Abstract
The traditional view holds that peroxisomes are autonomous organelles multiplying by growth and division. More recently, new observations have challenged this concept. Herein, we present evidence supporting the involvement of the endoplasmic reticulum (ER) in peroxisome formation by electron microscopy, immunocytochemistry and three-dimensional image reconstruction of peroxisomes and associated compartments in mouse dendritic cells. We found the peroxisomal membrane protein Pex13p and the ATP-binding cassette transporter protein PMP70 present in specialized subdomains of the ER that were continuous with a peroxisomal reticulum from which mature peroxisomes arose. The matrix proteins catalase and thiolase were only detectable in the reticula and peroxisomes. Our results suggest the existence of a maturation pathway from the ER to peroxisomes and implicate the ER as a major source from which the peroxisomal membrane is derived.
INTRODUCTION
Peroxisomes belong to the repertoire of subcellular membrane-bounded compartments of the eukaryotic cell. Depending on species, they participate in cellular metabolism in various ways.
A consistent feature is the presence of H2O2-producing oxidases, such as enzymes degrading fatty acids, and catalase decomposing H2O2. Peroxisomes are bounded by a single membrane and morphologically range from rather inconspicuous small vesicles in some cells to elaborate tubular networks (reticula) in others (De Duve and Baudhuin, 1966; Beevers, 1979; van den Bosch et al., 1992; Clayton et al., 1995; Subramani et al., 2000; Yamamoto and Fahimi, 1987).
Mutations in genes coding for peroxisomal proteins cause a number of diseases extending from relatively mild ones in which the catalytic activity of an enzyme is affected to a number of different neuropathies in which a protein is affected that contributes to organelle maintenance (Lazarow and Moser, 1995). Insight into this latter group of proteins was gained by studying peroxisome function and biogenesis in different yeasts and Chinese hamster ovary cells (Erdmann et al., 1989; Ghaedi et al., 1999). As a result, 25 PEX genes have been collected, 13 of which have orthologs in human and have been linked to disease (Gould and Valle, 2000; Purdue and Lazarow, 2001; Smith et al., 2002).
Peroxisomal matrix and membrane proteins are synthesized on free polyribosomes and delivered to the cytosol. Here, they are picked up by soluble receptors on the basis of peroxisomal targeting signals (PTSs) and guided to the peroxisomal membrane for import into the organelle or insertion into the membrane. Most matrix proteins are recognized by Pex5p via a short C-terminal tripeptide (PTS1), whereas a limited number have an N-terminal sequence (PTS2) and are recognized by Pex7p. Pex3p and Pex19p have been implicated in recognition of targeting signals (mPTS) in integral membrane proteins and their insertion into the membrane (Jones et al., 2001; Wang et al., 2001). The PTS1 (Pex5p) and PTS2 (Pex7p) import routes converge on the peroxisomal membrane, where a number of proteins (Pex10p, Pex12p, Pex13p, Pex14p, and Pex17p) are involved in the membrane translocation step.
An important and still unresolved question is how peroxisomes acquire their phospholipids for membrane formation. The source of most phospholipid biosynthesis is the endoplasmic reticulum (ER), but how the newly synthesized phospholipids reach peroxisomes is not known. In early observations, peroxisomes were often seen in close association with the ER, which might allow unilateral transfer of phospholipids from the ER to peroxisomal membranes (Novikoff and Shin, 1964). Even the idea of an ER origin of peroxisomes was entertained. However, with the authoritative review of Lazarow and Fujiki (1985) compelling experimental evidence was presented on the basis of targeting of newly synthesized proteins to peroxisomes, which supported the notion that peroxisomes were autonomous organelles multiplying by growth and division.
In recent years, the possibility that the ER contributes to peroxisome formation has received renewed attention, particularly after observations that certain peroxisomal proteins or their modified derivatives were found in the ER (Titorenko and Rachubinski, 2001; Faber et al., 2002). In most cases, these observations were made in cells in which the expression level of such proteins was raised to support detection or which had been manipulated otherwise. Under these circumstances, mistargeting is possible and indeed examples of such mistargeting have recently been reported, suggesting that proper targeting of proteins in cells, particularly membrane proteins, is easily thrown off balance (Stroobants et al., 1999; Borgese et al., 2001). A convincing observation that still stands is that in Yarrowia lipolytica two peroxins (Pex2p and Pex16p) are N-glycosylated, suggesting that they passed through the ER en route to peroxisomes (Titorenko et al., 1997). A critical summary of all the experiments relating to the ER–peroxisome connection was recently compiled by Purdue and Lazarow (2001).
In the present article, we report new observations in unmanipulated mouse dendritic cells showing the ER to be involved in peroxisome formation particularly with respect to the delivery of the peroxisomal membrane. With these observations, we substantiate already more than 30-year-old work done by Phyllis and Alex Novikoff in which they showed intimate contacts and membrane continuities between ER and peroxisomes in intestinal cells by electron microscopy (Novikoff and Novikoff, 1972).
MATERIALS AND METHODS
D1 cells were derived from immature splenic dendritic cells (DCs) from C57BL/6 mice and were cultured as described previously (Winzler et al., 1997). In one experiment, human bone marrow-derived DCs were used for comparison. For immunocytochemistry, we used polyclonal antibodies against mouse I-chain luminal part (S22; kind gift of Dr. N. Koch, University of Bonn, Bonn, Germany), mouse langerine (Valladeau et al., 1999), human protein disulfide isomerase (PDI; kindly provided by Dr I. Braakman, University of Utrecht, Utrecht, The Netherlands), rabbit anti-calreticulin was purchased from Affinity Bioreagents (Golden, CO), bovine catalase (Tager et al., 1985), rat thiolase (Heikoop et al., 1990), COPI (kindly provided by Dr. I. Majoul, University of Göttingen, Göttingen, Germany), Sec 13 (generously provided by Dr. W.J. Hong, Institute of Molecular and Cellular Biology, University of Singapore, Singapore), and rat PMP70 (Motley et al., 1994). A mouse monoclonal was against PDI tail (Vaux et al., 1992). Anti-Pex13p was raised against part of the SH3 domain (aa 275–454) of human Pex13p fused to the N terminus of dihydrofolate reductase. The antibody specifically cross-reacted with mouse Pex13p.
Immunocytochemistry
For immunocytochemistry, cells were washed by centrifugation in fetal calf serum-free medium and fixed in 2% paraformaldehyde and 0.2% glutaraldehyde. The cells were then processed as described previously (Liou et al., 1997). Briefly, the fixed cells were washed in phosphate-buffered saline with 50 mM lysine at room temperature to quench free aldehydes, embedded in 10% gelatin, and cryosectioned. For immunoelectron microscopy, ultrathin cryosections were indirectly single or double immunolabeled with 5 nm of gold, or 10- and 15-nm gold particles, respectively (Geuze et al., 1981; Liou et al., 1996).
Quantitation of immunogold for Pex13p and PMP70 on the peroxisomal system, i.e., peroxisomes, peroxisomal reticula, and lamellae, was done in 20 random electron micrographs with a final magnification of 25,000×. In total, 304 gold particles were counted for Pex13 and 232 for PMP70. Relative membrane surface areas of the subcompartments of the peroxisomal system were determined by putting a transparent overlay with a squared lattice of 1 cm spaced lines on top of the same micrographs as used for gold counting. Labeling densities of Pex13p and PMP70 in the peroxisomal complex subdomains were calculated by determining the ratio's of the respective percentages of gold particles over the percentages of intersections between the lines and the various membranes involved.
Three-dimensional (3-D) Reconstruction
Approximately 150-nm thin cryosections were immunogold labeled for catalase with 10 nm of gold to visualize peroxisomal complexes. Selected fields in the sections were imaged at 17,280× in a Tecnai (S) LaB6 electron microscope (FEI/Philips, Eindhoven, The Netherlands) operating at 200 kV. For 3-D imaging of the sections by means of electron tomography (Koster et al., 1997), the specimens were tilted at 10 intervals by using an ultra-high tilt specimen holder (model 670; Gatan, Pleasanton, CA) over a range of 1400 along two orthogonal axes (Mastronarde, 1997). All images were collected with a 2048 × 2048 pixel cooled slow-scan charge-coupled device camera (TemCam F214; Tietz Video and Image Processing Systems, Gauting, Germany). At the magnification used, the images represent a specimen area of 1659 × 1659 nm. For the tilt series, images of 1024 × 1024 pixels with 1.6 nm/pixel were collected (binning 2). Automated acquisition of the tilt series was carried out using the recently developed precalibration approach (Ziese et al., 2002). Postdata acquisition alignment of the tilt series, as well as the subsequent 3-D reconstructions (resolution-weighted back-projection) and modeling steps, were all carried out using the IMOD program (Kremer et al., 1996) running on a Unix workstation (Octane2 dual-processor 2 × 400 MHz R12000 with 2.5 GB RAM; Silicon Graphics, Mountain View, CA). The 10-nm gold beads at the surface of the sections were used as landmarks for alignment of the tilt series. The shrinkage of cryosections was ∼65% and we corrected for this in our model.
RESULTS
Dendritic cells exhibit a well-developed vacuolar system (Mellman and Steinman, 2001) and numerous peroxisomes, providing a good starting point for a study on possible relationships between peroxisomes and other organelles. We choose for localizing critical marker proteins with immunoelectron microscopy, taking advantage of our recently developed cryosectioning technique, which allows for an excellent delineation of membranes (Liou et al., 1997), together with electron tomography and 3-D reconstruction of peroxisomes and adjacent organelles. As markers, we studied the peroxisomal matrix proteins catalase and thiolase, which contain different targeting signals to enter the peroxisome (PTS1 and PTS2, respectively), and two peroxisomal integral membrane proteins: Pex13p, which is a component of the protein import complex (Elgersma et al., 1996; Erdmann and Blobel, 1996; Gould et al., 1996; Stein et al., 2002) and the ATP-binding cassette transporter PMP70 (Gartner et al., 1998). As markers for the ER, we localized protein disulfide isomerase (PDI), invariant chain (Ii), calreticulin, and for the vesicular transport route, the coat proteins COPI and COPII.
Peroxisomes Are Closely Associated with Lamellar Structures Enriched in Pex13p and PMP70
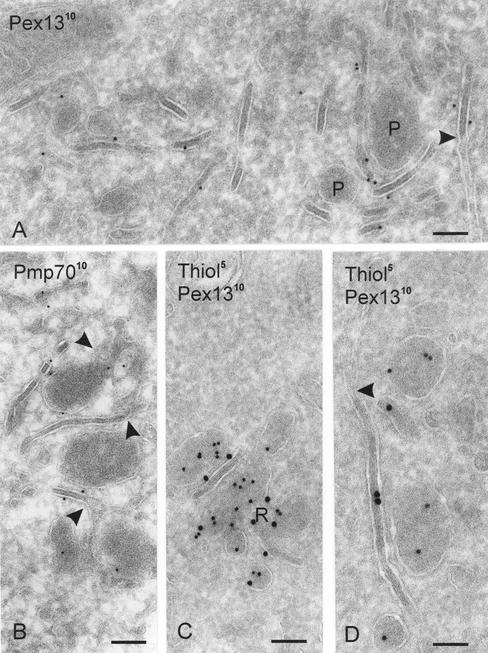
The peroxisomes occurred in clusters throughout the cells. In these clusters different structures could be distinguished: 1) peroxisomes (P in the figures); 2) a reticulate peroxisomal network with globular extensions (Figure 1, B and C), and 3) tubular structures encircling the peroxisomal clusters with a special density and striping that we will subsequently call lamellae (Figures 1, A, B, and D; 2A; and 3). Characteristic features of the lamellae were the tightly opposed limiting membranes in thin sections and the internal structure seen as striping (best seen in Figure 1, A and C). The lamellae resembled the so-called Birbeck granules in Langerhans cells. However, Birbeck granules are thicker, present a different striping pattern, and are associated with the plasma membrane and endosomes. In addition, Birbecks strongly labeled with an antibody against the Birbeck marker protein langerine, whereas the peroxisome-associated rigid lamellae did not (our unpublished data).
Figure 1.
Clusters of peroxisomes are associated with lamellae and peroxisomal reticula. (A) Survey of an area with lamellae and peroxisomes (P). Pex13p labeling is predominantly associated with the lamellae and is almost absent from the peroxisomes. Note that the limiting membranes of the striped lamella at the right are continuous with the ER at the arrowhead. Bar, 100 nm. (B) PMP70 labeling is present on both peroxisomes and lamellae. The upper and lower peroxisomes seem to be connected with the lamellae via a reticulum (arrowheads). Bar, 125 nm. (C) Peroxisomal cluster showing thiolase in its subcompartments, except in the lamella. Pex13p is present in the lamella and in the peroxisomal reticulum (R). Bar, 110 nm. (D) A Pex13p-positive and striped lamella shows a transition with the ER at the arrowhead. Only the peroxisomes are labeled for thiolase. Bar, 80 nm.
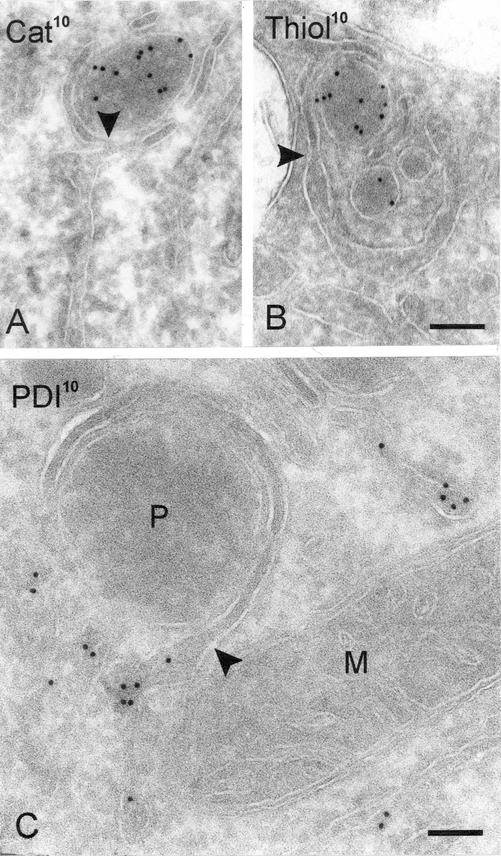
Figure 2.
Adjacent to peroxisomes the ER shows lamellar subdomains. A.Peroxisome immunolabeled for catalase is surrounded by a lamella that shows membrane continuity with the ER (arrowhead). At the transition, the ER lumen is narrowed. See also in B. B. Like in A, but immunolabeled for thiolase. Bar for A and B, 120 nm. (C) Peroxisome (P) encircled by a lamella that is continuous at the arrowhead with an ER cisterna labeled for the ER marker PDI. M, mitochondrion. Bar, 90 nm.
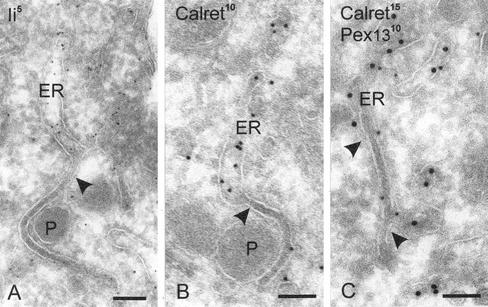
Figure 3.
ER that is continuous with lamellae contains ER markers. (A and B) ER cisternae with membrane connections to lamellae (arrowheads) and that are labeled for Ii and calreticulin, respectively. (C) Pex13p-positive lamella continuous with ER labeled for calreticulin. Bars, 100 nm.
Various marker proteins showed different distributions with respect to these structures. The peroxisomes and reticula, but not the lamellae were positive for catalase and thiolase (Figures 1C; 2, A and B). The rigid lamellae, on the other hand, contained abundant Pex13p (Figure 1, A, C, and D) and PMP70 (Figure 1B). Both membrane proteins were also present on the limiting membrane of peroxisomes and peroxisomal reticulum, albeit with different densities. A quantitative evaluation of the gold labeling patterns showed that of all Pex13p present in the peroxisomal system, 64% was present in the lamellae, 27% in the reticula, and only 9% in the peroxisomes. For PMP70, these figures were 12, 62, and 26%, respectively. Thus, by far the majority of Pex13p was located outside the typical globular peroxisomes, in particular in the lamellae. For PMP70, this distribution was reverse, i.e., shifted toward the reticula and peroxisomes (Table 1). When we related the gold labeling to the membrane surface areas in these structures to reveal labeling densities (see MATERIALS AND METHODS), the highest Pex13p density occurred in the lamellae and that of PMP70 in the peroxisomes. For both proteins, the peroxisomal reticula took an intermediate position (Table 1).
Table 1.
Distribution of Pex13p and PMP70 in peroxisomal clusters
|
Pex13p
|
PMP70
|
|||||
|---|---|---|---|---|---|---|
| Percent gold | Gold density | Percent gold | Gold density | |||
| Lamellae | 64 | 5.4 | 12 | 1.2 | ||
| Reticulum | 27 | 3.5 | 62 | 4.0 | ||
| Peroxisomes | 9 | 1.9 | 26 | 4.3 | ||
| Total gold | 304 | 232 | ||||
Peroxisome-associated Lamellae Represent a Subdomain of the ER
Interestingly, lamellae were also observed as protrusions from the ER. In all figures shown, these membrane continuities between the rough ER cisternae and lamellae have been marked with arrows (Figures 1, A and D; 2, A–C; and 3). Considering the importance of this observation, we have studied this aspect in further detail.
The lamellae lacked ribosomes and any visible cytoplasmic coat (best seen in Figures 1A, 2A, and 3A). That it was the ER with which the lamellae were continuous was further demonstrated by labeling for the ER markers PDI (Figure 2C) and calreticulin (Figure 3, B and C). As a powerful antigen-presenting cell type, dendritic cells contain large amounts of the major histocompatability complex class II chaperone Ii, especially in the ER. With all three ER markers, we demonstrate now that the lamellae are directly connected to ER cisternae. The absence of ribosomes from the lamellae and their continuity with the ER was reminiscent of the smooth transitional elements of the ER facing the cis-Golgi, which represent the COP II-coated exit sites for newly synthesized secretory, lysosomal, and membrane proteins for transport from the ER to the Golgi complex. However, the lamellae were negative for sec 13, a component of COP II. COP I, thought to be involved in retrograde transport from the Golgi to downstream elements of the secretory pathway, was also absent from the lamellae. Both COPs were present in the Golgi areas in the same sections (our unpublished data). Our observations show that the lamellae are subdomains of the ER, and the position of the marker proteins in the various structures suggests that the lamellae are engaged in peroxisome formation.
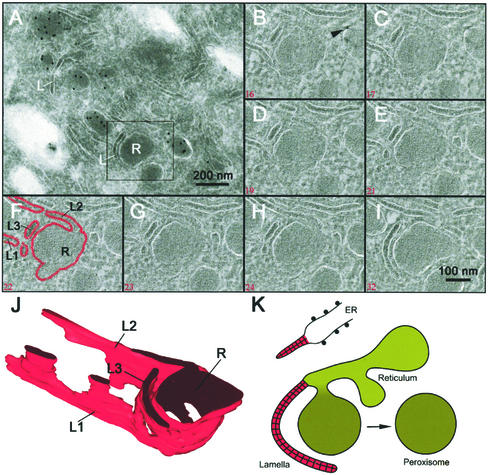
3-D Analysis Reveals an Anastomosing Membrane System Interconnecting ER and Peroxisomes
For a more comprehensive view on the intricate membranous relationships between the above-mentioned peroxisomal subcompartments, we made 3-D reconstructions of dual-axis tilt series of entire peroxisomal clusters. Figure 4A shows a peroxisomal cluster with lamellae, reticulum, and peroxisomes labeled for catalase. Of the squared area several tomographic slices (5 nm in thickness) were obtained as depicted in B–I. The membranes were manually traced as illustrated in F to generate the 3-D model shown in J. The lamellae seemed to be sheets that formed a complex network. The 3-D reconstructions confirmed their connection with the ER and the peroxisomal reticulae. Together, our data suggest a model for peroxisomal biogenesis as depicted in Figure 4K.
Figure 4.
A 3-D reconstruction of the spatial relationships between lamellae and peroxisomal reticulum. (A) Electron micrograph of a 150-nm-thick cryosection labeled for catalase with 10 nm of gold, showing lamellae (L) and reticula (R). (B–I) Several tomographic slices through the 3-D reconstruction (for orientation, see square in A) going from top of the cryosection to the bottom. A selection of slices (see numbers in red) is shown. Each slice represents 5 nm of volume in z direction. The arrowhead in B indicates a gold particle on top of the cryosection. F illustrates how membranes were manually traced to make the 3-D model. J shows a favorite view of the 3-D model. Note that lamellae (L) 1–3 are contiguous with the peroxisomal reticulum (R). K represents a simplified drawing of a peroxisomal system.
DISCUSSION
In this study, we present evidence in favor of a contribution of the ER to the formation of peroxisomes in an unmanipulated cell. The mouse dendritic cell line D1 can be propagated in vitro in the presence of granulocyte-macrophage colony-stimulating factor and fibroblast-derived growth factors (Lutz et al., 1996). When stocks of D1 cells were taken and put into culture for various periods, we observed special lamellar structures that were connected to the ER (“specialized ER”) and that contained the peroxisomal proteins Pex13p and PMP70. Similar structures were found in bone marrow-derived human dendritic cells. The reason for the abundance of the lamellar structures in dendritic cells is unknown. However, it is of interest to note that dendritic cells were reported to contain peroxisome proliferator-activated receptor (PPAR)γ, a member of the PPARs, which upon activation stimulate among others the formation of new peroxisomes (Gosset et al., 2001). It is conceivable that the special growth medium required for the in vitro cultivation of dendritic cells contains ligands that put PPARγ into action. Pex13p, an integral membrane protein, which is involved in import of matrix proteins into peroxisomes, was abundantly present in the lamellae. The lamellae were often seen as extensions of the ER. The identity of the ER was confirmed by the localization of three different marker proteins: PDI, calreticulin, and Ii. This specialized ER was morphologically different from the ER itself. It lacked associated ribosomes and its narrow lumen showed a characteristic striping. The lamellar extensions of the ER contained only very small amounts of the luminal ER markers PDI, calreticulin, and Ii. We concluded that the lamellar extensions of the ER are specialized subcompartments of the ER. Our 3-D reconstructions showed that free lamellae that were not connected to the ER were continuous with the peroxisomal reticulum, the precursor compartment of the globular peroxisomes (Yamamoto and Fahimi, 1987; Schrader et al., 2000). Quantification of immunogold indicated the existence of a protein gradient with respect to these structures, with Pex13p highest in the lamellar ER domains and free lamellae, the ATP-binding cassette transporter protein PMP70 highest in the reticula and peroxisomes, and thiolase and catalase exclusively present in wide portions of the reticula and globular peroxisomes. Considering the fact that components of the protein import machinery must be assembled first, before the import of PTS1 or PTS2 containing matrix proteins can start, the marker proteins combined with their subcellular locations suggest the existence of a maturation pathway. The observations favor a model in which certain integral peroxisomal membrane proteins are recruited to the specialized ER subdomains, or are sorted toward these domains from other regions of the ER to form the lamellar ER extensions. Once formed, additional integral membrane proteins are recruited for building up the protein-import machinery. With the start of matrix protein import, the formation of reticula and globular peroxisomes is concluded. How Pex13p finds its position in the specialized ER is difficult to answer at the moment. Studies in yeast argue that peroxisomes can still be formed in the absence of a functional ER protein import complex (South et al., 2001). However, the mandatory application of mutants (ts sec61 and Δssh1), the use of GFP-PTS1 as a late read out for peroxisome formation and the presence of residual fluorescent, unidentified punctate structures may still leave room for alternative explanations. It is likely that at a certain stage cytosolic factors are involved in severing the specialized ER compartment from its donor compartment. In the past, much effort has been invested in attempts to find clues for vesicular trafficking from ER to peroxisomes that possibly involved components of the well-characterized COPI- and COPII-dependent processes in membrane traffic (South et al., 2000; Voorn-Brouwer et al., 2001). Thus far, these attempts have all been negative. Also in our present study, we were unable to detect COP coats on the lamellar extensions of the ER, although in the same preparations COPI and COPII were found in the Golgi region. Our results suggest an explanation for this, because they indicate that large pieces of ER membrane are selected for peroxisome formation, which obviates the need for proteins required for small vesicle formation. More likely candidates for this severing process, therefore, might be components involved in organelle fission events such as the dynamins and/or SNARE proteins. This is borne out by our recent observations in Saccharomyces cerevisiae that Vps1p, a dynamin-like protein involved in routing of vesicles from Golgi to vacuole, is also responsible for keeping a constant number of peroxisomes per cell. The involvement of Myo2p in peroxisome partitioning, a motor protein that is also responsible for the delivery of the vacuole to the emerging bud additionally argues in favor of shared features between peroxisomes and the vacuole compartment (Hoepfner et al., 2001). Together, our results challenge the concept that peroxisomes are autonomous organelles and rather support a semiautonomous character. To form the preperoxisome, the organelles are dependent on a priming event that occurs in the ER. The ER provides the membrane and one or a few integral membrane proteins. From then on further maturation takes place in an autonomous manner.
Our results are based on morphological observations. It will be important to confirm these data and to further characterize the implications of this new concept by biochemical means. Considering the fact that the lamellar intermediate structures represent a minority within the variety of membranous compartments of a cell, it will be a difficult task to enrich them, particularly in view of the special growth conditions required for cultivation and the inherent biological complexity of dendritic cells. We hope that our observations provide a stimulus to readdress the controversial issue of ER involvement in peroxisome formation from a new perspective using new genetic screens and biochemical approaches.
Acknowledgments
We are highly indebted to Rene Scriwanek and Marc van Peski for printing the micrographs, Ulrike Ziese for helping with the tomography software, Misjaël N. Lebbink for combining the two tomograms in the reconstruction, Dr. Paola Ricciardi-Castagnoli (Department of Biotechnology and Bioscience, University of Milano-Bicocca, Milan, Italy) for providing the D1 cells, and Tineke Voorn-Brouwer for preparing the anti-Pex13p antibodies. The research of A.J.K. has been made possible by a fellowship of the Royal Netherlands Academy of Arts and Sciences (KNAW). A.K.S. was supported by the Medical Science department of the Netherlands Foundation for Scientific Research (NWO-MW)
Abbreviations used: COP, coatomer protein; DC, dendritic cell; ER, endoplasmic reticulum; Ii, invariant chain; PDI, protein disulfide isomerase; Pex, peroxin; PEX, gene coding for peroxin; PMP, peroxisomal membrane protein; PTS, peroxisomal targeting signal.
References
- Beevers, H. (1979). Microbodies in higher plants. Annu. Rev. Plant Physiol. 8, 159-193. [Google Scholar]
- Borgese, N., Gazzoni, I., Barberi, M., Colombo, S., and Pedrazzini, E. (2001). Targeting of a tail-anchored protein to endoplasmic reticulum and mitochondrial outer membrane by independent but competing pathways. Mol. Biol. Cell 12, 2482-2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton, C., Hausler, T., and Blattner, J. (1995). Protein trafficking in kinetoplastid protozoa. Microbiol. Rev. 59, 325-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Duve, C., and Baudhuin, P. (1966). Peroxisomes (microbodies and related particles). Physiol. Rev. 46, 323-357. [DOI] [PubMed] [Google Scholar]
- Elgersma, Y., Kwast, L., Klein, A., Voorn-Brouwer, T., van den Berg, M., Metzig, B., America, T., Tabak, H.F., and Distel, B. (1996). The SH3 domain of the Saccharomyces cerevisiae peroxisomal membrane protein Pex13p functions as a docking site for Pex5p, a mobile receptor for the import PTS1-containing proteins. J. Cell Biol. 135, 97-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann, R., and Blobel, G. (1996). Identification of Pex13p a peroxisomal membrane receptor for the PTS1 recognition factor. J. Cell Biol. 135, 111-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann, R., Veenhuis, M., Mertens, D., and Kunau, W.H. (1989). Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 86, 5419-5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber, K.N., Haan, G.J., Baerends, R.J., Kram, A.M., and Veenhuis, M. (2002). Normal peroxisome development from vesicles induced by truncated Hansenula polymorpha Pex3p. J. Biol. Chem. 277, 11026-11033. [DOI] [PubMed] [Google Scholar]
- Gartner, J., Brosius, U., Obie, C., Watkins, P.A., and Valle, D. (1998). Restoration of PEX2 peroxisome assembly defects by overexpression of PMP70. Eur. J. Cell Biol. 76, 237-245. [DOI] [PubMed] [Google Scholar]
- Geuze, H.J., Slot, J.W., van der Ley, P.A., and Scheffer, R.C. (1981). Use of colloidal gold particles in double-labeling immunoelectron microscopy of ultrathin frozen tissue sections. J. Cell Biol. 89, 653-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaedi, K., Kawai, A., Okumoto, K., Tamura, S., Shimozawa, N., Suzuki, Y., Kondo, N., and Fujiki, Y. (1999). Isolation and characterization of novel peroxisome biogenesis-defective Chinese hamster ovary cell mutants using green fluorescent protein. Exp. Cell Res. 248, 489-497. [DOI] [PubMed] [Google Scholar]
- Gosset, P., Charbonnier, A.-S., Delerive, P., Fontaine, J., Staels, B., Pestel, J., Tonnel, A.-B., and Trottein, F. (2001). Peroxisome proliferator-activator receptor γ activators affect the maturation of human monocyte-derived dendritic cells. Eur. J. Immunol. 31, 2857-2865. [DOI] [PubMed] [Google Scholar]
- Gould, S.J., Kalish, J.E., Morrell, J.C., Bjorkman, J., Urquhart, A.J., and Crane, D.I. (1996). Pex13p is an SH3 protein of the peroxisome membrane and a docking factor for the predominantly cytoplasmic PTs1 receptor. J. Cell Biol. 135, 85-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould, S.J., and Valle, D. (2000). Peroxisome biogenesis disorders: genetics and cell biology. Trends Genet 16, 340-345. [DOI] [PubMed] [Google Scholar]
- Heikoop, J.C., van Roermund, C.W., Just, W.W., Ofman, R., Schutgens, R.B., Heymans, H.S., Wanders, R.J., and Tager, J.M. (1990). Rhizomelic chondrodysplasia punctata. Deficiency of 3-oxoacyl-coenzyme A thiolase in peroxisomes and impaired processing of the enzyme. J. Clin. Investig. 86, 126-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner, D., van Den Berg, M., Philippsen, P., Tabak, H.F., and Hettema, E.H. (2001). A role for Vps1p, actin, and the Myo2p motor in peroxisome abundance and inheritance in Saccharomyces cerevisiae. J. Cell Biol. 155, 979-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, J.M., Morrell, J.C., and Gould, S.J. (2001). Multiple distinct targeting signals in integral peroxisomal membrane proteins. J. Cell Biol. 153, 1141-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koster, A.J., Grimm, R., Typke, D., Hegerl, R., Stoschek, A., Walz, J., and Baumeister, W. (1997). Perspectives of molecular and cellular electron tomography. J. Struct. Biol. 120, 276-308. [DOI] [PubMed] [Google Scholar]
- Kremer, J.R., Mastronarde, D.N., and McIntosh, J.R. (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71-76. [DOI] [PubMed] [Google Scholar]
- Lazarow, P.B., and Fujiki, Y. (1985). Biogenesis of peroxisomes. Annu. Rev. Cell Biol. 1, 489-530. [DOI] [PubMed] [Google Scholar]
- Lazarow, P.B., and Moser, H.W. (1995). Disorders in peroxisome biogenesis. In: The Metabolic and Molecular Bases of Inherited Disease, ed. C.R. Scriver, A.L. Beadet, W.S. Sly, and D. Valle, New York: McGraw-Hill Companies, 2287-2324.
- Liou, W., Geuze, H.J., Geelen, M.J., and Slot, J.W. (1997). The autophagic and endocytic pathways converge at the nascent autophagic vacuoles. J. Cell Biol. 136, 61-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou, W., Geuze, H.J., and Slot, J.W. (1996). Improving structural integrity of cryosections for immunogold labeling. Histochem. Cell Biol. 106, 41-58. [DOI] [PubMed] [Google Scholar]
- Lutz, M.B., Girolomoni, G., and Ricciardi-Castagnoli, P. (1996). The role of cytokines in functional regulation and differentiation of dendritic cells. Immunobiology 195, 431-455. [DOI] [PubMed] [Google Scholar]
- Mastronarde, D.N. (1997). Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol. 120, 343-352. [DOI] [PubMed] [Google Scholar]
- Mellman, I., and Steinman, R.M. (2001). Dendritic cells: specialized and regulated antigen processing machines. Cell 106, 255-258. [DOI] [PubMed] [Google Scholar]
- Motley, A., Hettema, E., Distel, B., and Tabak, H. (1994). Differential protein import deficiencies in human peroxisome assembly disorders. J. Cell Biol. 125, 755-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novikoff, A., and Shin, W.Y. (1964). The endoplasmic reticulum in the Golgi zone and its relation to microbodies, Golgi apparatus and autophagic vacuoles in rat liver cells. J. Microsc. 3, 187-206. [Google Scholar]
- Novikoff, P.M., and Novikoff, A.B. (1972). Peroxisomes in absorptive cells of mammalian small intestine. J. Cell Biol. 53, 532-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purdue, P.E., and Lazarow, P.B. (2001). Peroxisome biogenesis. Annu. Rev. Cell Dev. Biol. 17, 701-752. [DOI] [PubMed] [Google Scholar]
- Schrader, M., King, S.J., Stroh, T.A., and Schroer, T.A. (2000). Real time imaging reveals a peroxisomal reticulum in living cells. J. Cell Sci. 113, 3663-3671. [DOI] [PubMed] [Google Scholar]
- Smith, J.J., Marelli, M., Christmas, R.H., Vizeacoumar, F.J., Dilworth, D.J., Ideker, T., Galitski, T., Dimitrov, K., Rachubinski, R.A., and Aitchison, J.D. (2002). Transcriptome profiling to identify genes involved in peroxisome assembly and function. J. Cell Biol. 158, 259-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South, S.T., Baumgart, E., and Gould, S.J. (2001). Inactivation of the endoplasmic reticulum protein translocation factor, Sec61p, or its homolog, Ssh1p, does not affect peroxisome biogenesis. Proc. Natl. Acad. Sci. USA 98, 12027-12031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South, S.T., Sacksteder, K.A., Li, X., Liu, Y., and Gould, S.J. (2000). Inhibitors of COPI and COPII do not block PEX3-mediated peroxisome synthesis. J. Cell Biol. 149, 1345-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, K., Schell-Steven, A., Erdmann, R., and Rottensteiner, H. (2002). Interactions of Pex7p and Pex18p/Pex21p with the peroxisomal docking machinery: implications for the first steps in PTS2 protein import. Mol. Cell. Biol. 22, 6056-6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroobants, A.K., Hettema, E.H., van den Berg, M., and Tabak, H.F. (1999). Enlargement of the endoplasmic reticulum membrane in Saccharomyces cerevisiae is not necessarily linked to the unfolded protein response via Ire1p. FEBS Lett. 453, 210-214. [DOI] [PubMed] [Google Scholar]
- Subramani, S., Koller, A., and Snyder, W.B. (2000). Import of peroxisomal matrix and membrane proteins. Annu. Rev. Biochem. 69, 399-418. [DOI] [PubMed] [Google Scholar]
- Tager, J.M., Van der Beek, W.A., Wanders, R.J., Hashimoto, T., Heymans, H.S., Van den Bosch, H., Schutgens, R.B., and Schram, A.W. (1985). Peroxisomal beta-oxidation enzyme proteins in the Zellweger syndrome. Biochem. Biophys. Res. Commun. 126, 1269-1275. [DOI] [PubMed] [Google Scholar]
- Titorenko, V.I., Ogrydziak, D.M., and Rachubinski, R.A. (1997). Four distinct secretory pathways serve protein secretion, cell surface growth, and peroxisome biogenesis in the yeast Yarrowia lipolytica. Mol. Cell. Biol. 17, 5210-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko, V.I., and Rachubinski, R.A. (2001). The life cycle of the peroxisome. Nat. Rev. Mol. Cell. Biol. 2, 357-368. [DOI] [PubMed] [Google Scholar]
- Valladeau, J., et al. (1999). The monoclonal antibody DCGM4 recognizes Langerin, a protein specific of Langerhans cells, and is rapidly internalized from the cell surface. Eur. J. Immunol. 29, 2695-2704. [DOI] [PubMed] [Google Scholar]
- van den Bosch, H., Schutgens, R.B., Wanders, R.J., and Tager, J.M. (1992). Biochemistry of peroxisomes. Annu. Rev. Biochem. 61, 157-197. [DOI] [PubMed] [Google Scholar]
- Vaux, D., Tooze, J., and Fuller, S. (1992). Identification by antiidiotype antibodies of an intracellular membrane protein that recognizes a mammalian endoplasmic reticulum retention signal. Nature 360, 372. [DOI] [PubMed] [Google Scholar]
- Voorn-Brouwer, T., Kragt, A., Tabak, H.F., and Distel, B. (2001). Peroxisomal membrane proteins are properly targeted to peroxisomes in the absence of COPI- and COPII-mediated vesicular transport. J. Cell Sci. 114, 2199-2204. [DOI] [PubMed] [Google Scholar]
- Wang, X., Unruh, M.J., and Goodman, J.M. (2001). Discrete targeting signals direct Pmp47 to oleate-induced peroxisomes in Saccharomyces cerevisiae. J. Biol. Chem. 276, 10897-10905. [DOI] [PubMed] [Google Scholar]
- Winzler, C., Rovere, P., Rescigno, M., Granucci, F., Penna, G., Adorini, L., Zimmermann, V.S., Davoust, J., and Ricciardi-Castagnoli, P. (1997). Maturation stages of mouse dendritic cells in growth factor-dependent long-term cultures. J. Exp. Med. 185, 317-328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, K., and Fahimi, H.D. (1987). Three-dimensional reconstruction of a peroxisomal reticulum in regenerating rat liver: evidence of interconnections between heterogeneous segments. J. Cell Biol. 105, 713-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziese, U., Janssen, A.H., Murk, J.L., Geerts, W.J., Van der Krift, T., Verkleij, A.J., and Koster, A.J. (2002). Automated high-throughput electron tomography by pre-calibration of image shifts. J. Microsc. 205, 187-200. [DOI] [PubMed] [Google Scholar]