, plain latex beads in HL5. Phg1b overexpression did not complement the phagocytosis defect of PHG1a knockout cells, whereas the expression of the B/A fusion protein led to a partial complementation of the phagocytosis defect (compare with Figure 5C). These data represent the average of four independent experiments and the SD is indicated.
A first indication on the function of TM9 proteins arose from a reverse genetics approach in the amoeba Dictyostelium discoideum. The TM9 protein Phg1a was found to be essential for cellular adhesion and phagocytosis (Cornillon et al., 2000), and it was then proposed that TM9 proteins might represent a family of adhesion receptors. Their variable extracellular domains could confer distinct receptor specificities, whereas the conserved membrane-associated domain would establish the link with the intracellular machinery. However, this interpretation remains highly hypothetical and a regulatory role in cellular adhesion was not excluded (Cornillon et al., 2000).
To gain further insight into the function of proteins belonging to the TM9 family, knockouts of two genes encoding distinct members of the family were obtained and characterized in D. discoideum. Together, our results indicate that TM9 proteins play synergistic roles and strongly support a model where they function as regulators of cellular adhesion by regulating the protein composition of the cell surface.
MATERIALS AND METHODS
Cell Culture, Media, and Antibodies
The wild-type DH1 cells and PHG1a knockout cells used in this study were described by Cornillon et al. (2000). Cells were grown at 20°C in HL5 medium (Cornillon et al., 1998) and subcultured twice a week to maintain a density of <106 cells/ml. D. discoideum cell transfection was performed as described previously (Cornillon et al., 2000). For experiments at 25°C, cells were incubated overnight at this temperature in HL5 medium before the experiment.
For developmental analysis, cells were plated on 0.45-μm membrane filters laid onto two layers of Whatman paper grade 1 soaked with phosphate buffer (5 mM Na2HPO4, 5 mM KH2PO4, 1 mM CaCl2, 2 mM MgCl2, pH 6.5) (Whatman, Maidstone, United Kingdom) (Sussman, 1987). For each mutant used herein, three independent mutant strains were tested and identical phenotypes were observed. Thereafter, only one clone was chosen for extensive characterization.
Rabbit polyclonal antiserum against a Phg1a peptide (YC1) was described previously (Cornillon et al., 2000). An anti-peptide rabbit polyclonal antibody against Phg1b was directed to a sequence in the lumenal domain of the Phg1b protein (GEYYYAEMIYDDLP).
Nucleotide Sequences and Plasmids
PHG1b Sequence and Expression Plasmid. The full-length PHG1b open reading frame is represented in cDNA clone SSE715 from the D. discoideum cDNA project in Japan (Morio et al., 1998) (GenBank accession no. C92378). This clone was obtained and fully sequenced (accession no. AJ507828). PHG1b was subcloned into a derivative of pDXA-3C (Manstein et al., 1995; Cornillon et al., 2000) to obtain the pSC3B vector that upon transfection into D. discoideum cells drives overexpression (actin 15 promoter) of Phg1b protein.
PHG1c Sequence. The full-length PHG1c open reading frame is represented in cDNA clone dda25m17 (GenBank accession no. BJ329614) from the D. discoideum cDNA project in Japan (Morio et al., 1998). This clone was obtained and fully sequenced (accession no. AJ507829).
PHG1b Knockout Plasmid. The pSC4 plasmid was produced by introducing into pDXA-3C (Manstein et al., 1995), two fragments of the PHG1b gene: XbaI/ClaI DNA fragment and a polymerase chain reaction DNA fragment amplified with the two following oligonucleotides, 5′-GGTACAGTTGGTTTCTAC-3′ and 5′-ACCCTATTTGTAGCACCA-3′.
A/B Fusion Protein. The pMB25 plasmid harbors a fusion gene encoding a chimera made of the hydrophilic extracellular domain of Phg1a (M1 to V276) and the nine transmembrane domains of Phg1b (H221–D587).
B/A Fusion Protein. The pMB24 plasmid harbors a fusion gene encoding a chimera made of the hydrophilic extracellular domain of Phg1b (M1 to I220) and the nine transmembrane domains of Phg1a (H277 to N642). pMB24 and pMB25 were linearized with pvUI and used to transfect PHG1a knockout cells to obtain the G418 resistant cells expressing the corresponding fusion protein.
Phg1a-GST Fusion Protein. The pMB30 plasmid harbors a hybrid gene encoding an N-terminal fusion of glutathione transferase to the full-length Phg1a protein and allows its expression in D. discoideum cells. pMB30 was linearized with pvUI and used to transfect PHG1a knockout cells to obtain the G418-resistant cells expressing the fusion protein.
Multiple alignment and phylogenetic analysis were performed at the Pôle Bio-Informatique Lyonnais Web site with ClustalW. Homology searches and sequences submission were performed at the European Molecular Biology Laboratory Web site.
Phagocytosis and Fluid Phase Uptake Assays
Rhodamine-labeled bacteria were produced as described previously (Cornillon et al., 2000). Fluoresbrite YG 1-μm carboxylate microspheres, fluoresbrite YG 1-μm plain microspheres, and fluospheres 1-μm amine-modified microspheres were purchased from Polysciences (Warrington, PA).
Measurements of phagocytic substrate internalizations were carried out as described previously (Cornillon et al., 2000). Phagocytosis assays were done at 20°C and 25°C in either HL5 medium or phosphate buffer (SB; 2 mM Na2HPO4, 14.7 mM KH2PO4, pH 6.5). When the phagocytosis assay using latex beads in HL5 medium was carried out at 25°C with cells grown at 20°C, a 50% defect in phagocytosis by PHG1b knockout cells relative to wild-type cells was observed (our unpublished data). Because the thermosensitive growth phenotype of PHG1b knockout cells became significant after 48 h, a 16-h preincubation at 25°C was chosen to assess the effect of temperature on phagocytosis in PHG1b knockout cells. Longer incubations at 25°C, however, did not enhance the defect in phagocytosis (our unpublished data). Fluid-phase uptake was measured using fluorescein isothiocyanate-dextran (Molecular Probes, Eugene, OR) as described previously (Cornillon et al., 2000).
Adhesion of Cells to Substrate
Cell adhesion to glass surfaces was probed using the radial flow detachment assay, performed at 20°C and 25°C in either HL5 or phosphate buffer, according to Decave et al. (2002). The flow was applied during 7 min. Adhesion was quantified by measuring the value of the shear stress detaching 50% of the cells.
Affinity Purification of Phg1a-GST Fusion Protein
D. discoideum cells (5 × 106) expressing the Phg1a-GST fusion protein were lysed with either 0.5% Triton X-100 or 0.5% 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate (CHAPS) in 20 mM phosphate buffer, pH 7.4, containing 150 mM of NaCl. Under these conditions, Phg1b as well as Phg1a are found in the cellular lysate and absent from the insoluble pellet (our unpublished data). Lysates were cleared by centrifugation, 15 min at 10,000 × g at 4°C, and applied to either glutathione S-transferase GST or FLAG affinity resin (Amersham Biosciences AB, Uppsala, Sweden, and Sigma-Aldrich, St. Louis, MO, respectively) for 1 h at 4°C. Both resins were washed five times with the corresponding lysate buffer and bound proteins were eluted with sample buffer (0.103 g/ml sucrose, 5 × 10–2 M Tris, pH 6.8, 5 × 10–3 M EDTA, 0.5 mg/ml bromophenol blue, 2% SDS). D. discoideum cell equivalent (2 × 106) was loaded on SDS-PAGE for Western blot analysis.
Protein Electrophoresis and Immunodetection
To determine the presence of Phg1a and Phg1b proteins, cells were washed once in HL5 medium, resuspended at 2 × 105 cells/10 μl in sample buffer (0.103 g/ml sucrose, 5 × 10–2 M Tris, pH 6.8, 5 × 10–3 M EDTA, 0.5 mg/ml bromophenol blue, 2% SDS), and each sample was run on a 9% acrylamide gel under nonreducing conditions. The proteins were then transferred to a nitrocellulose BA 85 membrane (Schleicher & Schuell, Dassel, Germany). The membrane was incubated with a primary rabbit antibody to Phg1a or Phg1b and then a horseradish peroxidase-coupled donkey antiserum to rabbit Ig (Amersham Biosciences AB), washed, and revealed by enhanced chemiluminescence.
Cell Surface Protein Biotinylation
Cells were grown in HL5 medium at a concentration of 5 × 105 cells/ml. After cooling down to 4°C, cells were collected and washed once with ice-cold SB, pH 7.8, by centrifugation. Cells were then resuspended in ice-cold SB containing 2 mg/ml NHS-Sulfobiotin (Pierce Chemical, Rockford, IL). After 30-min incubation on ice, the cells were washed twice with ice-cold SB containing 100 mM glycine, pH 7.2, and resuspended in nonreducing sample buffer. Lysate of 2.5 × 105 cells was loaded onto a 9% acrylamide gel and electrophoresed under nonreducing conditions. The lane was cut out of the gel, incubated in reducing sample buffer for 20 min, and loaded onto a second 9% reducing acrylamide gel. Biotinylated proteins were detected with ImmunoPure Avidin horseradish peroxidase conjugate (Pierce Chemical).
Fluorescence Microscopy
Cells were grown on glass coverslips in HL5 medium for 3 d at 20°C and were processed for p80 immunofluorescence analysis as described previously (Ravanel et al., 2001). Briefly, cells were allowed to internalize a fluid-phase marker (fluoresbrite YG 50-nm microspheres; Polysciences) in HL5 for 90 min and then fixed in 4% paraformaldehyde, permeabilized with saponin, and incubated successively with H161 monoclonal antibody specific for the p80 endosomal protein and the corresponding fluorescent secondary antibody. Cells were visualized with a confocal microscope (LSM510; Carl Zeiss, Jena, Germany).
RESULTS
TM9 Family in D. discoideum
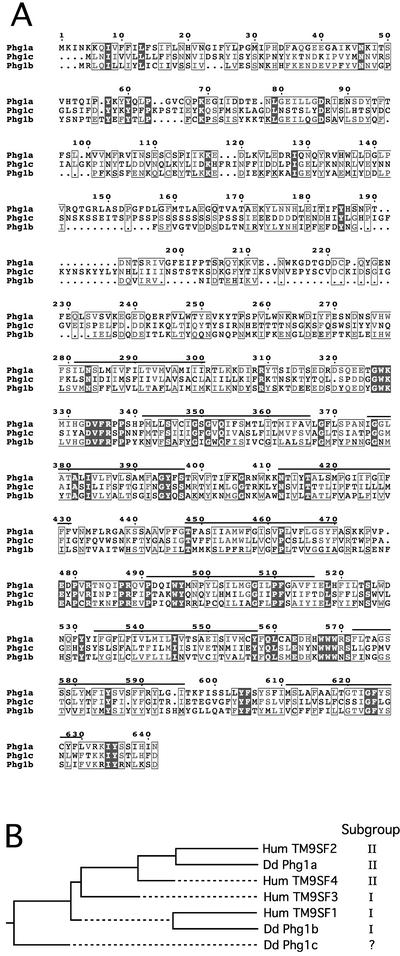
In addition to Phg1a, two other D. discoideum TM9 proteins designated Phg1b and Phg1c were identified in databases. The two corresponding cDNA clones were fully sequenced. PHG1a, b, and c encode proteins of similar length (642, 587, and 654 amino acids, respectively) (Figure 1A). The general organization of these proteins is also conserved and presents the characteristic features of members of the TM9 family: an N-terminal signal peptide followed by a large extracellular domain and a hydrophobic C terminus containing nine putative transmembrane domains.
Figure 1.
TM9 family in D. discoideum. (A) Protein alignment of three members of the TM9 in D. discoideum. Putative transmembrane domains are underlined. (B) Phylogenetic tree of TM9 proteins in D. discoideum (Phg1a, b, and c) and in human (TM9SF1, 2, 3, and 4). Sequences were TM9SF1 (hEMP70; accession no. O15321), TM9SF2 (p76; accession no. Q99805), TM9SF3 (hSMBP; accession no. Q9HD45), TM9SF4 (accession no. Q92544), Phg1a (accession no. AJ318760), Phg1b (accession no. AJ507828), and Phg1c (accession no. AJ507829).
Phg1b and Phg1c are 24.5 and 21.2% homologous to the Phg1a protein, respectively (16.3 and 12.9% in the extracellular domain, respectively; 33 and 31.3% in the membrane domain, respectively). A previous report classified the TM9 proteins into two subgroups, referred to herein as I and II (Sugasawa et al., 2001). Subgroup I is characterized by a shorter (∼220 amino acids) hydrophilic N-terminal domain and a characteristic motif at position 50 (VGPYxNxQETY), whereas subgroup II contains a conserved sequence [FY(V/L)PG(VL)AP] immediately after the signal peptide and a longer N-terminal domain (∼280 amino acids). Phg1a belongs to subgroup II (FYLPGMIP at amino acid position 25) and Phg1b to subgroup I (VGPYSNPTETY at amino acid position 42) (Figure 1B). The Phg1c sequence does not contain either of these two distinctive motifs and therefore cannot be classified unambiguously into either subgroup. Phylogenetic analysis shows that Phg1a and b proteins are distantly related members of the TM9 family in D. discoideum, differing to the same extent as the most divergent members of the human family TM9SF1 and TM9SF2 (Figure 1B). For comparison, TM9SF1 and TM9SF2 are 27.2% homologous (15% in the extracellular domain, 35.8% in the membrane domain). Because Phg1a and b are divergent members of the TM9 family and belong to two distinct subclasses, analysis of their function(s) may provide a general view of the function of TM9 proteins. Functional analysis was therefore focused on these two proteins.
TM9 Proteins Play Synergistic Roles in Growth and Development
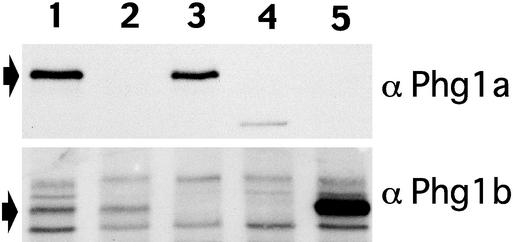
To investigate the respective roles of the Phg1a and Phg1b proteins, the PHG1b gene was inactivated by homologous recombination in wild-type cells to obtain the PHG1b knockout or in PHG1a knockout cells to obtain the PHG1a/PHG1b double knockout. Expression of Phg1a and Phg1b proteins in the resulting knockout strains was assessed by Western blot (Figure 2). In wild-type and PHG1a knockout cells, the polyclonal antiserum directed to the Phg1b protein recognized one major band at a molecular weight of ∼57 kDa (Figure 2, lanes 1 and 2). This band was absent in PHG1b and PHG1a/PHG1b knockout cells (Figure 2, lanes 3 and 4) and clearly more intense in cells overexpressing Phg1b (Figure 2, lane 5), confirming that it corresponds to the Phg1b protein. Several other bands represented nonspecific cross-reactivity of the antiserum with unrelated proteins. The Phg1a antiserum detected one band at 65 kDa in wild-type cells and in PHG1b knockout cells (Figure 2, lanes 1 and 3) that was absent in PHG1a and PHG1a/PHG1b knockout cells (Figure 2, lanes 2, 4, and 5).
Figure 2.
Targeted disruption of PHG1a and PHG1b in D. discoideum. Cell lysates were analyzed by 9% SDS-PAGE, processed for protein gel blotting and probed with anti-αPhg1a-specific antibody (top) or anti-Phg1b-specific antibody (bottom). Arrows indicate bands corresponding to the Phg1a and Phg1b proteins. Lane 1 shows proteins from wild-type cells; lane 2, PHG1a knockout cells; lane 3, PHG1b knockout cells; lane 4, PHG1a/PHG1b double knockout cells; and lane 5, PHG1a knockout cells overexpressing PHG1b. The low molecular weight Phg1a species visible in PHG1a/PHG1b knockout cells corresponds to an unstable truncation product of Phg1a produced in PHG1a knockout cells (Cornillon et al., 2000).
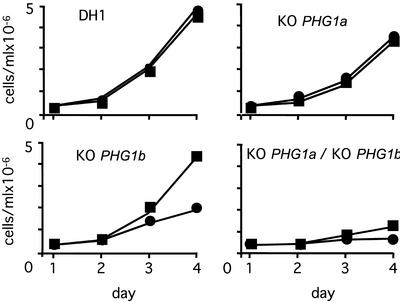
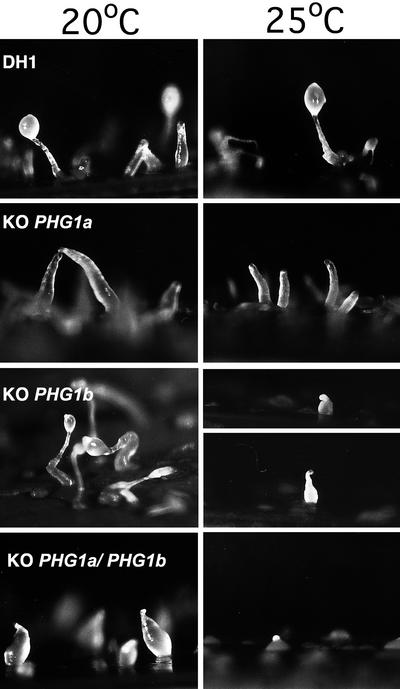
Although both PHG1a and PHG1b knockout cells grew almost as well as wild-type cells in HL5 medium at 20°C, growth of PHG1a/PHG1b double knockout cells was severely reduced (Figure 3). Furthermore, a thermosensitive growth phenotype was observed for PHG1b knockout cells: growth was normal at 20°C (usual growth temperature for D. discoideum) but severely inhibited at 25°C (Figure 3). A temperature shift to 25°C also further impaired the growth of PHG1a/PHG1b double knockout cells (Figure 3). Both PHG1a and PHG1b knockout cells also showed defects in development upon starvation (Figure 4). At 20°C, development of PHG1a knockout cells was arrested at the finger stage, whereas PHG1b knockout cells formed normal fruiting bodies. The double knockout strain exhibited a more severe phenotype, development of these cells stopped at the early culmination stage. At 25°C the development of PHG1a knockout cells was similar to that observed at 20°C (arrest at finger stage), whereas the development of PHG1b knockout cells was stopped at the early culmination stage (Figure 4).
Figure 3.
Growth kinetics show a synergistic effect of PHG1a and PHG1b mutations. Growth of wild-type cells, PHG1a, PHG1b, and PHG1a/PHG1b knockout cells was measured in HL5 medium at 20°C (triangles) and at 25°C (circles) over a period of 3 d, by counting cells.
Figure 4.
The developmental phenotypes of PHG1 knockout cells reveal a synergistic effect of PHG1a and PHG1b mutations. Wild-type cells, PHG1a, PHG1b, and PHG1a/PHG1b knockout cells were deposited on filters at the indicated temperature and observed for development under a dissection microscope. PHG1a knockout cells were arrested at the finger stage at 20°C or 25°C. PHG1b knockout cells exhibited a normal development at 20°C but were arrested at the early culmination stage at 25°C. PHG1a/PHG1b double knockout cells were stopped at the early culmination stage at 20°c and at the tipped mound stage at 25°C.
Development of PHG1a/PHG1b double knockout cells was blocked at an even earlier stage: the tipped mound stage (Figure 4).
In summary the combination of PHG1a and PHG1b mutations led to more than additive phenotypes on D. discoideum growth. This synergistic effect strongly suggests that the Phg1 proteins are involved in the same cellular function(s).
Different TM9 Knockout Cells Exhibit Similar Phagocytosis and Adhesion Defects
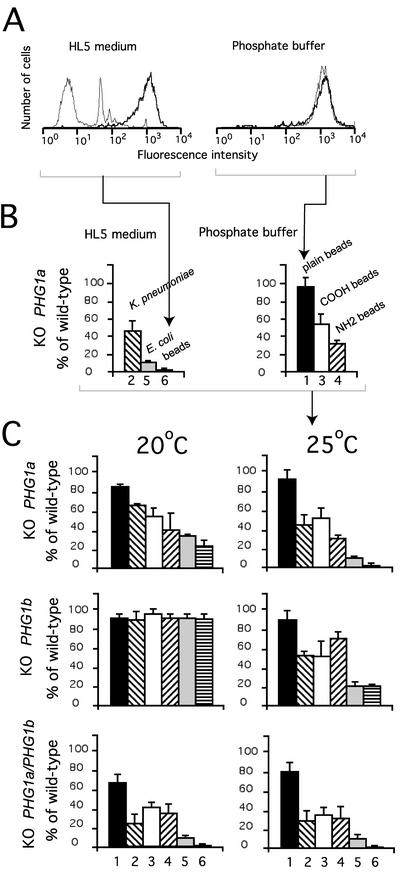
A previous study established that PHG1a knockout cells are specifically defective for adhesion to and phagocytosis of hydrophilic particles (latex beads coated with HL5 medium) but show no defect in phagocytosis of hydrophobic particles (plain latex beads in phosphate buffer) (Cornillon et al., 2000) (Figure 5A). Herein, this selectivity was further documented by showing that in phosphate buffer phagocytosis of other hydrophilic beads (carboxylate or amino group substituted beads) was also inhibited in PHG1a knockout cells (Figure 5B, columns 3 and 4). Phagocytosis rates of Klebsiella pneumoniae and Escherichia coli bacteria were also decreased in PHG1a knockout cells (Figure 5B, columns 2 and 5). At least three different types of phagocytosis receptors are present in D. discoideum: a lectin, a receptor for hydrophobic surfaces, and a receptor for hydrophilic surfaces (Vogel et al., 1980). Because PHG1a knockout cells are specifically defective in adhesion to hydrophilic substrates, Phg1a protein might represent the hydrophilic receptor or be specifically involved in its regulation. Herein, we also report that PHG1a knockout cells displayed a slight temperature-sensitive phenotype that increased the phagocytosis defect at 25°C compared with 20°C (Figure 5C).
Figure 5.
PHG1 knockout cells display a similar phagocytosis defect. (A) Fluorescent-activated cell sorter analysis. Wild-type cells (thick lane) or PHG1a knockout cells (thin lane) were incubated for 20 min in the presence of fluorescent plain latex beads in HL5 or phosphate buffer at 25°C. The amount of internalized fluorescence was measured as relative fluorescence intensity. Arrowheads indicate the corresponding bar graph in B. (B) Phagocytosis rates of three substrates by PHG1a knockout cells were measured at 25°C in HL5 medium or phosphate buffer and expressed as percentage of wild-type cells: 1, plain latex beads in phosphate buffer (hydrophobic surface); 2, K. pneumoniae in HL5; 3, carboxylate (COOH)-substituted latex beads in phosphate buffer; 4, amino (NH2)-substituted latex beads in phosphate buffer; 5, E. coli in HL5; and 6, plain, carboxylate, or amino-latex beads in HL5 (hydrophilic surface). These data were combined in C to provide a general view of the phagocytosis defects in the different mutants. (C) Specificity of the phagocytosis defect observed for PHG1a, PHG1b, and PHG1a/PHG1b knockout cells at 20°C or 25°C is represented. These data represent the average of four independent experiments and the SD is indicated.
At 20°C, PHG1b knockout cells phagocytosed all of the substrates tested as efficiently as wild-type cells (Figure 5C). On the contrary, at 25°C, PHG1b knockout cells displayed a strong phagocytosis defect (Figure 5C). Remarkably, PHG1a and PHG1b knockout cells showed a similar specificity in their phagocytosis defect at 25°C (Figure 5C). Interestingly, even at 20°C PHG1a/PHG1b double knockout cells showed a strong phagocytosis defect even more severe than PHG1a knockout cells alone (Figure 5C). This is again suggestive of a functional synergy between Phg1a and Phg1b proteins.
To confirm that the phagocytosis defect observed was due to a defect in adhesion, the ability of knockout cells to adhere to a substrate was measured and is presented in Table 1: PHG1a knockout cells showed a strong decrease in adhesion to a hydrophilic surface (glass coated with HL5 components), but not to a more hydrophobic surface (glass in phosphate buffer). At 20°C, PHG1b cells adhered normally to both substrates but at 25°C they exhibited a significant defect in adhesion to hydrophilic surfaces. PHG1a/PHG1b double knockout cells did not adhere to a hydrophilic substrate and showed a small decrease in adhesion to a more hydrophobic surface. Together, these results demonstrate that Phg1a and Phg1b proteins both play a role in adhesion to and phagocytosis of hydrophilic particles.
Table 1.
Inactivation of PHG1a or PHG1b genes result in cellular adhesion defect
|
Cellular adhesion (% of wild-type)
|
|||
|---|---|---|---|
| Cell line | Temperature | HL5 medium | Phosphate buffer |
| KO PHG1a | 20°C | <7.5 | 100.0 ± 5.9 |
| KO PHG1b | 20°C | 114.3 ± 14.3 | 108.0 ± 20 |
| 25°C | 66.7 ± 16.7 | 85.7 ± 8.5 | |
| KO PHG1a/KO PHG1b | 20°C | <7.5 | 35.3 ± 11.8 |
| KO PHG1a + PHG1a | 20°C | 125.0 ± 25 | Not done |
| KO PHG1a + PHG1b | 20°C | <7.5 | 82.2 ± 8.8 |
As observed previously, PHG1a knockout cells did not show any defect in fluid phase internalization (Cornillon et al., 2000). We did not observe any fluid phase uptake defect in PHG1b or PHG1a/PHG1b double knockout cells (our unpublished data). Because fluid phase uptake in Dictyostelium discoideum cells is mostly accomplished by macropinocytosis, the basic machinery for controlling rearrangement of the actin cytoskeleton is still functional in the PHG1 knockout cells. Consequently, in these cells the phagocytosis defect is limited to the first step in phagocytosis, i.e., adhesion to the particles.
Together, these results are compatible with a receptor function of Phg1a and Phg1b for phagocytosis of hydrophilic particles.
TM9 Proteins Are Not Functionally Redundant
To test further the functional relationship between Phg1a and Phg1b proteins, we performed complementation analysis. Expression of Phg1a protein in PHG1a knockout cells restores phagocytosis (Cornillon et al., 2000) and adhesion (Table 1). In contrast, overexpression of Phg1b protein in PHG1a knockout cells did not complement the defect in phagocytosis (Figure 6B) or in adhesion (Table 1). These results clearly demonstrate that the Phg1a and Phg1b proteins do not share redundant functions.
TM9 proteins differ most significantly in their N-terminal extracellular domain. To investigate the nature of the functional difference between Phg1a and Phg1b, we generated two hybrid proteins in which the extracellular domains were swapped (Figure 6A). Whereas expression of the B/A fusion largely complemented the phagocytosis defect of PHG1a knockout cells (Figure 6B), overexpression of the A/B fusion had no detectable effect (our unpublished data). Surprisingly, these results suggest that the main functional difference between the Phg1a and Phg1b proteins resides in the conserved hydrophobic domain, whereas the more divergent N-terminal domain is functionally similar.
No Detectable Association between the TM9 Proteins
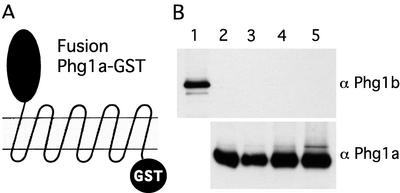
The results described so far demonstrate that the functions of the Phg1a and Phg1b proteins are similar but not redundant. A possible interpretation is that the two proteins interact physically to form a Phg1a-Phg1b complex or are part of a multisubunit complex essential for cellular adhesion and phagocytosis. To test this possibility, a fusion protein encoding the full-length Phg1a fused to GST and tagged with FLAG epitope (Figure 7A) was expressed in PHG1a knockout cells. This hybrid protein fully complemented the adhesion and phagocytosis defect of PHG1a knockout cells (our unpublished data). Two different cell solubilization conditions were used (Triton-X 100 or CHAPS), and coimmunoprecipitation was then performed using either a GST or a FLAG affinity resin. Western blot analysis revealed that Phg1a was efficiently immunoprecipitated in both cases. Although the amount of material loaded (2 × 106 cells) corresponded to 10 times more cells than in Figure 2, Phg1b was not detected in these samples even after prolonged exposure (Figure 7B). Although it is virtually impossible to rule out a weak or detergent-sensitive interaction, this result argues against the existence of a stable protein complex between Phg1a and Phg1b.
Figure 7.
No detectable interaction between Phg1a and Phg1b proteins. (A) PHG1a knockout cells were transfected with a construct encoding Phg1a C-terminal fusion to GST tagged with a FLAG epitope, Phg1a-GST. (B) Triton X-100 (lanes 2 and 3) or CHAPS (lanes 4 and 5) lysates from cells expressing the Phg1a-GST fusion were incubated with Sepharose beads coupled to glutathione (lanes 2 and 4) or to an antibody against the FLAG epitope (lanes 3 and 5). The adsorbed proteins were processed for Western blotting and revealed with an antiserum to the Phg1b protein (top) or the Phg1a protein (bottom). To indicate the position of the Phg1b protein, an aliquot of lysate from a cell overexpressing Phg1b was comigrated (lane 1).
Composition of the Cell Surface Is Altered in TM9 Knockout Cells
The defective adhesion properties of PHG1 knockout cells prompted us to examine the protein cell surface composition. The cell surface was biotinylated and total protein lysates were then resolved by two-dimensional nonreduced/reduced gels. This technique allows visualization of transmembrane proteins and resolution of those containing disulfide bridges because they migrate differently in nonreducing and reducing conditions.
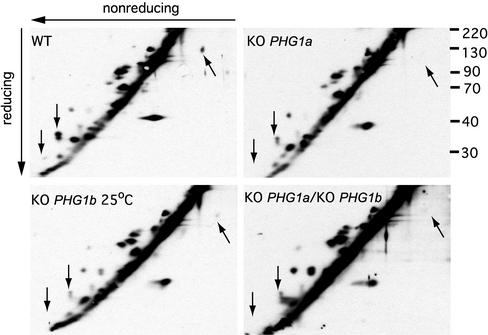
Approximately 20 proteins were detected outside the diagonal in the wild-type cells (Figure 8), whereas only a weak background was observed only on the diagonal for nonbiotinylated wild-type cells (our unpublished data). None of these proteins corresponded to Phg1a or Phg1b as both migrated on the diagonal (our unpublished data). Interestingly, the same analysis performed in PHG1a, PHG1b, and PHG1a/PHG1b knockout cells revealed a number of differences (Figure 8). In particular, in conditions where cellular adhesion is defective, at least three proteins are clearly down-regulated in all PHG1 knockout cells compared with wild-type cells (Figure 8, arrowheads). In addition, a number of other changes were observed, for example up-regulated proteins in PHG1b and PHG1a/PHG1b knockout cells (Figure 8).
Figure 8.
Pattern of cell surface proteins is altered in PHG1 knockout cells. After cell surface biotinylation, cells were lysed and total proteins resolved on a two-dimensional nonreduced/reduced gel. Biotinylated proteins were revealed after incubation with horseradish peroxidase-avidin conjugate. The cell surface expression of at least three proteins (indicated by arrows) was markedly reduced in mutants exhibiting a phagocytosis defect.
These results demonstrate that deletion of PHG1 genes changes dramatically the cell surface composition and this might be the cause of the loss of cellular adhesion in the knockout cells.
p80 Endosomal Protein Is Shifted to the Cell Surface in PHG1a/PHG1b Knockout Cells
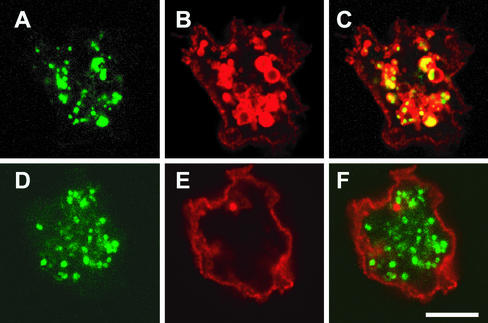
The p80 membrane protein was characterized previously in D. discoideum as an integral membrane protein, mostly localized to the endocytic compartments (Ravanel et al., 2001). Minor amounts of p80 can also be found at the cell surface, but it is absent from the contractile vacuole (Ravanel et al., 2001). Subcellular localization of p80 was determined to detect any modifications in the intracellular transport. Although in wild-type cells p80 was mostly localized in endosomes (Figure 9), in PHG1a/PHG1b knockout cells p80 was mainly present at the cell surface and often absent from intracellular compartments (Figure 9). To quantify this, the number of cells with no visible intracellular p80 was counted for wild-type cells and PHG1a/PHG1b knockout cells. In two independent experiments only very few wild-type cells seemed devoid of intracellular p80 (1.3 and 0%, n = 304 and 100, respectively). On the contrary, almost half of the PHG1a/PHG1b knockout cells exhibited pure p80 surface labeling (42.4 and 48%, n = 309 and 100, respectively). Interestingly in PHG1a/PHG1b knockout cells exhibiting no internal p80, endosomes were still visible as evidenced by the presence of an endocytosed fluid-phase marker (Figure 9). Together, these experiments indicate that p80 is relocalized from endosomal membranes to the cell surface in PHG1a/PHG1b knockout cells. In single PHG1a or PHG1b knockouts, no major p80 relocalization was observed (our unpublished data), again suggesting a synergistic role of the Phg1a and Phg1b proteins. To conclude alteration in the intracellular transport, in particular in the endocytic pathway, might account for the modified cell surface composition in PHG1 knockout cells.
Figure 9.
p80 endosomal protein is shifted to the cell surface in the PHG1a/PHG1b knockout cells. Dictyostelium cells were incubated for 90 min in HL5 containing 50-nm fluorescent latex beads (A and D) and then fixed and processed for immunofluorescence with anti-p80 monoclonal antibody (B and E). (A–C) Wild-type cells. (D–F) PHG1a/PHG1b knockout cells. Bar, 5 μM.
DISCUSSION
The hallmark of proteins belonging to the TM9 family is the presence of a large variable extracellular N-terminal domain and nine C-terminal transmembrane domains (Chluba-de Tapia et al., 1997). The presence of TM9 proteins in evolutionary distant organisms (four members in human, three in yeast, and several in plants) raises several interesting questions regarding their function. For instance, do these proteins carry out a common evolutionary conserved function in these organisms? In the same organism do different members of the family play redundant roles? Does the variable extracellular domain confer distinct properties to each member of the family? The D. discoideum genome contains at least three distinct genes encoding TM9 proteins: Phg1a, Phg1b, and Phg1c. The function of two divergent TM9 members, Phg1a and Phg1b, was investigated by analyzing the phenotype of the corresponding knockout cells.
Synergy in phagocytosis, adhesion, growth, and development suggests strongly that Phg1a and Phg1b proteins function in the same cellular process. Compared with the non-selective phagocytosis defect of LvsA mutant cells (Cornillon et al., 2002), PHG1a and PHG1b knockout cells display a selective phagocytosis defect. This is compatible with the hypothesis that Phg1a and Phg1b proteins represent receptors necessary for adhesion to hydrophilic particles, whereas the LvsA protein is only involved at later stages in the phagocytic process (Cornillon et al., 2002). However, the fact that overexpression of PHG1b failed to complement PHG1a knockout cells argues against a role for Phg1a and Phg1b proteins as two phagocytosis receptors sharing similar specificity. Similarly, PHG1a knockout complementation by the B/A fusion protein suggests that the variable extracellular domain of TM9 proteins does not confer a specific role in cellular adhesion to each member of the family. Biochemical experiments also excluded the possibility that Phg1a and Phg1b proteins might together form one oligomeric phagocytosis receptor.
We are thus brought to propose that TM9 proteins function as regulators of cellular adhesion rather than as adhesion molecules. Inactivation of two converging regulatory pathways would indeed be expected to result in similar and synergistic effects but no complementation. Interestingly, regulator role for a TM9 protein (SMBP) has recently been suggested in rat: SMBP binds to a β3-adrenergic agonist and is involved in regulation of eosinophil chemotaxis and relaxation of depolarized rat colon (Sugasawa et al., 2001). This suggests that TM9 proteins could act as receptors for as yet unidentified ligands and regulate a number of important cellular functions, such as adhesion in D. discoideum or chemotaxis in eosinophil. This model would also account for the presence of TM9 proteins in nonphagocytic species such as Saccharomyces cerevisiae and plants. Presumably in these organisms, TM9 proteins regulate other cellular functions.
At least two mechanisms by which regulators modulate cellular adhesion can be proposed: first, modulation of the adhesive properties of adhesion molecules; second, modulation of their cell surface expression. In support of the second mechanism, we observed an alteration of the protein composition at the cell surface in PHG1 knockout cells. Any down-regulated cell surface protein in PHG1 knockout cells might represent a potential receptor for hydrophilic substrates and at least three of them were down-regulated. Hence, TM9 proteins might control cellular adhesion in D. discoideum by determining the cell surface expression of as yet unidentified receptor(s). The selective phagocytosis defect of PHG1 knockout cells might then reflect the specificity of the regulated receptor(s). Additionally, the large number of up- or down-regulated cell surface proteins in PHG1 knockout cells points to a more general function of TM9 proteins, i.e., regulation of the cell surface composition. Interestingly, at least three TM9 proteins have been found in the endosomal or phagosomal pathways: the human p76 was predominantly localized in endosomes (Schimmoller et al., 1998); the yeast Emp70 cleavage product p24 has been identified in endosome-enriched membranes (Singer-Kruger et al., 1993); the D. discoideum Phg1a was reported to be present in phagosomes (Cornillon et al., 2000). It is therefore tempting to postulate that TM9 proteins regulate the cell surface composition by modulating intracellular transport of proteins in the endocytic pathway. This hypothesis is further supported by the observation that the intracellular localization of an endosomal marker protein such as p80 is affected in PHG1 knockout cells.
In conclusion, both TM9 proteins Phg1a and Phg1b are required for cellular adhesion. Our results strongly suggest a regulatory role of TM9 proteins rather than a receptor role in adhesion and phagocytosis. Additional studies, such as analysis of Phg1c function in D. discoideum or TM9 proteins in mammalian system are necessary to gain further insight into the regulatory function of the TM9 proteins and to establish their putative function in intracellular transport.
Acknowledgments
We thank Gisou Van der Goot for critical reading of the manuscript. This work was supported in part by a START fellowship of the Fonds National Suisse dela Recherche Scientifique and a grant from the Fondation Gabriella Giorgi-Cavaglieri (both to P.C.); by a grant from the Association pour la Recherche contre le Cancer and a grant from the Fondation pour la Recherche Médicale (both to F.L.); and by the Japan Society for the Promotion of Science grant RFTF96L00105 and Ministry of Education, Science, Sports and Culture of Japan grant 08283107, both for the Dictyostelium discoideum cDNA project in Japan.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E02–11–0724. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E02-11-0724.
References
- Chluba-de Tapia, J., de Tapia, M., Jaggin, V., and Eberle, A.N. (1997). Cloning of a human multispanning membrane protein cDNA: evidence for a new protein family. Gene 197, 195–204. [DOI] [PubMed] [Google Scholar]
- Cornillon, S., Dubois, A., Bruckert, F., Lefkir, Y., Marchetti, A., Benghezal, M., De Lozanne, A., Letourneur, F., and Cosson, P. (2002). Two members of the beige/CHS (BEACH) family are involved at different stages in the organization of the endocytic pathway in Dictyostelium. J. Cell Sci. 115, 737–744. [DOI] [PubMed] [Google Scholar]
- Cornillon, S., Olie, R.A., and Golstein, P. (1998). An insertional mutagenesis approach to Dictyostelium cell death. Cell Death Differ. 5, 416–425. [DOI] [PubMed] [Google Scholar]
- Cornillon, S., Pech, E., Benghezal, M., Ravanel, K., Gaynor, E., Letourneur, F., Bruckert, F., and Cosson, P. (2000). Phg1p is a nine-transmembrane protein superfamily member involved in Dictyostelium adhesion and phagocytosis. J. Biol. Chem. 275, 34287–34292. [DOI] [PubMed] [Google Scholar]
- Decave, E., Garrivier, D., Brechet, Y., Fourcade, B., and Bruckert, F. (2002). Shear flow-induced detachment kinetics of Dictyostelium discoideum cells from solid substrate. Biophys. J. 82, 2383–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manstein, D.J., Schuster, H.P., Morandini, P., and Hunt, D.M. (1995). Cloning vectors for the production of proteins in Dictyostelium discoideum. Gene 162, 129–134. [DOI] [PubMed] [Google Scholar]
- Morio, T., et al. (1998). The Dictyostelium developmental cDNA project: generation and analysis of expressed sequence tags from the first-finger stage of development. DNA Res 5, 335–340. [DOI] [PubMed] [Google Scholar]
- Ravanel, K., de Chassey, B., Cornillon, S., Benghezal, M., Zulianello, L., Gebbie, L., Letourneur, F., and Cosson, P. (2001). Membrane sorting in the endocytic and phagocytic pathway of Dictyostelium discoideum. Eur. J. Cell Biol. 80, 754–764. [DOI] [PubMed] [Google Scholar]
- Schimmoller, F., Diaz, E., Muhlbauer, B., and Pfeffer, S.R. (1998). Characterization of a 76 kDa endosomal, multispanning membrane protein that is highly conserved throughout evolution. Gene 216, 311–318. [DOI] [PubMed] [Google Scholar]
- Singer-Kruger, B., Frank, R., Crausaz, F., and Riezman, H. (1993). Partial purification and characterization of early and late endosomes from yeast. Identification of four novel proteins. J. Biol. Chem. 268, 14376–14386. [PubMed] [Google Scholar]
- Sugasawa, T., Lenzen, G., Simon, S., Hidaka, J., Cahen, A., Guillaume, J.L., Camoin, L., Strosberg, A.D., and Nahmias, C. (2001). The iodocyanopindolol and SM-11044 binding protein belongs to the TM9SF multispanning membrane protein superfamily. Gene 273, 227–237. [DOI] [PubMed] [Google Scholar]
- Sussman, M. (1987). Cultivation and synchronous morphogenesis of Dictyostelium under controlled experimental conditions. Methods Cell Biol. 28, 9–29. [DOI] [PubMed] [Google Scholar]
- Vogel, G., Thilo, L., Schwarz, H., and Steinhart, R. (1980). Mechanism of phagocytosis in Dictyostelium discoideum: phagocytosis is mediated by different recognition sites as disclosed by mutants with altered phagocytotic properties. J. Cell Biol. 86, 456–465. [DOI] [PMC free article] [PubMed] [Google Scholar]