Abstract
Arfophilin is an ADP ribosylation factor (Arf) binding protein of unknown function. It is identical to the Rab11 binding protein eferin/Rab11-FIP3, and we show it binds both Arf5 and Rab11. We describe a related protein, arfophilin-2, that interacts with Arf5 in a nucleotide-dependent manner, but not Arf1, 4, or 6 and also binds Rab11. Arfophilin-2 localized to a perinuclear compartment, the centrosomal area, and focal adhesions. The localization of arfophilin-2 to the perinuclear compartment was selectively blocked by overexpression of Arf5-T31N. In contrast, a green fluorescent protein-arfophilin-2 chimera or arfophilin-2 deletions were localized around the centrosome in a region that was also enriched for transferrin receptors and Rab11 but not early endosome markers, suggesting that the distribution of the endosomal recycling compartment was altered. The arfophilins belong to a conserved family that includes Drosophila melanogaster nuclear fallout, a centrosomal protein required for cellularization. Expression of green fluorescent protein-nuclear fallout in HeLa cells resulted in a similar phenotype, indicative of functional homology and thus implicating the arfophilins in mitosis/cytokinesis. We suggest that the novel dual GTPase-binding capacity of the arfophilins could serve as an interface of signals from Rab and Arf GTPases to regulate membrane traffic and integrate distinct signals in the late endosomal recycling compartment.
INTRODUCTION
Eukaryotes have developed highly sophisticated means of sorting and delivering intracellular membranes to/from different subcellular compartments. This trafficking is crucial for maintaining cell homeostasis and polarity. Small-molecular-weight GTPases of the ADP-ribosylation factor (Arf) and Rab families are central regulators of membrane trafficking pathways, the former playing a critical role in vesicle biogenesis and the latter controlling various aspects of vesicle targeting.
Arf1 is the best-studied of the Arf family. It is required for the recruitment and assembly of numerous coat protein complexes needed for vesicle formation. These include COPI/coatomer, and the clathrin adaptors AP-1, AP-3, AP-4, and GGAs, all of which function in the Golgi and trans-Golgi network (TGN) systems (Boman and Kahn, 1995; Boman et al., 2000; Donaldson and Jackson, 2000; Boehm and Bonifacino, 2001). Arf6, the sole class III member, is the most divergent Arf family member in mammalian cells and it functions in the plasma membrane/endocytic compartments where it seems to regulate endosomal recycling and actin dynamics (D'Souza-Schorey et al., 1995, 1998; Song et al., 1998). In contrast, virtually nothing is known about the function of the class II Arfs, Arf4 and Arf5.
The endocytic pathway is a complex, highly dynamic system that serves many functions, including the internalization of extracellular components and their subsequent sorting to different intracellular destinations (Mukherjee and Maxfield, 2000; Gruenberg, 2001). Rab GTPases regulate discrete membrane-trafficking events throughout the entire secretory/endocytic systems. It has been suggested that each Rab assembles a multitude of effectors into a “Rab domain,” thus coordinating numerous events (Sonnishsen et al., 2000; Zerial and McBride, 2001). For example Rab5-GTP binding partners include Rabaptin-5, EEA1, and PI3 kinase, proteins involved in early-endosome fusion (Woodman, 2000; Zerial and McBride, 2001). In addition, Rab5-GTP also interacts with a motor protein to stimulate endosome motility and may regulate soluble N-ethylmaleimide sensitive factor attachment protein receptor-mediated fusion (Zerial and McBride, 2001). Other Rabs are likely to have similarly pleiotropic roles.
Rab11 is implicated in regulating traffic through the endosomal recycling compartment (ERC). Transferrin receptors (TfRs) are internalized via clathrin-coated vesicles before entering early endosomes from where the majority rapidly recycle back to the cell surface after releasing their cargo (Hopkins, 1983). However, some TfRs traffic further into the cell to the perinuclear recycling endosomes adjacent to the centrosomal area before returning to the plasma membrane (the “slow” recycling route) (Ullrich et al., 1996; Ren et al., 1998; Wilcke et al., 2000). Distinct domains along this pathway can be distinguished based on the presence or absence of the small GTPases Rab4, Rab5, and Rab11 (Sonnishsen et al., 2000). The latter is found predominantly in the ERC and regulates the exit of traffic from this compartment (Ullrich et al., 1996; Ren et al., 1998; Wilcke et al., 2000). Although recent work has uncovered molecules such as RME-1 and myosin Vb (Grant et al., 2001; Lapierre et al., 2001) as potential regulators of the ERC, exactly how its structure, function, and integrity are maintained is still poorly understood.
The search for Arf effectors has yielded several candidate proteins, including the Golgi-localized, gamma adaptin ear homology domain containing Arf binding proteins (GGAs) (Boman et al., 2000; Dell'Angelica et al., 2000; Hirst et al., 2000), the arfaptins (Shin and Exton, 2001), and arfophilin. With the exception of the GGAs, the function of these putative Arf effectors is unknown. Arfophilin interacts specifically with GTP-bound class II Arfs via its C-terminal domain (Shin et al., 1999). This finding was particularly interesting given class II Arfs were assumed to play only supplementary roles to the more abundant class I Arfs. Arfophilin can also interact with Arf6 (Shin et al., 2001). Very recently, arfophilin has been rediscovered in a search for Rab 11 effectors; two laboratories have reported that arfophilin binds Rab 11 GTP and termed this protein Eferin and Rab 11-FIP3 (Hales et al., 2001; Prekeris et al., 2001). Despite this interest, neither the subcellular distribution nor the function of arfophilin/eferin/Rab11-FIP3 is known.
Herein, we describe the identification and characterization of a homolog of arfophilin that we term arfophilin-2 that unlike arfophilin interacts strongly with GTP-bound Arf5 and not with GDP-bound Arf5 or other Arf family members.
Like arfophilin, arfophilin-2 also binds Rab11. We find that endogenous arfophilin-2 localizes to the TGN/perinuclear region of HeLa cells with minor staining at both the centrosomal area and focal adhesions. Expression of GFP-arfophilin-2 chimera's results in a dramatic and specific accumulation of TfR and Rab11 in a pericentrosomal compartment, but does not affect TfR recycling. We further show that the arfophilins are homologous to Drosophila nuclear fallout (Nuf), a protein which functions at the centrosome where it plays a key role in cellularization and cytokinesis (Rothwell et al., 1998). We show that arfophilin-2 and Nuf are functionally related based on overexpression studies in mammalian cells. These data suggest that the arfophilin family may represent a point of convergence of both Rab and Arf signals in membrane traffic.
MATERIALS AND METHODS
Yeast Two-Hybrid Screening
Murine Arf5Q71L was subcloned into the BamHI and SalI sites of pGBT9 (BD Biosciences Clontech, Palo Alto, CA). This was then transformed into PJ69-2A to produce the bait strain that was then transformed with a human testis cDNA library in pACT2. Transformants were grown on synthetic media lacking Leu, Trp, and His at 30°C, and colonies developing within 5 d were picked and grown on media also lacking adenine. Prey vectors allowing growth under these conditions were rescued by electroporation of yeast miniprep DNA into KC8 Escherichia coli and screened against Arf5T31N to select only those that interacted with active Arf.
Molecular Biology
5′ Rapid amplification of cDNA ends polymerase chain reaction was performed using Expand High-Fidelity polymerase (Roche Diagnostics, Indianapolis, IN), Marathon-Ready human testis cDNAs (BD Biosciences Clontech) and nested gene specific primers (5′-CCTCGACCCTTTCATGGCGAACACCC-3′ and 5′-CCTTGAAGTTGATTCTCCCAGGTCGTTGG-3′) designed against extended sequence tag accession number AI570483, yielding two distinct but specific arfophilin-2 5′ ends. Full-length open reading frame of arfophilin-2 was gel purified and TA-cloned. Green fluorescent protein (GFP)-tagged arfophilin-2 was constructed by subcloning into pEGFP-C2 (BD Biosciences Clontech). GFP-arfophilin-2ΔN was produced by ligating a BglII fragment directly into the BamH1 site of pEGFP-C2. Dr. Ohara (Riren, Yokohama, Japan) supplied KIAA0665 clone (arfophilin-1). Arf cDNAs were either provided by K. Nakayama (University of Tsukuba, Tsukuba, Japan), J.M. Tavare, or J. Donaldson (National Institutes of Health, Bethesda, MD), or amplified from mouse cDNA. A recombinant adenovirus designed to overexpress myc-tagged Rab11-S25N was constructed using Adeno-X expression system (BD Biosciences Clontech) as described in the manufacturer's protocols.
Northern Blotting
Random primed [32P]CTP-labeled cDNA probes corresponding to the 3′ 1 kb of arfophilin-1 and -2 open reading frames were used to probe human MTN I and II Northern blots (BD Biosciences Clontech). Blots were probed for arfophilin-2 first, stripped, and then probed for arfophilin-1.
Antibodies
Antibodies specific for arfophilin-1 and -2 were raised using bacterially expressed recombinant protein. For arfophilin-2, a 1.2-kb BglII fragment originating from the prey vector was subcloned into pQE32 (QIAGEN, Valencia, CA). This fragment encodes the C-terminal 330 amino acids of arfophilin-2 in frame with an N-terminal hexahistidine tag. In the case of arfophilin-1, a 900-base pair BglII fragment (also from the prey vector) encoding the C-terminal 200 amino acids was subcloned into pQE31. Antibodies to arfophilin-1 and arfophilin-2 were raised in rabbits and sheep, respectively, by Diagnostics Scotland (Law Hospital, Carluke, Scotland) and affinity purified using recombinant antigen immobilized on CH-Sepharose 4B.
Other Antibodies
Anti-GRASP55 or GRASP65 were those described in Barr et al., (1997). Antibodies against AP-1 (γ-adaptin), Arf5, and mRME-1 were from M.S. Robinson (University of Cambridge, Cambridge, United Kingdom), anti-CI-MPR was from G.E. Lienhard (Dartmouth Medical School, Hannover, NH), and CD-MPR from A. Hille-Rehfeld (Universitat Göttingen, Göttingen, Germany). Antibodies against Rab4, Rab5, and Rab11 were from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-TfR was from Zymed (Cambridge, United Kingdom) and anti-α- and γ-tubulin and anti-β COP from Sigma Chemical (Poole, Dorset, United Kingdom).
Cell Culture and Transfection
HeLa cells were grown in minimal essential media containing with 10% fetal bovine serum and nonessential amino acids. Cells were transfected using LipofectAMINE reagent (Invitrogen, Paisley, Scotland) according to manufacturer's instructions and used 18 h later. Chinese hamster ovary (CHO) cells were grown in Ham's F-12 media containing fetal bovine serum and transfected using LipofectAMINE at ∼60% confluence. When cells were infected with adenovirus, the virus was diluted in low-serum-containing media and incubated on the cells for 3 h. Thereafter, an equal volume of normal media was added and the incubation continued for 24 h. After this time, cells were fixed for immunofluorescence as outlined below.
Pull-Down Assays
Dishes (100 mm) of CHO cells expressing GFP-tagged arfophilin-1 or arfophilin-2 were washed with ice-cold phosphate-buffered saline (PBS) and scraped in 250 μl of ARF binding buffer (50 mM HEPES, 100 mM KCl, 5 mM NaCl, 1 mM MgCl2, 0.5 mM EGTA, 1 mM EDTA, 1 mM dithiothreitol, pH 7.2). Lysates were prepared by passing suspension through a 27-gauge needle before centrifugation at 21,000 × g for 20 min at 4°C. Arf4Q71L, Arf5-Q71L, Arf6-Q71L, and wild-type Arf5 were subcloned into pGEX5X1 and expressed by 3-h isopropyl β-d-thiogalactoside induction in E. coli strain Bl21 DE3 (pLysS). Cells were lysed in 10 ml of 50 mM Tris pH 7.5, 1 mM EDTA, 100 mM NaCl, 5% glycerol, 0.1% Triton X-100, 1 mM dithiothreitol with protease inhibitors per 100-ml culture, and sonicated for 3 × 30 s. GST-ARF proteins were affinity purified using glutathione-Sepharose 4B, eluted with 10 mM reduced glutathione, and dialyzed against PBS.
GST-Arf protein (20–40 μg) was mixed with 25 μl of glutathione-Sepharose 4B for 30 min. CHO cell lysate (500 μl) was then added and rotated at 4°C for 1 h. The Sepharose was washed and bound proteins eluted by boiling in 30 μl of SDS-PAGE loading buffer. The same protocol was used for GST-Rab11 pull-downs.
Immunocytochemistry and Immunofluorescence Microscopy
Cells on coverslips were fixed either in methanol at -20°C for 4 min or fixed in 3.7% p-formaldehyde in PBS for 10 min followed by permeabilization in 0.1% Triton X-100 in PBS for 1 min. The cells were washed, blocked and incubated with antibodies as described previously (Millar et al., 1999). Confocal microscopy was performed on either a Zeiss 410 or Zeiss Pascal confocal system using a microscope fitted with 63× 1.4-numerical aperture plan apochromat objectives. Cells labeled with red fluorophores were excited at 543 nm, and a 590-nm long-pass filter was used for collecting emissions. Green fluorophores were excited at 488 nm with a 505- to 520-nm band-pass detection filter. The fraction of cross-over was determined to be <3%. Images were analyzed using MetaMorph software (Universal Imaging, Downington, PA).
Transferrin Pulse-Chase Experiments
HeLa cells on coverslips were incubated in serum-free media containing 0.6% (wt/vol) bovine serum albumin for 2 h, with 2.5 μg/ml Texas Red transferrin (Molecular Probes, Leiden, Holland) added for the final 10–60 min. This was chased by incubation with 100 μg/ml iron-loaded human transferrin. Cells were washed in ice-cold buffer, fixed, and analyzed. Cell surface transferrin receptor levels were quantified exactly as outlined in Millar et al. (1999. For assay of the rate of transferrin (Tf) release, cells were incubated for 2 h in 3 nM 125I-Tf, washed in ice-cold citrate buffer to remove cell surface-bound Tf, and then warmed to 37°C and the rate of Tf externalization assayed as outlined in Millar et al. (1999).
RESULTS
Arfophilin-2 Is a Novel Arf-interacting Protein and a Homolog of Arfophilin/Eferin/Rab11-FIP3
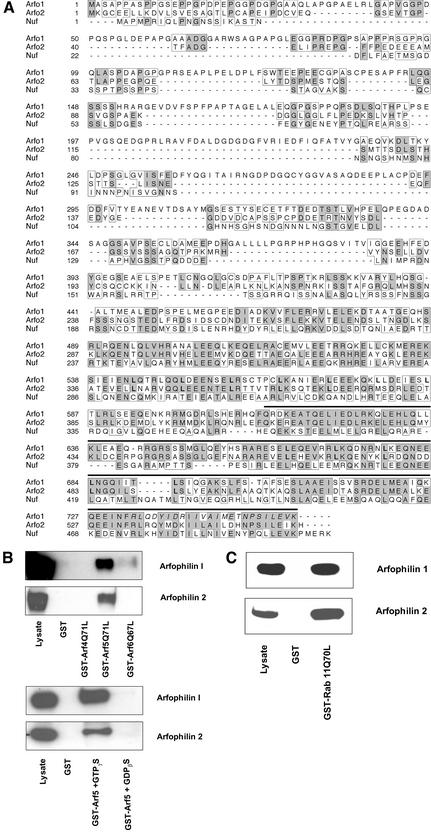
In an attempt to identify novel class II Arf effectors, we performed a yeast two-hybrid screen of a human testis library with the GTPase-defective mutant Arf5Q71L. From a small-scale screen of 60,000 transformants, eight clones were isolated that conferred a specific interaction with Arf5Q71L but not with dominant negative Arf5T31N. One of these encoded full-length arfaptin 2/POR1, a previously reported Arf-interacting protein (Shin and Exton, 2001). Five clones contained cDNA inserts of 0.9 to 2.1 kb that were all derived from the 3′ end of the same gene on human chromosome 17. 5′ Rapid amplification of cDNA ends amplified a full-length arfophilin-2 open-reading frame that contained in frame upstream stop codons. The protein encoded by this gene shared significant homology in its C termini with the Arf-binding domain of arfophilin (63% identity, 79% homology >146 residues) (Figure 1A). We therefore named this protein arfophilin-2 and will subsequently refer to arfophilin (KIAA0665) as arfophilin-1.
Figure 1.
Arfophilin-2 is a novel Arf-interacting protein, homologous to nuclear fallout. Shown in A is a ClustalW alignment of the predicted amino acid sequences of human arfophilin-1 (Arfo1), arfophilin-2 (Arfo2), and the Drosophila protein Nuf. Shaded boxes highlight residues that are identical in at least two of the sequences while unshaded boxes highlight homologous residues. Note the minimal Arf binding domain (bold line) of arfophilin-1 as reported previously (Shin et al., 1999), the minimal Rab 11 binding domain (italics) (Prekeris et al., 2001) and putative leucine zipper motifs (bold). (B) In the top panel, beads loaded with GST, GST-Arf4Q71L, GST-Arf5Q71L, or GST-Arf6Q67L were mixed with lysates of CHO cells overexpressing either arfophilin-1 or arfophilin-2. The beads were recovered and bound proteins eluted and immunoblotted with anti-arfophilin-1 or -2 (as indicated) together with an aliquot of the CHO cell lysate. The bottom panel shows the effect of 10 μM GTP and GDP on the interaction of GST-wild-type Arf5 with both arfophilins. (C) The result of a typical experiment by using GST or GST-Rab11-Q70L to pull-down arfophilin-1 or -2.
Analysis of the predicted sequence revealed that arfophilin-2 is hydrophilic with no potential signal sequences or transmembrane domains and a molecular mass of ∼61 kDa. Arfophilin-1 and -2 shared significant homology with the Caenorhabditis elegans protein F55C12.1a/F55C12.1b and Drosophila melanogaster nuclear fallout (Figure 1A) (Rothwell et al., 1998). The major similarities between the arfophilins and Nuf lie in the C-terminal ∼300 amino acids in regions predicted to form coiled-coils, whereas the N termini are more divergent (Figure 1A). The homologous regions of all the proteins belong to a domain of ∼70 amino acids present in the ProDom database (designated PD031147). Arfophilins-1 and -2 also have leucine zipper motifs in their C termini (Figure 1A). Neither of the arfophilins possess the sec7 domains characteristic of Arf guanine nucleotide exchange factors, nor do they have the characteristics of Arf GTPase activating proteins, suggesting that they are unlikely to be direct regulators of the Arf GTPase cycle.
Interestingly, arfophilin-1 is identical to the recently described Rab 11 binding protein eferin/Rab 11-FIP3 (Hales et al., 2001; Prekeris et al., 2001). Analysis of the predicted amino acid sequence of several recently identified Rab 11 binding proteins has identified a 20-amino acid domain involved in Rab 11 binding that is present at the extreme C termini of arfophilin-1 (sequence in italics in Figure 1A) (Prekeris et al., 2001). Arfophilin-2 is strikingly similar to arfophilin-1 over this region, suggesting that this protein is also likely to interact with Rab 11 (see below). However, the N-terminal half of arfophilin-1 and -2 are quite distinct. This suggests that although both of these proteins may be predicted to interact with the same sets of small GTPases, they may subserve distinct functions and/or be differentially regulated.
To confirm that arfophilin-2 was a bona fide Arf and Rab binding protein, we examined the ability of GST-Arf proteins to bind both arfophilin-1 and arfophilin-2. GST-Arf5-Q71L effectively pulled down both overexpressed arfophilin-1 and arfophilin-2 from CHO cell lysates. In contrast, neither GST-Arf6-Q71L, GST-Arf1Q67L, nor GST-Arf4-Q71L seemed to interact with arfophilin-2 (Figure 1B). These data are subtly different to that reported for arfophilin-1, which was shown to interact weakly with Arf6-Q71L (Figure 1B). Neither of the arfophilins interact with Arf1 (our unpublished data). To further confirm the nucleotide specificity of the Arf5-arfophilin-2 interaction, we performed a series of pull-down experiments by using wild-type Arf5 expressed as a GST fusion, including either GTPγS or GDPβS in the buffer (Figure 1B). The data clearly demonstrate that the interaction of Arf5 with arfophilin-2 is dependent upon the nucleotide status of the Arf. These data was further confirmed using two-hybrid assays (our unpublished data). Hence, arfophilin-1 and -2 exhibit some subtle differences in their ability to bind activated Arf proteins. Using the same approach, we showed that GST-Rab11-Q71L was able to effectively pull-down both arfophilin-1 or -2 expressed in CHO cells (Figure 1C). Hence, both of these proteins are capable of binding directly to two distinct classes of small molecular weight GTPases.
Tissue Expression of Arfophilin-2: A Protein Abundant in Testes
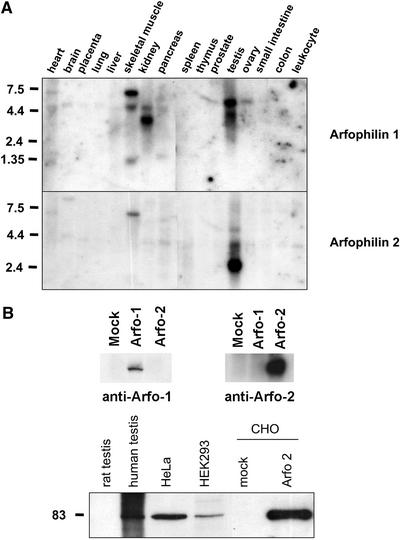
Northern blot analysis of a panel of 16 human tissues was performed to study the pattern of expression of arfophilin-2. The highest levels of expression were observed in testis (Figure 2A). Prolonged exposure of the blot revealed a 4.4-kb transcript present in all tissues examined (our unpublished data). After stripping, an analogous arfophilin-1 probe was also used on the same blots and this showed no crosshybridization with the major arfophilin-2 transcripts. Arfophilin-1 is also highly expressed in testes with major 5.2-kb and minor 4.2-kb transcripts. Arfophilin-1 expression was also noted in ovary, skeletal muscle, and kidney. Skeletal muscle and heart also express large (∼7-kb) arfophilin-1 mRNAs.
Figure 2.
Tissue distribution of human arfophilin-1 and -2. (A) Northern blot analysis of the tissue distribution of the arfophilin-1 and -2 mRNAs. A 1-kb fragment of the 3′ end of arfophilin-2 was used as a probe (bottom panel), revealing high expression of a 2.4-kb transcript in testis, and lower expression of a 4-kb transcript in most other tissues. An analogous arfophilin-1 probe (top) was used on the same blot after stripping. (B) Top, illustrates the specificity of affinity purified antiarfophilin-1 and anti-arfophilin-2 antibodies. The three lanes shown contain 16 μg of mock-transfected HeLa cells, cells expressing GFP-arfophilin-1, or cells expressing GFP-arfophilin-2. The anti-arfophilin-2 blot is overexposed to illustrate the lack of cross-reactivity with arfophilin-1. Bottom, an immunoblot showing that endogenous arfophilin-2 protein from human testis and HeLa cells migrates with an apparent molecular mass of 82 kDa, as does full-length untagged arfophilin-2 expressed in CHO cells. This immunoblot is typical of four experiments of this type.
We raised polyclonal antibodies to recombinant proteins comprising either the C-terminal 330 amino acids of arfophilin-2 or the C-terminal 200 amino acids of arfophilin-1. In both cases, the antigen was recognized specifically by the immune but not the preimmune serum, and the signals were blocked by prior incubation of the serum with the appropriate recombinant protein (our unpublished data). Control immunoblots in which the antiserum were used to probe either purified recombinant arfophilin-1 or -2, or the protein overexpressed in cells revealed that these antibodies specifically recognized the protein of interest and did not exhibit cross-reactivity (Figure 2B, top). Immunoblot analysis identified a major band of ∼85 kDa in human testis, HeLa, and human embryonic kidney 293 cells. Rat testis and a CHO cell lysate, however, did not exhibit any immunoreactivity, presumably due to a lack of species cross-reactivity (Figure 2B, bottom). When overexpressed in CHO cells, arfophilin-2 exhibited identical electrophoretic mobility compared with the endogenous protein in testis and HeLa cells (Figure 2B).
Localization of Arfophilin-2
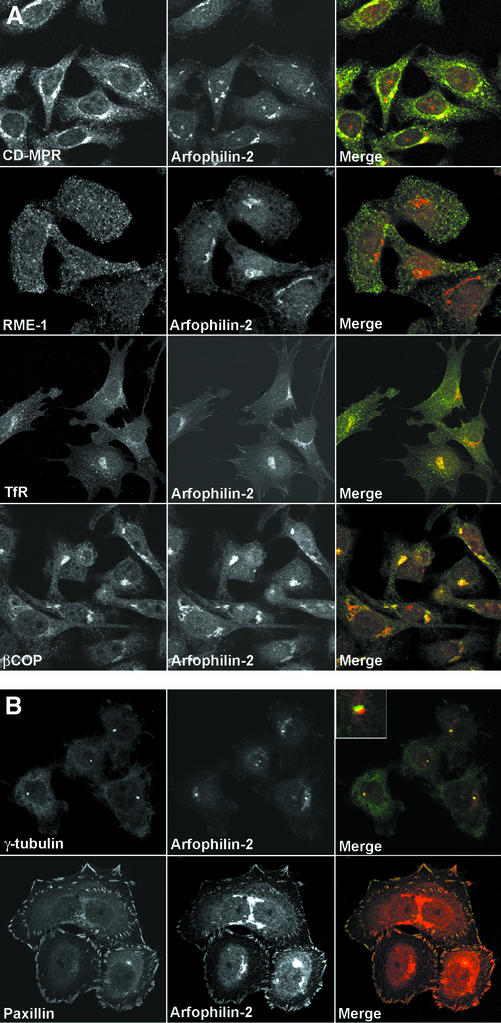
We next used confocal microscopy to determine the subcellular localization of endogenous arfophilin-2 in HeLa cells. Staining was observed primarily in a perinuclear region that partially overlapped with staining for the cation-independent mannose-6-phosphate receptor (CI-MPR), which traffics between the TGN, lysosomes, and plasma membrane (Kornfeld, 1992) (Figure 3A). Arfophilin-2 staining was also partially coincident with βCOP (Figure 3A) and with TfRs in the perinuclear region. In contrast, overlap with early endosome markers such as EEA1 or RME1 was limited (Figure 3A). In addition to this major pool, arfophilin-2 immunoreactivity was also detected at the centrosomal area, where it was coincident with, although slightly offset from, γ-tubulin staining (Figure 3B). Finally, we also observed arfophilin-2 staining at the periphery of cells, in structures that resembled focal adhesions. Indeed, this staining was found to overlap with staining for paxillin, a protein known to localize at focal adhesions (Figure 3B) (Norman et al., 1998). Arfophilin-2 staining at all of these sites was abolished by prior incubation with recombinant arfophilin-2 but not arfophilin-1 and no staining was observed using preimmune serum (our unpublished data). We therefore conclude that there are at least three pools of arfophilin-2 in HeLa cells, a major one around the TGN and minor pools around the centrosome and at focal adhesions.
Figure 3.
Intracellular localization of endogenous arfophilin-2. (A) Antibodies specific for arfophilin-2 were used to localize the endogenous protein in HeLa cells (middle, red in merged images) and to compare its distribution with marker proteins (left, green in merged images). Arfophilin-2 staining was predominantly detected in a perinuclear region partially coincident with the CI-MPR and βCOP but with little overlap with the TfR or RME-1. (B) Minor arfophilin-2 staining (red in the merged images) in the centrosome region, adjacent to γ-tubulin, and also in focal adhesions where it colocalized with paxillin. Data are typical of eight experiments. Note that to reveal the staining at focal adhesions, significantly increased laser power was used to collect this image with respect to that shown in A.
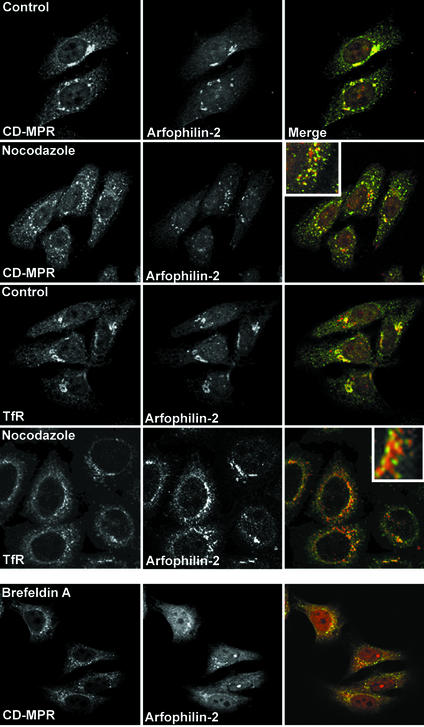
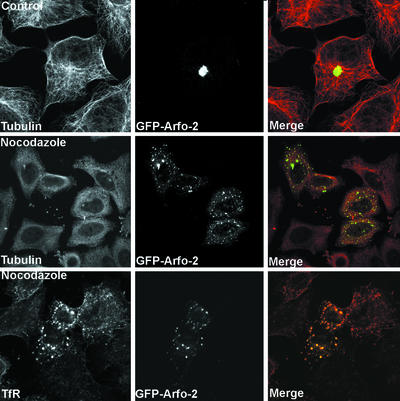
The effects of the drugs nocodazole and brefeldin A (BFA) on arfophilin-2 distribution were examined by immunofluorescence microscopy (Figure 4). The former is a microtubule-disrupting agent, whereas the latter disrupts the Golgi by inhibiting Arf activation. In control cells arfophilin-2 partially colocalizes with the CI-MPR and TfR in the TGN region. On BFA treatment (20 μg/ml for 10 min) arfophilin-2 staining became very diffuse and cytosolic, with little colocalization with either TfR (our unpublished data) or CI-MPR (Figure 4). Nocodazole treatment (50 ng/ml for 1 h) also disrupted the localization of both proteins, although arfophilin-2 remained in punctate structures. Under these conditions, the extent of overlap of arfophilin-2 and CI-MPR was greatly reduced, whereas a significant degree of colocalization of arfophilin-2 and TfR was evident in nocodazole-treated cells. Conversely, in our hands neither BFA nor nocodazole treatment seemed to have any effect on the distribution of arfophilin-2 at focal adhesion, suggesting that the localization/maintenance at these sites does not require functional microtubules (our unpublished data).
Figure 4.
Effect of nocodazole and BFA on endogenous arfophilin-2 distribution. HeLa cells treated with or without (control) brefeldin A (20 μg/ml for 10 min) or nocodazole (50 ng/ml for 1 h) were fixed and stained with antibodies against arfophilin-2 (red in merged images), tubulin, TfR, or the CI-MPR (green in merged images). The experiment was repeated four times with similar results.
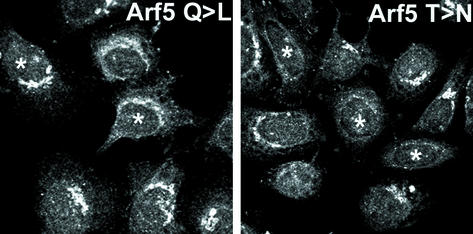
Finally, we set out to determine whether the overexpression of mutants of either Rab11 or Arf5 could modulate the distribution of endogenous arfophilin-2. The results (Figure 5) show that overexpression of Arf5-T31N displaced endogenous arfophilin-2 from the perinuclear localization. Such data offer the hypothesis that the localization of arfophilin-2 to the perinuclear region of the cells is mediated by an interaction with Arf5-GTP. This contention is further supported by our data showing that BFA-treatment displaced arfophilin-2 from the TGN area (Figure 4). In contrast, the GTP-locked mutant of Arf5 had little discernible effect on arfophilin-2 distribution (Figure 5), neither did the GDP or GTP mutants of Arf6 (our unpublished data). To determine the effect of Rab11 mutants, we infected cells with a recombinant adenovirus engineered to overexpress myc-tagged Rab11-S25N. Despite high level overexpression, we observed no effect on the distribution of arfophilin-2 at either the TGN or focal adhesions (our unpublished data).
Figure 5.
Overexpression of Arf5-T31N displaces arfophilin-2 from the perinuclear area. HeLa cells were transiently transfected with HA-tagged Arf5-T31N or Arf5-Q71L. Twenty-four hours after transfection, cells were fixed and stained using anti-HA (to identify the Arf-overexpressing cells, indicated here by * on the figure) and anti-arfophilin-2 (shown). In the cells shown, note that the perinuclear arfophilin-2 staining is greatly reduced in cells expressing Arf5-T31N. Data from a representative experiment is shown, repeated four times with similar results.
GFP-Arfophilin-2 Overexpression Alters Recycling Endosome Morphology
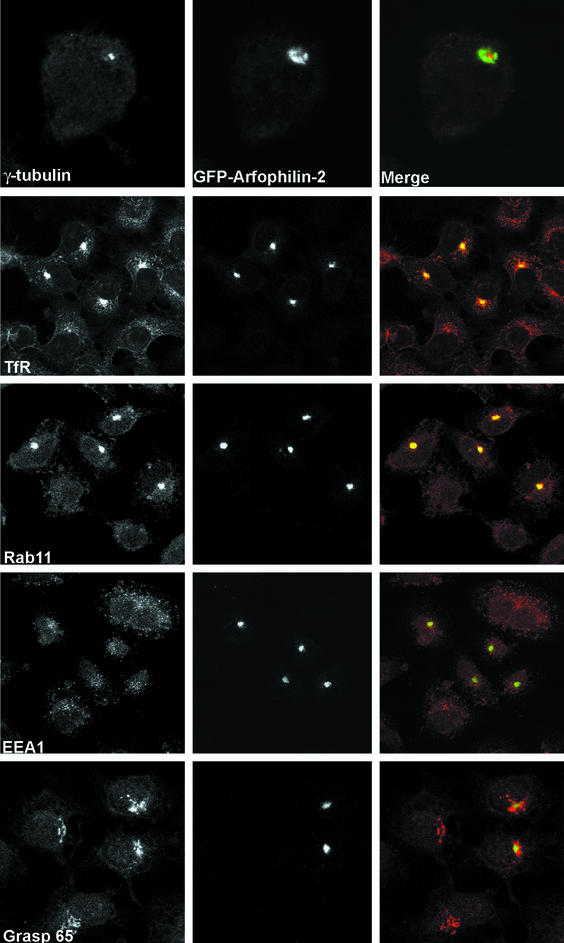
To examine the cellular function of arfophilin-2, we generated full-length and N-terminally truncated arfophilin-2 constructs fused to the C termini of GFP. In contrast to the endogenous proteins, these chimeric proteins were localized tightly around γ-tubulin staining at the centrosome (Figure 6), with little or no staining observed in the Golgi region (compare Figure 6b with Figure 3b). More strikingly, transfected cells displayed a dramatic accumulation of TfR and the recycling endosomal marker Rab 11 in the same pericentriolar region, an effect absent in adjacent nontransfected cells (Figure 6). The data shown is for GFP-tagged full-length arfophilin-2, but similar results were obtained using GFP-fused to the C-terminal 330 amino acids of arfophilin-2 (our unpublished data). This altered morphology of the endosomal system seemed to be confined to the perinuclear recycling endosomes. Expression of GFP-arfophilin-2 had no discernible effect on the distribution of early endosome markers such as EEA1 (Figure 6), or on the distribution of another endosomal associated protein, RME1 (Figure 6). Similarly, the localization of a range of other proteins such as MPR, AP-1, and AP-3, and GRASP65 and 55 were all unaffected (Figure 6; our unpublished data). This suggests that arfophilin-2 is controlling either some aspect of the flux of TfR traffic through the Rab11-positive perinuclear recycling endosomal system or the three-dimensional distribution of the recycling compartment. Note that the low levels of expression of endogenous Arf5 have precluded analysis of the distribution of Arf5 in cells expressing GFP-arfophilin-2.
Figure 6.
Expression of GFP-arfophilin-2 in HeLa cells alters recycling endosome morphology. GFP-arfophilin-2 was expressed in HeLa cells and its distribution compared with that of known marker proteins. GFP-arfophilin-2 localized to a compartment closely associated with the centrosome, as evidenced by γ-tubulin staining. This compartment was enriched in TfR, indicative of an endosomal origin. The recycling endosome marker Rab11 also colocalized strongly with GFP-arfophilin-2, whereas the early endosome marker RME1 did not. The data shown are for full-length GFP-tagged arfophilin-2, although similar data were obtained using a GFP-fusion to the C-terminal 330 residues of arfophilin-2 (our unpublished data). The data presented are typical of six experiments.
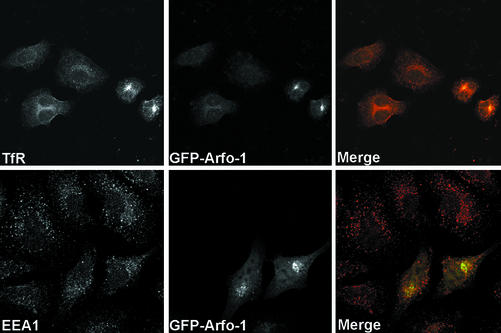
Given the homology between arfophilin-1 and arfophilin-2 (Figure 1A), we also expressed the C-terminal 330 amino acids of arfophilin-1 in HeLa cells as a GFP fusion protein (Figure 7). GFP-arfophilin-1 was predominantly localized to a perinuclear compartment. In cells expressing GFP-arfophilin-1, we again observed accumulation of both TfR and Rab 11 into this compartment, consistent with the previously reported ability of arfophilin/eferin/Rab11-FIP3 to bind Rab 11. In contrast, the distribution of EEA1 was unaffected by GFP-arfophilin-1 overexpression.
Figure 7.
GFP-tagged arfophilin-1/eferin/Rab 11-FIP3 behaves similarly to GFP-arfophilin-2. GFP-arfophilin-1 (C-terminal 330 residues fused to the C termini of GFP) was expressed in HeLa cells and the effect on the distribution of TfR and EEA1 examined by immunofluorescence microscopy. This revealed a similar phenotype to that observed for GFP-arfophilin-2. GFP-arfophilin-1 fluorescence (middle, green in merged images) is compared with TfR and EEA1. A representative experiment is shown.
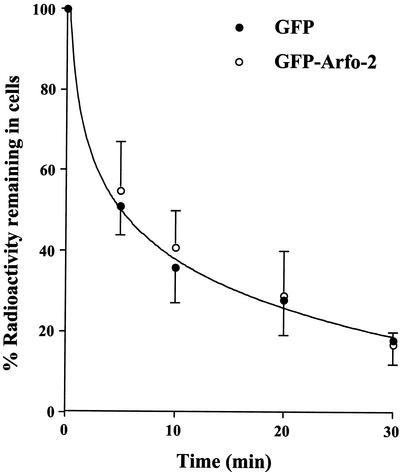
We sought to determine whether the profound effects on TfR distribution resulting from GFP-arfophilin expression were due to changes in the rate of TfR entry/exit from the ERC or whether the effects were a consequence of altered morphology of the compartment. To this end, we determined whether the GFP-arfophilin-2-/TfR-positive compartment was accessible to fluorescent Tf added to the culture medium, and if so, whether this could be chased out of the compartment with unlabeled Tf. Within 20 min of uptake at 37°C, significant colocalization between the GFP-arfophilin-2 and Texas-Red Tf was observed, demonstrating that TfR from the cell surface are able to traffic into the GFP-arfophilin-2 compartment (our unpublished data). After a 60-min uptake and subsequent chase with nonlabeled Tf, a steady decrease in Tf fluorescence was observed overtime in both transfected and nontransfected cells. By 90 min, nearly all the labeled Tf had disappeared, suggesting that exit from the compartment is not compromised. These experiments suggest that there is not a quantitative block in TfR trafficking to the GFP-arfophilin-2 compartment. Similar data were observed in cells overexpressing GFP-arfophilin-1. Such data are difficult to quantify, and thus in an attempt to examine transferrin trafficking in more detail, we examined the rate or Tf externalization and levels of cell surface TfR in HeLa cells transiently expressing GFP-arfophilin-2. Neither levels of TfR at the cell surface (our unpublished data) nor the rate of Tf externalization (Figure 8) were affected by overexpression of GFP-arfophilin-2. We next examined the effect of nocodazole and BFA on the pericentrosomal localization of GFP-arfophilin-2. Figure 9 shows that treatment with nocodazole induced a fragmentation of the GFP-arfophilin-2 compartment, which became punctate and dispersed throughout the cell. Interestingly, when the GFP-arfophilin-2 compartment was dispersed with nocodazole, TfR remained tightly associated with the GFP-arfophilin-2 compartment (Figure 9). In contrast, BFA had minimal effect on the localization of GFP-arfophilin-2 localization (our unpublished data; see DISCUSSION).
Figure 8.
TfR traffic is not significantly perturbed in cells overexpressing GFP-arfophilin-2. HeLa cells expressing GFP-arfophilin-2 were incubated with 125I-Tf for 2 h at 37C, washed in ice-cold citrate buffer at 4°C to remove cell surface-associated Tf, and then rewarmed into buffer at 37°C. The rate of Tf externalization was assayed by measurement of radioactivity associated with the cells at the times shown. The data shown are from triplicate determinations at each time point from a typical experiment, repeated three times with similar results.
Figure 9.
Nocodazole disrupts the pericentrosomal distribution of GFP-arfophilin-2. GFP-arfophilin-2 was expressed in HeLa cells as in Figure 6, which were then treated with or without (control) nocodazole (50 ng/ml for 1 h), fixed, and stained with antibodies against arfophilin-2, tubulin or TfR as indicated. The data shown are for GFP-fused to the C-terminal 330 residues of arfophilin-2 and are typical of four experiments.
Arfophilins Are Mammalian Homologs of Drosophila Nuclear Fallout
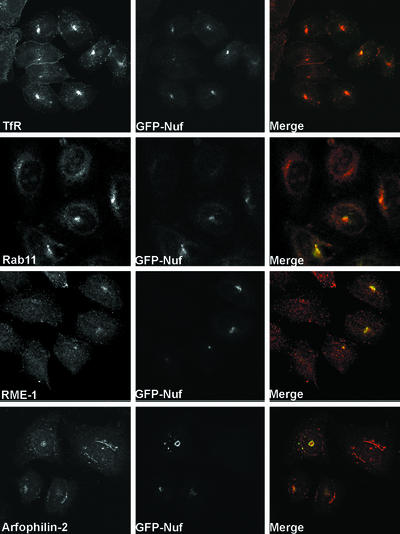
Analysis of the databases for proteins with homology to arfophilins-1 or -2 identified the Drosophila protein Nuf (Figure 1) (Rothwell et al., 1998). We therefore expressed GFP-Nuf in mammalian cells and examined its localization and effects on TfR distribution. Like GFP-arfophilin-2, GFP-Nuf localized to a region immediately adjacent to the γ-tubulin–positive centrosome (Figure 10). Moreover, the effect of expression of GFP-Nuf on TfR and Rab11 distribution is strikingly similar to that seen for arfophilins in that both molecules become tightly localized to the Nuf-positive region. In addition, endogenous arfophilin-2 became clustered in the same GFP-Nuf–positive region (Figure 10). Similar data were obtained using a construct encoding GFP fused to the C-terminal (arfophilin-homologous) 300 amino acids of Nuf (our unpublished data). The experiment shown in Figure 8 was repeated in cells expressing GFP-Nuf. The data (our unpublished data) revealed that Tf added to the media of cells expressing GFP-Nuf rapidly entered the pericentrosomal compartment and could be chased out with unlabeled Tf with kinetics similar to that shown for GFP-arfophilin-2. These data argue strongly that the arfophilins, by virtue of their conserved C termini, are mammalian homologs of Nuf. These data are further supported by data showing that Nuf directly binds mammalian Rab11 in a nucleotide-dependent manner (Matheson and Gould, unpublished data).
Figure 10.
Nuclear fallout exhibits a similar phenotype when expressed in HeLa cells. GFP-Nuf was transiently expressed in HeLa cells and its distribution compared with that of known marker proteins. As with the GFP-tagged arfophilins, GFP-Nuf (middle, green in the merged images) localized around the centrosomal region, where it was coincident with TfR and Rab11, but not with RME1 (red in merged images). GFP-Nuf expression also altered the localization of endogenous arfophilin-2 that became clustered in the centrosomal region in GFP-Nuf–positive cells. These data are representative of three experiments of this type.
DISCUSSION
Characterization of Arfophilin-2 and Comparison with Arfophilin-1/Eferin/Rab11-FIP3.
Despite considerable research, definitive functions for class II Arfs remain unresolved. Arfophilin-1 was identified by virtue of its GTP-dependent interaction with Arf5, but no information as to its subcellular localization or function was provided (Shin et al., 1999, 2001). During the course of this work, we noted that arfophilin-1 is identical to the Rab11 effector protein eferin/Rab11-FIP3 (Hales et al., 2001; Prekeris et al., 2001). Herein, we describe the identification of a second member of the arfophilin family, arfophilin-2. The arfophilin-2 cDNA encodes a protein of 557 amino acids and predicted molecular mass of 61 kDa. Arfophilins-1 and -2 exhibit most homology in the C-terminal 300 amino acids in regions predicted to form coiled-coils (Figure 1A). This region interacts with both the Arf (Shin et al., 1999, 2001) and Rab GTPases (Prekeris et al., 2001), but at distinct sites. The binding site for Rab11 in arfophilin-1 is within the C-terminal 20 residues. Arfophilin-1 and -2 are well conserved over this region, and we have shown that both of these proteins directly bind Rab11. We further show that arfophilin-2 interacts with Arf5 in a GTP-dependent manner (Figure 1B). Hence, the arfophilins physically interact with both Rab11 and Arf5, thus suggesting the existence of a family of dual Rab/Arf effectors expressed in mammalian cells. Divalent Rab effectors such as Rabenosyn-5 (which interacts with Rab4 and Rab5; Nielsen et al., 2000) and RCP (Rab4 and Rab11; Lindsay et al., 2002) have been described, as have dual Arf/Rho family binders (arfaptin-2/POR1; Shin and Exton, 2001). However, this is the first indication of direct Rab/Arf cross talk.
Tissue Distribution of Arfophilin-2
The arfophilin-2 gene is moderately large (∼200 kb), and there are homologs in Musca musculus, Rattus norvegicus, Xenopus laevis, Orizayas latipes, C. elegans, and D. melanogaster but not in the Saccharomyces cerevisiae genome. Northern blotting revealed a 4.4-kb arfophilin-2 transcript in all tissues, albeit at low levels (Figure 2A). Both arfophilins are highly expressed in testes, the significance of this is not presently known but may reflect the high degree of mitotic and meiotic cell division occurring in testes compared with the other tissues examined. The notion of a role in cell division is attractive given the homology with Nuf (see below).
Subcellular Distribution and Function of Arfophilin-2
Immunofluorescence microscopy identified a major BFA-sensitive pool of arfophilin-2 localized to the TGN region of the cell, consistent with a role for arfophilin-2 in Arf5-mediated events. Minor but specific staining was also observed at the centrosomal area and in focal adhesions. This latter observation is interesting given that membrane traffic to sites of plasma membrane protrusion seems to involve traffic from Rab11-positive recycling endosomes and may involve Arfs. The staining at each of these sites was specific, because it was selectively blocked by prior incubation of the antibody with recombinant arfophilin-2 but not arfophilin-1, and was not observed using preimmune serum (our unpublished data).
We have further shown that overexpression of tagged and/or N-terminally truncated members of the arfophilin family in HeLa cells causes a dramatic and specific effect on the distribution of TfRs, implicating arfophilin-2 in the regulation of the endosomal system. Such dramatic effects on the endosomal compartment have been observed when other Rab-11 binding proteins are overexpressed (Hales et al., 2001). Furthermore, the fact that Rab 11, but not EEA1 was similarly affected strongly argues that arfophilin-2 specifically affects the ERC and is consistent with arfophilin-2 interacting with Rab11 (Ullrich et al., 1996; Ren et al., 1998). Several lines of evidence argue that perturbations of endosomal morphology and the centrosomal accumulation of GFP-arfophilin-2 are not artifacts of either GFP-tagging or overexpression. First, overexpression of hemaggultinin (HA)-tagged truncated arfophilin-2 mutants also accumulate at the centrosomal area of cells (our unpublished data). In contrast, expression of full-length, untagged arfophilin-2 results in markedly enhanced staining in all three subcellular locations described for the endogenous protein in Figure 3 (our unpublished data). This suggests that the overexpressed full-length protein seems to localize in an identical manner to the endogenous protein, but the truncated protein is localized to the centrosomal area only. (The inability of our antibodies to distinguish between endogenous and overexpressed proteins precludes a detailed analysis of this.) Second, the localization of a fraction of the endogenous protein to the centrosomal region implies that at some stage during its functional cycle, endogenous arfophilin-2 is located at this region. Finally, overexpression of GFP-Nuf has an almost identical phenotype and also recruits a significant fraction of the endogenous protein to the centrosomal area. Our interpretation of this data is that the presence of the bulky GFP at the N terminus, or the deletion of the N terminus, result in perturbation of some aspect of either the localization or the function of arfophilin-2. Although this pericentrosomal location of GFP-arfophilin-2 could represent aggresome formation, we suggest for the reasons indicated above that this is unlikely.
The fact that transferrin can still enter and leave the GFP-arfophilin-2–positive compartment (Figure 8) argues that the changes in TfR distribution are not due to a quantitative trafficking block (and also mitigates against aggresome formation). Such a recycling block was recently described upon overexpression of GFP-myosin Vb tail, which caused internalized transferrin to accumulate in a similar Rab 11-positive compartment, which could not be chased out (Lapierre et al., 2001). Although this is clearly not the case with GFP-arfophilin-2 overexpression, subtle changes in the kinetics of entry/exit from the recycling endosomes may still be occurring that are below the limit of resolution of the assays used herein, which used either optical assays or Tf-trafficking assays in transfected cells (Figure 8), both of which are limited by sensitivity. Difficulties in obtaining stable cell lines overexpressing these arfophilin-2 deletions or GFP-tagged species have precluded a more detailed analysis of TfR-trafficking thus far. These caveats notwithstanding, the significant change in distribution of TfR upon GFP-arfophilin expression suggests that these molecules are exerting an effect on TfR distribution and/or compartment morphology.
Exactly how arfophilin-2 could regulate the function and/or distribution of the ERC is unclear. One potential explanation could be that arfophilin-2 is involved in motor protein recruitment or the regulation of other microtubule associated proteins. In this regard, Sullivan and colleagues have demonstrated that Nuf interacts physically with the motor protein dynein (Riggs, Rothwell, Mische, Debec, Hickson, Gould, Hays, and Sullivan (unpublished data) that may be involved in the regulation of ERC morphology. The nocodazole sensitivity of GFP-arfophilin-2 localization (Figure 9) clearly demonstrates the microtubule dependency of the compartment that is characteristic of the ERC. The fact that it is reversible implies that minus-end–directed motor protein activity is still functional. It may be that plus-end–directed motor recruitment or function is inhibited by GFP-arfophilin-2/Nuf overexpression. This sits well with the model proposed for Nuf function (Rothwell et al., 1998, 1999). Indeed, it is striking that the localization of GFP-Nuf to the centrosome is dependent upon functional microtubules.
The localization of GFP-arfophilin-2 around the centrosome is interesting. Studies in other cells have shown that the ERC collapses into this region upon BFA treatment (Reaves and Banting, 1992). The significant overlap of GFP-arfophilin-2 with markers for the ERC supports the notion that these arfophilin mutants become stably associated with Rab11-positive ERC membranes and induce their redistribution to the centrosomal area. Because endogenous arfophilin-2 is BFA sensitive (Figure 4), it is conceivable that endogenous arfophilin-2 traffics between a major BFA-sensitive pool in the TGN area and a BFA-resistant recycling endosome pool and that it gets trapped in the latter upon expression of the GFP-tagged full-length or truncated form of arfophilin-2. Such a model could place two distinct GTPases at each location, Rab11 in the ERC and Arf5 in the TGN, which function in a concerted manner to regulate traffic between the two compartments. This hypothesis is also supported by our data showing that the dominant negative Arf5 mutant could effectively displace endogeous arofphilin-2 from the perinuclear area, suggesting that the localization of the molecule in the perinuclear area requires active Arf5 (Figure 5).
Arfophilins and Nuclear Fallout: Homologous Proteins with Similar Functions?
We have also identified a conserved family of proteins in metazoans that includes the arfophilins, hypothetical C. elegans protein F55C12.1a/F55C12.1b and D. melanogaster Nuf. Nuclear fallout is an essential maternal effect gene, whose product is required for cellularization in Drosophila (Rothwell et al., 1998, 1999). The subcellular localization of Nuf changes during the cell cycle. During the late syncytial divisions, it is cytoplasmic but concentrates at the centrosomes during prophase. It is also found at the centrosomes throughout cellularization where it is believed to load vesicles onto microtubules for transport to the furrows. In nuf-mutant embryos, the recruitment of cortical actin is disrupted (Rothwell et al., 1999; Zhang et al., 2000).
The observation that GFP-Nuf overexpression in HeLa cells results in largely the same phenotype as that of GFP-arfophilin-2 overexpression suggests that these proteins are functionally related. This has several important implications. First, it suggests a potential role for the arfophilins in cell division, particularly cytokinesis, because Nuf is required for the formation of cellularization furrows in the Drosophila embryo. Because testis is a tissue undergoing a high degree of cell division, such a role may help to explain the high level of expression observed in this tissue. The findings also implicate the ERC in cytokinesis. This is consistent with recent studies in C. elegans where RNA interference-induced suppression of Rab 11 leads to specific regression of the cleavage furrow at the final stage of abscission (Skop et al., 2001). The significance of the role of Rab 11 should not be overlooked, particularly given the interaction of arfophilins with this GTPase. Arfs have also been implicated in cellularization in Drosophila and cytokinesis in C. elegans (Sisson et al., 2000; Skop et al., 2001). Such observations suggest the hypothesis that arfophilin-2, and by extension the ERC, may be involved in traffic to the midbody during cytokinesis. Moreover, the identification of arfophilin-2 as a Rab 11 and Arf binding protein offers the tantalizing suggestion that this protein may integrate the Rab 11 and Arf signals to membrane traffic during cytokinesis. Such a hypothesis is being actively pursued in the laboratory.
In summary, we describe the functional characterization of a novel member of the arfophilin family, termed arfophilin-2. The arfophilins share significant sequence homology with the Drosophila protein Nuclear Fallout, and our data suggests that Nuf and the arfophilin family are functionally related. Arfophilin-1 and 2 protein, together with Nuf, define a new group of molecules involved in membrane traffic that are capable of integrating signals from both class II Arfs and also from Rab11, an entirely distinct GTPase. The challenge ahead will be to define the relative importance of the GTPase inputs, and to define the precise functional role of these interesting dual GTPase effectors.
Acknowledgments
We thank those colleagues who supplied antibodies and cDNAs for this study; S. Winder, K. Ayscough, and all members of the G.W.G. laboratory for helpful discussions; and C. McInerny for help with Northern blots. G.R.X.H. and A.B.F. thank Diabetes UK and the Wellcome Trust for Ph.D. studentships, and F.A.B. thanks the Wellcome Trust for a Career Development Award. This work was supported by National Institutes of Health grant R01 GM-46409 (to W.S.), grants from American Diabetes Association and Howard Hughes Medical Institute (to R.P.) and by grants 17/C13723 and 17/REI18423 from the Biotechnology and Biological Sciences Research Council (to G.W.G.).
Abbreviations used: Arf, ADP-ribosylation factor; BFA, Brefeldin A; CI-MPR, cation-independent mannose-6-phosphate receptor; EEA1, early endosomal autoantigen-1; ERC, endosomal recycling compartment; GFP, green fluorescent protein; GGA, Golgi localized, gamma adaptin ear homology domain containing Arf-binding protein; Nuf, nuclear fallout; Tf, transferrin; TfR, transferrin receptor; TGN, trans-Golgi network.
References
- Barr, F.A., Puype, M., Vanderkerchove, J., and Warren, G. (1997). GRASP65, a protein involved in the stacking of Golgi cisternae. Cell 91, 253-262. [DOI] [PubMed] [Google Scholar]
- Boehm, M., and Bonifacino, J.S. (2001). Adaptins The final account. Mol. Biol. Cell 12, 2907-2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boman, A.L., and Kahn, R.A. (1995). Arf proteins: the membrane traffic police? Trends Biochem. Sci. 20, 147-150. [DOI] [PubMed] [Google Scholar]
- Boman, A.L., Zhang, C.-L., Zhu, X., and Kahn, R.A. (2000). A family of ADP-ribosylation factor effectors that can alter membrane transport though the trans Golgi. Mol. Biol. Cell 11, 1241-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dell'Angelica, E.C., Puertollano, R., Mullins, C., Aguilar, R.C., Vargas, J.D., Hartnell, E.M., and Bonifacino, J.S. (2000). GGAs: a family of A.D.P. ribosylation factor-binding proteins related to adaptors and associated with the Golgi complex. J. Cell Biol. 149, 81-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson, J.G., and Jackson, C.L. (2000). Regulators and effectors of the ARF GTPases. Curr. Opin. Cell Biol. 12, 475-482. [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey, C., Li, G., Columbo, M.I., and Stahl, P.D. (1995). A regulatory role for Arf6 in receptor-mediated endocytosis. Science 267, 1175-1178. [DOI] [PubMed] [Google Scholar]
- D'Souza-Schorey, C., van Donselaar, E., Hsu, V.W., Yang, C., Stahl, P.D., and Peters, P.J. (1998). ARF6 targets recycling vesicles to the plasma membrane: insights from an ultrastructural investigation. J. Cell Biol. 140, 603-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, B., Zhang, Y., Paupard, M.-C., Lin, S.X., Hall, D., and Hirsh, D. (2001). Evidence that Rme-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nat. Cell Biol. 3, 573-579. [DOI] [PubMed] [Google Scholar]
- Gruenberg, J. (2001). The endocytic pathway: a mosaic of domains. Nat. Rev. Mol. Cell. Biol. 2, 721-730. [DOI] [PubMed] [Google Scholar]
- Hales, C.M., Griner, R., Hobdy-Henderson, K.C., Dorn, M.C., Hardy, D., Kumar, R., Navarre, J., Chan, E.K.L., Lapierre, L.A., and Goldenring, J.R. (2001). Identification and characterisation of a family of Rab11-interacting proteins. J. Biol. Chem. 276, 29067-29075. [DOI] [PubMed] [Google Scholar]
- Hirst, J., Lui, W.W.Y., Bright, N.A., Totty, N., Seaman, M.N.J., and Robinson, M.S. (2000). A family of proteins with γ-adaptin and VHS domains that facilitate trafficking between the trans Golgi reticulum and the vacuole/lysosome. J. Cell Biol. 149, 67-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins, C.R. (1983). Intracellular routing of transferrin receptors in epidermoid carcinoma A431 cells. Cell 35, 321-330. [DOI] [PubMed] [Google Scholar]
- Kornfeld, S. (1992). Structure and function of the mannose-6-phosphate/insulin-like growth factor II receptors. Annu. Rev. Biochem. 61, 307-330. [DOI] [PubMed] [Google Scholar]
- Lapierre, L.A., Kumar, R., Hales, C.M., Navarre, J., Bhatur, S.G., Burnette, J.O., Provance, D.W., Mercer, J.A., Bahler, M., and Goldenring, J.R. (2001). Myosin Vb is associated with plasma membrane recycling systems. Mol. Biol. Cell 12, 1843-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay, A.J., Hendrick, A.G., Cantalupo, G., Senic-Matuglia, F., Goud, B., Bucci, C., and McCaffrey, M.W. (2002). Rab coupling protein (RCP), a novel Rab4 and Rab11 effector protein. J. Biol. Chem. 277, 12190-12199. [DOI] [PubMed] [Google Scholar]
- Millar, C.A., Sherwan, A., Hickson, G.R.X., James, D.E., and Gould, G.W. (1999). Differential regulation of secretory compartments containing the insulin-responsive glucose transporter, GLUT4, in 3T3–L1 adipocytes. Mol. Biol. Cell 10, 3675-3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee, S., and Maxfield, F.R. (2000). Role of membrane organisation and membrane domains in endocytic lipid trafficking. Traffic. 1, 203-211. [DOI] [PubMed] [Google Scholar]
- Nielsen, E., Christoforidis, S., Uttenweiler-Joseph, S., Miaczynska, M., Dewitte, F., Wilm, M., Hoflack, B., and Zerial, M. (2000). Rabenosyn-5, a novel Rab5 effector, is complexed with hVP.S45 and recruited to endosomes through a FYVE finger domain. J. Cell Biol. 151, 601-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman, J.C., Jones, D., Barry, S.T., Holt, M.R., Cockcroft, S., and Critchley, D.R. (1998). ARF1 potentiates paxillin recruitment to focal adhesions and potentiates Rho-stimulated stress fibre formation in intact and permeabilised Swiss 3T3 fibroblasts. J. Cell Biol. 143, 1981-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prekeris, R., Davies, J.M., and Scheller, R.H. (2001). Identification of a novel Rab11/25 binding domain present in Eferin and Rip proteins. J. Biol. Chem. 276, 38966-38970. [DOI] [PubMed] [Google Scholar]
- Reaves, B., and Banting, G. (1992). Perturbation of the morphology of the trans Golgi network following brefeldin A treatment: redistribution of a TGN-specific integral membrane protein, TGN38. J. Cell Biol. 116, 85-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren, M., Xu, G., Zeng, J., De Lemos-Chiarandini, C., Adesnik, M., and Sabatini, D.D. (1998). Hydrolysis of GTP on rab11 is required for the direct delivery of transferrin from the pericentriolar recycling compartment to the cell surface, but not from sorting endosomes. Proc. Natl. Acad. Sci. USA 95, 6187-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell, W.F., Fogarty, P., Field, C.M., and Sullivan, W. (1998). Nuclear-fallout, a Drosophila protein that cycles from the cytoplasm to the centrosomes, regulates cortical microfilament organisation. Development 125, 1295-1303. [DOI] [PubMed] [Google Scholar]
- Rothwell, W. F., Zhang, C. X., Zelano, C., Hsieh, T.-s., and Sullivan, W. 1999. The Drosophila centrosomal protein Nuf1 is required for recruiting Dah, a membrane associated protein, to furrows in the early embryo. J. Cell Sci. 112, 2885-2893. [DOI] [PubMed] [Google Scholar]
- Shin, O.H., Couvillon, A.D., and Exton, J.H. (2001). Arfophilin is a common target of both class II and class III ADP ribosylation factors. Biochemistry. 40, 10846-10852. [DOI] [PubMed] [Google Scholar]
- Shin, O.-H., and Exton, J.H. (2001). Differential binding of arfaptin 2/POR1 to ADP-ribosylation factors and Rac1. Biochem. Biophys. Res. Commun. 285, 1267-1273. [DOI] [PubMed] [Google Scholar]
- Shin, O.-H., Ross, A.H., Mihai, I., and Exton, J.H. (1999). Identification of arfophilin, a target protein for GTP-bound class II ADP-ribosylation factors. J. Biol. Chem. 274, 26609-26615. [DOI] [PubMed] [Google Scholar]
- Sisson, J.C., Field, C., Ventura, R., Royou, A., and Sullivan, W. (2000). Lava lamp, a novel peripheral Golgi protein is required for Drosophila melanogaster cellularisation. J. Cell Biol. 151, 905-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop, A.R., Bergmann, D., Mohler, W.A., and White, J.G. (2001). Completion of cytokinesis in C. elegans requires brefeldin A-sensitive membrane accumulation at the cleavage furrow apex. Curr. Biol. 11, 735-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J., Khachikian, Z., Radhakrishna, H., and Donaldson, J.G. (1998). Localization of endogenous ARF6 to sites of cortical actin rearrangement and involvement of ARF6 in cell spreading. J. Cell Sci. 111, 2257-2267. [DOI] [PubMed] [Google Scholar]
- Sonnishsen, B., De Renzis, S., Neilsen, E., Reitdorf, J., and Zerial, M. (2000). Distinct membrane domains on endosomes in the recycling pathway visualised by multicolour imaging of Rab4, Rab5 and Rab11. J. Cell Biol. 149, 901-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich, O., Reinsche, S., Urbe, S., Zerial, M., and Parton, R.G. (1996). Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 135, 913-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcke, M., Johannes, L., Galli, T., Mayau, V., Goud, B., and Salamero, J. (2000). Rab11 regulates the compartmentalisation of early endosomes required for efficient transport from early endosomes to the trans Golgi network. J. Cell Biol. 151, 1207-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodman, P.G. (2000). Biogenesis of the sorting endosome: the role of Rab5. Traffic 1, 695-701. [DOI] [PubMed] [Google Scholar]
- Zerial, M., and McBride, H. (2001). Rab proteins as membrane organizers. Nat. Rev. Mol. Cell. Biol. 2, 107-117. [DOI] [PubMed] [Google Scholar]
- Zhang, C.X., Rothwell, W.F., Sullivan, W., and Hsieh, T.S. (2000). Discontinuous actin hexagon, a protein essential for cortical furrow formation in Drosophila, is membrane associated and hyperphosphorylated. Mol. Biol. Cell 11, 1011-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]