Abstract
Of the >20 epithelial keratins, keratin 20 (K20) has an unusual distribution and is poorly studied. We began to address K20 function, by expressing human wild-type and Arg80→His (R80H) genomic (18 kb) and cDNA K20 in cells and mice. Arg80 of K20 is conserved in most keratins, and its mutation in epidermal keratins causes several skin diseases. R80H but not wild-type K20 generates disrupted keratin filaments in transfected cells. Transgenic mice that overexpress K20 R80H have collapsed filaments in small intestinal villus regions, when expressed at moderate levels, whereas wild-type K20-overexpressing mice have normal keratin networks. Overexpressed K20 maintains its normal distribution in several tissues, but not in the pancreas and stomach, without causing any tissue abnormalities. Hence, K20 pancreatic and gastric expression is regulated outside the 18-kb region. Cross-breeding of wild-type or R80H K20 mice with mice that overexpress wild-type K18 or K18 that is mutated at the conserved K20 Arg80-equivalent residue show that K20 plays an additive and compensatory role with K18 in maintaining keratin filament organization in the intestine. Our data suggest the presence of unique regulatory domains for pancreatic and gastric K20 expression and support a significant role for K20 in maintaining keratin filaments in intestinal epithelia.
INTRODUCTION
Most mammalian cells contain a complex cytoskeleton composed of three major protein families: actin-containing microfilaments, tubulin-containing microtubules, and intermediate filaments (IF), and their associated proteins (Fuchs and Cleveland, 1998; Ku et al., 1999). The so-called “soft” keratins (i.e., those excluding the “hard' keratins found in epidermal appendages such as hair) make up the IF proteins of epithelial cells. Soft keratins (K) consist of >20 members (K1–20) that include types I (K9-K20) and II (K1-K8) IF proteins. All epithelial cells express one or more type I and type II keratins as noncovalent heteropolymers. Regardless of the number of keratins found in a given epithelial cell, the ratio of type I to type II keratins is 1:1 (reviewed by Moll et al., 1982; Fuchs and Weber, 1994; Herrmann and Aebi, 2000; Coulombe and Omary, 2002). For example, hepatocytes express K8/18 exclusively, whereas intestinal epithelial cells express K7/8 (type II) and K18/19/20 (type I) at variable levels.
Because cell culture models have not been revealing in understanding keratin function, transgenic animal models were generated for several keratins as a handle toward understanding their function. These models provided clear evidence for keratins as serving an essential role in protecting cells from a variety of mechanical and nonmechanical forms of stress, particularly in the skin and liver (Fuchs and Weber, 1994; Magin et al., 2000; Coulombe and Omary, 2002; Omary et al., 2002; Oshima, 2002). The disease phenotypes of these mouse models also led to the identification of several human diseases as keratin-associated diseases (Fuchs and Cleveland, 1998; Omary et al., 2002). For example, transgenic mice (R89C mice) that overexpress human (h) K18 that is mutated at a highly conserved Arg (Arg89→Cys; R89C) develop mild chronic hepatitis and a marked predisposition to drug-induced liver injury, in association with hepatocyte and pancreatic acinar cell keratin filament disruption (Ku et al., 1995, 1996; Toivola et al., 1998, 2000a; Omary et al., 2002). Arg89 of K18 was chosen because it is mutated in K14 and K5 (the equivalent residue is Lys in some type II keratins) in some patients with the severe form of the blistering skin disorder epidermolysis bullosa simplex (Fuchs and Coulombe, 1992). The phenotype of the R89C mice led to the search and subsequent identification of K8/18 mutations in patients with cryptogenic forms of liver disease (Ku et al., 1997, 2001). Dramatic keratin filament collapse in R89C mice was noted in the liver and pancreas but not in the small or large intestine, likely due to the compensatory presence of other type I keratins (K19 and K20) that are either absent in the liver (Ku et al., 1995) or present in small amounts in the pancreas (Toivola et al., 2000b).
We focused on studying K20, which is the most recently identified keratin protein (Moll et al., 1990, 1993; Quaroni et al., 1991; Calnek and Quaroni, 1993). Human K20 has several unique properties, including its relatively greater sequence divergence compared with other type I keratins, with only 58% identical amino acids in the conserved α-helical “rod” domain with the closest keratin, K14; unique distribution that is nearly confined to the gastric and intestinal epithelium, urothelium, bladder, and Merkel cells; expression in the more differentiated enterocytes along the crypt-villus differentiation axis; and lack of immunological cross-reaction of most anti-keratin antibodies (Moll et al., 1993). Human K20 has 79% amino acid identity with rat K20 (Chandler et al., 1993). Although nothing is known regarding K20 function, it has been used as a marker for the histological classification of tumors (Moll et al., 1992; Harnden et al., 1999; Wildi et al., 1999; Leech et al., 2001). The observation that K20 is coexpressed in the same intestinal cell type as other type I keratins, including K18 and K19 suggest that these three type I keratins may have redundant or complementary functions.
In this study, we generated transgenic mice that overexpress wild-type (WT) and Arg→His (R80H) K20 (corresponding to the conserved Arg89 in hK18) to begin addressing K20 function in digestive epithelia. We focused on the highly conserved Arg because it has provided a useful target in studying K18 (Ku et al., 1995, 1996).
MATERIALS AND METHODS
Cell Culture and Antibodies
NIH-3T3 (mouse fibroblast), HT29 (human colon), and BHK-21 (baby hamster kidney) cells (American Type Culture Collection, Manassas, VA) were cultured as recommended by the supplier. The antibodies used include mouse monoclonal antibody (mAb) L2A1 (Chou et al., 1993), which recognizes human K18 without cross-reacting with mouse keratins; rat mAbs Troma-I (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), Troma-II and -III [kindly provided by Drs Helene Baribault at Tularik (South San Francisco, CA) and Thomas Magin at the University of Bonn (Bonn, Germany), respectively], which recognize mouse K8, K18, at Tularik (South San Fracnisco, CA) and K19, respectively; rabbit antibody 4668, which is an anti-K18 peptide antibody (raised against the peptide RPVSSAASVYAGA; G; Ku et al., 1998) that recognizes human and mouse K18; mouse mAb LF6, which recognizes human K8 and is generated by immunizing mice with high salt extracts of human K8/18 isolated from HT29 cells (our unpublished observations); mouse anti-K20 mAb (clone Ks20.8), which recognizes mouse and human K20 and anti-chromogranin A mAb (NeoMarkers, Fremont, CA); mouse anti-K20 mAb (clone ITKs20.10), which recognizes human but not mouse K20 and mouse anti-K7 mAb (RCK105), which recognizes mouse and human K7 (Progen; Heidelberg, Germany); and rabbit antibody 8592 (Ku et al., 1995), which recognizes human and mouse K8/18.
Transgene Construct and Generation of Transgenic Lines
The human K20 genomic DNA (18 kb) and cDNA (1.35 kb) were mutated at codon CGT→CAT to convert Arg80→His. The mutation was first introduced into K20 cDNA by using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) and was confirmed by DNA sequencing. For the 18-kb genomic DNA, an XhoI fragment (15 kb) was first subcloned in pBluescript SK+ and then an internal BstI fragment (8 kb) was cut out to generate a workable DNA fragment (10 kb, including the vector) to introduce the mutation. After mutation, the DNA was religated to regenerate the 18-kb gene. Mutant and WT K20 genes were injected into pronuclei of fertilized FVB/n eggs. Four heterozygous K20 transgenic lines that manifest germline expression of WT (WT1 and WT2) and K20 R80H (M1 and M2) K20 were selected. The transgene copy numbers were estimated using tail vein DNA of test animals and a densitometric standard curve generated using a plasmid that expresses genomic human K20, as described previously (Ku et al., 1995). Presence of the transgene was verified by polymerase chain reaction screening with hK20-specific primers that generated a 550-base pair fragment corresponding to the first intron of the K20 gene: (+) 5′CCGGAATAAAGTTCATCCTCTG-3′ and (-) 5′-GACATTTCTTTTAGCCCCTGTG-3′. The PCR conditions were 94°C for 2 min and then 35 cycles at 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min. The WT1, WT2, M1, and M2 heterozygous K20-overexpressing lines were also cross-bred with homozygous transgenic mice that overexpress WT hK18 (TG2 mice) or hK18 R89C mice (F22 and F30 lines) that were previously described in detail (Ku et al., 1995, 1996; Toivola et al., 1998, 2000a).
Sequencing of Mouse K20 and Generation of Antibodies
A database search (www.ncbi.nlm.nih.gov), by using the human K20 cDNA sequence, indicated the presence of an expressed sequence tag clone that likely represents mouse K20. This clone was purchased (American Type Culture Collection), sequenced, and identified as a full-length mouse K20 cDNA. The sequence was deposited in GenBank as “MuK20” no. AF473907. The sequence of mouse K20, in alignment with rat and human K20, is shown in Table 1. MuK20 was subcloned into a PET23A(+) bacterial expression system, followed by partial purification of the mouse K20 protein by using HiTrap Q and SP columns (Amersham Biosciences AB, Uppsala, Sweden). The partially purified protein was used to immunize BALB/c mice followed by generation of mAbs by using standard procedures. One clone that recognized mouse but not human K20, termed XQ1, was further subcloned and used for immunoblotting.
Table 1.
Amino acid alignment of human, rat, and mouse K20 sequences
| Human | MDFSRRSFHRSLSSSLQAPVVSTVG-------MQRLGTTPSVYGGAGG |
| Rat | MDFSRRSFHRSLSSSSQGPALSTSGSLYRKGTMQRLGLH-SVYGG-WR |
| Mouse | MDFSRQSFHRSLSSSSQGPALSMSGSLYRKGTVQRLGAAPSVYGGAGG |
| ***** ********* * * * * **** ***** | |
| Human | RGIRISNSRHTVNYGSDLTGGGDLFVGNEKMAMQNLNDRLASYLEKVR |
| Rat | HGTRISVSKTTMSYGNHLSNGGDLFGGNEKLAMQNLNDRLASYLEKVR |
| Mouse | HGTHISVSKAVMSYGGDLSNGSDLFGGNGKLAMHNLNDRLANYLEKVR |
| * ** * ** * * *** ** * ** ******* ****** | |
| Human | TLEQSNSKLEVQIKQWYETNRPRAGRDYSAYYRQIEELRSQIKDAQLQ |
| Rat | SLEQSNSKLEAQIKQWYETNAPSTIRDYSSYYAQIKELQDQIKDAQIE |
| Mouse | SLEQSNCRLEAQIKQRYETNAPSTIRDYSSYYAQIKELQNQVKDAQVQ |
| ***** ** **** **** * **** ** ** ** * **** | |
| Human | NARCVLQIDNAKLAAEDFRLKYETERGIRLTVEADLQGLNKVFDDLTL |
| Rat | NARCVLQIDNAKLAAEDFRLKFETERGMRITVEADLQGLSKVYDDLTL |
| Mouse | NAQCVLRIDNAKLAAEDFRLKFETERGMRIAVEADLQGLSKVYDNLTL |
| ** *** ************** ***** * ******** ** * *** | |
| Human | HKTDLEIQIEELNKDLALLKKEHQEEVDGLHKHLGNTVNVEVDAAPGLN |
| Rat | QKTDLEIQIEELNKDLALLKKEHQEEVEVLRRQLGNNVNVEVDAAPGLN |
| Mouse | QKTDLEIQIEELNKDLALLKKEHQEEVEVLRRQLGNNVNVEVDAAPGLN |
| ************************** * *** ************ | |
| Human | LGVIMNEMRQKYEVMAQKNLQEAKEQFERQTAVLQQQVTVNTEELKGTE |
| Rat | LGEIMNEMRQKYEILAQKNLQEAKEQFERQTQTLEKQVTVNIEELRGTE |
| Mouse | LGEIMNEMRQRYEVLAQKNLQEAKEQFERQSQTLQQQVTVNTEELKGFE |
| ** ******* ** *************** * ***** *** * * | |
| Human | VQLTELRRTSQSLEIELQSHLSMKESLEHTLEETKARYSSQLANLQSLL |
| Rat | VQVTELRRSYQTLEIELQSQLSMKESLERTLEETKARYASQLAAIQEML |
| Mouse | VQVTELRRTYQNLEIELQSHLSMKESLERNLEDVKARYASQLAAIQEML |
| ** ***** * ******* ******** ** **** **** * * | |
| Human | SSLEAQLMQIRSNMERQNNEYHILLDIKTRLEQEIATYRRLLEGEDVKT |
| Rat | SSLEAQLMQIRSDTERQNQEYNILLDIKTRLEQEIATYRRLLEGEDIKT |
| Mouse | SSLEAQLMQIRSDTERQNQEHNILLDIKTRLEQEIATYRRLLEGEDIKT |
| ************ **** * ************************ ** | |
| Human | TEYQLSTLEERDIKKTRKIKTVVQEVVDGKVVSSEVKEVEENI |
| Rat | TEYQLNTLEAKDIKKTRKIKTVVEEVVDGKVVSSEVKEIEENI |
| Mouse | TEYQLSTLEMKDIKKTRKIKTVVEEVVDGKVVSSEVKEIEESV |
| ***** *** ************ ************** ** |
The mouse sequence is presented as part of this study. The human and rat sequences were described previously (Moll et al., 1990; Chandler and Quaroni, 1993). * indicates identical amino acids. The percentage of identities at the amino acid level among the K20 sequences are human and rat, 80%; human and mouse, 78%; rat and mouse, 89%
Transfection and Immunofluorescence Staining
The genomic and cDNA constructs of hK20 were cotransfected with hK8 cDNA, by using LipofectAMINE (Invitrogen, Carlsbad, CA), into BHK-21 cells (to allow relatively high yield biochemical extraction of keratins) or NIH-3T3 cells (for cell staining). NIH-3T3 cells were cultured on chamber slides for 2–3 d after transfection and then fixed in methanol (3 min, -20°C). Fixed cells were “blocked” for 15 min with phosphate-buffered saline (PBS) containing 2% bovine serum albumin (buffer B) and then coincubated with anti-K20 mAb in buffer B (30 min). After washing three times with PBS, cells were blocked with buffer B containing 2% normal goat serum (15 min) followed by incubating with Texas Red-conjugated goat anti-mouse antibody for 20 min and then washing with PBS.
Mouse tissues were frozen in optimum cutting temperature compound, sectioned and fixed in acetone (-20°C, 10 min) and then stained with mouse or rabbit anti-keratin antibodies as described above. For double staining, mouse and rabbit primary antibodies were used, followed by Texas Red and fluorescein isothiocyanate-conjugated secondary antibodies. Images of single confocal sections of stained cells and tissues were obtained with a Nikon TE300 microscope coupled to a Bio-Rad MRC1024ES confocal microscope.
Keratin Isolation, Western and Northern Blotting
High salt extraction (HSE) was used to isolate keratins from cultured transfected cells or from tissues of transgenic and nontransgenic mice (Ku et al., 1995). SDS-PAGE was done using 8 or 10% acrylamide gels. Immunoblotting was carried out after transferring HSE to polyvinylidene difluoride membranes followed by blocking with 5% nonfat dry milk and then incubating with anti-keratin antibodies, washing, incubating with peroxidase-conjugated goat anti-mouse, anti-rabbit, or anti-rat immunoglobulins. Immune-reactive bands were visualized using enhanced chemiluminescence (PerkinElmer Life Sciences, Boston, MA).
Northern blot analysis was done with total tissue RNA isolated from the small intestine and colon of various mouse lines by using an RNeasy Midi kit (QIAGEN, Chatsworth, CA). Total RNA (20 μg/lane) was loaded then separated using 1% denaturing formaldehyde gels and transferred to nylon membranes. Blots were UV cross-linked, prehybridized for 2 h, and then hybridized with 32P-labeled cDNA probes for hK20, mouse K20, and mouse glyceraldehyde-3-phosphate dehydrogenase (overnight, 68°C). The blots were then washed and visualized by autoradiography.
RESULTS
Expression of K7, K8, K18, K19, and K20 in Normal Mouse Small Intestine and Colon
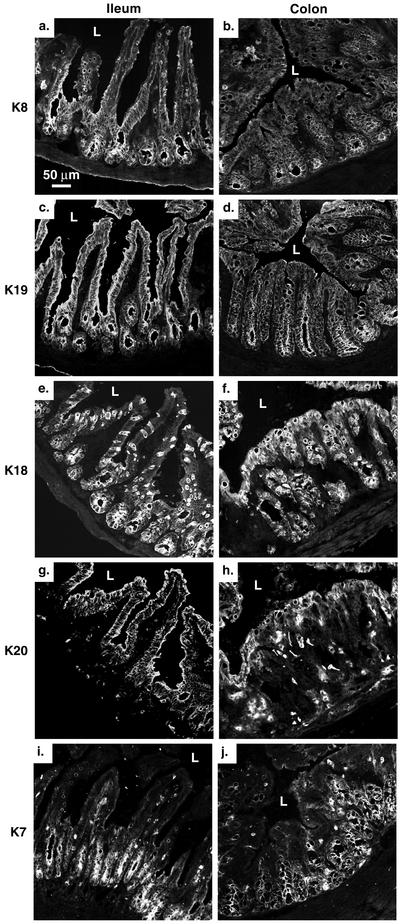
Because our emphasis was to study K20 in the small intestine and colon of K20-overexpressing transgenic mice, we first used immunofluorescence staining to examine the expression of the individual K7, K8, K18, K19, and K20 proteins in nontransgenic FVB/n mice (Figure 1). K8 and K19 are nearly evenly distributed throughout the small intestinal and colonic epithelium (Figure 1, a–d). K7 is preferentially found at the crypt base of the small intestine and colon (Figure 1, i and j) K18 is distributed uniformly throughout the tubular crypt axis in the colon, whereas in the small intestine it is preferentially located in the lower half of the crypt-villus axis in glandular enterocytes with scattered goblet cell staining within the upper one-half of the villus (Figure 1, e and f). In contrast, K20 expression in the small intestine is essentially absent from the crypt base but is found in the more differentiated suprabasal regions. In the colon, K20 is found mainly in the upper surface aspect and in scattered cells (some with extended processes) throughout the remaining epithelium (Figure 1, g and h). These process-associated cells are likely enteroendocrine-related cells as determined by costaining with anti-chromogranin A antibody (our unpublished data). In quantitative terms, K8 and K19 are the major keratins of mouse small intestine, whereas K7, K18, and K20 are far less abundant (Figure 2A, lane 3). However, in the colon, K18 and K19 are nearly equally abundant, whereas K7 and K20 expression is limited (our unpublished data, but see Figure 3 for summary).
Figure 1.
Immunofluorescence staining of K7, K8, K18, K19, and K20 in the ileum and colon of nontransgenic FVB/n mice: The ileum and colon of FVB/n mice were stained using Troma-I (anti-mouse K8; a and b), Troma-III (anti-mouse K19; c and d), Troma-II (anti-mouse K18; e and f), Ks20.8 (anti-mouse/human K20; g and h), and anti-K7 (i and j). L, lumen.
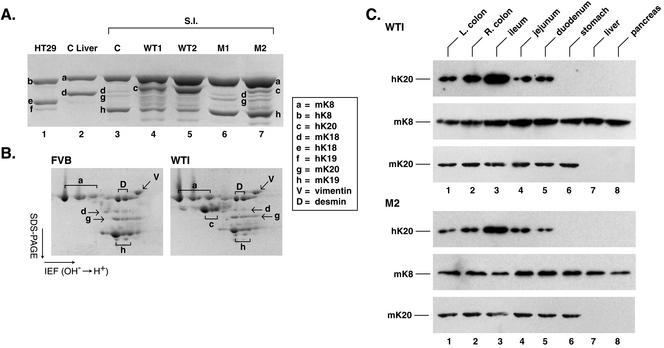
Figure 2.
Biochemical characterization of keratin expression in transgenic lines that overexpress K20. (A) HSE of the small intestines that were used in Figure 5 were separated by SDS-PAGE followed by Coomassie staining. Also shown are HSE obtained from nontransgenic mouse control “C” liver (which expresses only K8 and K18) and from human HT29 cells, which express high levels of K8/18, limited amount of K19, and barely detectable levels of K20 (our unpublished data). (B) HSE of the small intestine from nontransgenic (FVB/n) and WT1 mice were analyzed by isoelectric focusing in the horizontal dimension followed by SDS-PAGE in the vertical dimension. Letters highlight the various IF proteins (key corresponds to A and B), which was confirmed by immunoblotting (our unpublished data). (C) HSE were prepared from the various indicated tissues of the WT1 and M2 transgenic mice followed by blotting with mAb ITKs20.10 for hK20, Troma-I for mK8, and XQ1 for mK20. The analyzed tissues were left (L) colon, right (R) colon, small intestine (ileum, jejunum, or duodenum), stomach, liver, and pancreas.
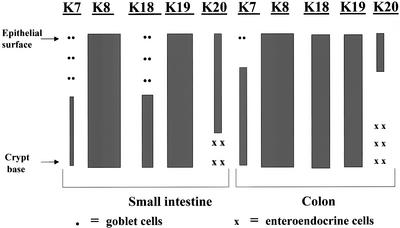
Figure 3.
Schematic of keratin distribution in epithelial cells of the small and large intestine. The qualitative distribution and expression levels of keratins is shown. The major type II enterocyte keratin is K8, with low levels of K7 that are present in goblet cells and the crypt base. Three type I keratins (K18, K19, and K20) are found in enterocytes. K18 and K19 are distributed uniformly throughout the villus-crypt axis in the colon, while K18 is preferentially located in the lower half of this axis with scattered K18-containing goblet cells suprabasally. Quantitatively, K8 and K19 are the major keratins of the small intestine, while K18 and K20 are far less abundant. In the colon, K18 and K19 levels are similar. Strongly positive-staining K20 cells, which are basally located, likely represent enteroendocrine cells.
Effect of the K20 Arg80→His (R80H) Mutation on Keratin Filament Assembly in Transfected Cells
We targeted Arg80 of K20 for mutation, given the conserved nature of this arginine among IF proteins and the significant impact of its mutation in causing several skin diseases involving epidermal keratins (Fuchs and Coulombe, 1992). Furthermore, mutation of the equivalent residue in K18 (Arg89→Cys) and expression in transfected cells, or in mice as a transgene, results in keratin filament collapse (Ku et al., 1995). We generated a K20 Arg80→His mutation, and chose His because an Arg-to-His mutation entailed introducing a single nucleotide substitution (CGT→CAT) and because the equivalent K14 residue has a similar mutation (Arg125→His) in patients with epidermolysis bullosa simplex (Coulombe et al., 1991).
We used cDNA (c) and genomic (g) human K20 constructs to generate the K20 R80H mutation (Figure 4). Transfection of various combinations of cK8/cK18, cK8/cK20, or cK8/gK20 (WT or R80H K20) into baby hamster kidney (BHK) cells showed that the transfected proteins can be detected by immunoblotting of HSE of the transfected cell and confirmed the predicted reactivity of the keratin-specific antibodies (Figure 4A). Immunofluorescence staining of NIH-3T3 cells cotransfected with WT K8 and either WT or R80H K20 (genomic and cDNA) showed that WT K20 generates filaments, whereas R80H K20 (both genomic and cDNA) result in collapse of the keratin filaments into distinct dots (Figure 4B). Thus, the R80H residue of K20 is critical for keratin filament organization in cultured cells.
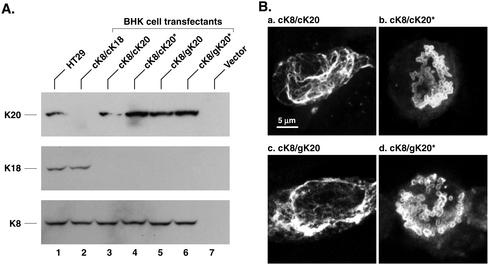
Figure 4.
Analysis of WT and R80H hK20 in transfected cells: (A) BHK cells were transfected with vector alone or cotransfected with WT K8 cDNA (cK8) and one of the following constructs: WT K18 cDNA (cK18), WT cK20, R80H cK20 (cK20*), genomic WT K20 (gK20), and R80H gK20 (gK20*). HSE of nontransfected HT29 cells and the transfected BHK cells was carried out followed by immunoblotting with antibodies to hK20, hK18, and hK8. (B) NIH-3T3 cells were cotransfected with cK8/cK20, cK8/cK20*, cK8/gK20, and cK8/gK20* followed by immunofluorescence staining with anti-K20 antibody. Note the collapse of keratin filaments into prominent dots in cells that are transfected with genomic or cDNA K20 R80H in b and d.
Characterization of Transgenic Mice that Overexpress Human WT and R80H K20
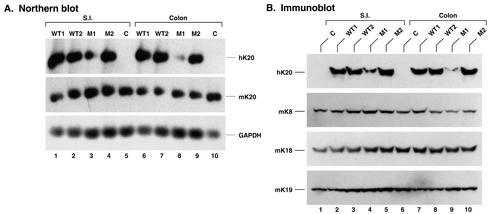
We generated four K20 transgenic mouse lines, two that overexpress WT hK20 (WT1 and WT2) and two that overexpress hK20 R80H (M1 and M2). The transgene copy number was 30 and 35 for WT1 and WT2, and 2 and 20 for M1 and M2, respectively. hK20 mRNA (Figure 5A) and protein (Figure 5B) expression in the small intestine and colon of the transgenic lines were confirmed. As expected, hK20 was not detected in control nontransgenic mice (Figure 5A, lanes 5 and 10; B, lanes 1 and 6), whereas endogenous mouse keratins were detected in all the mouse lines (Figure 5, A and B). The hK20 protein is present at low levels in the small intestine and colon of M1 mice compared with the other transgenic mouse lines (Figure 5B), in a manner that parallels the transgene copy number.
Figure 5.
Expression of WT and R80H hK20 in transgenic mice: (A) Total RNA was extracted from the small intestine (S.I.) and colon of the indicated transgenic lines that overexpress WT hK20 (WT1/WT2) or R80H hK20 (M1/M2) and from nontransgenic mice “C” as control. After transferring (20 μg of total RNA/lane) to nylon membranes, the blots were hybridized with 32P-labeled human cDNA K20, mouse cDNA K20, and mouse glyceraldehyde-3-phosphate dehydrogenase cDNA probes. (B) Immunoblotting by using antibodies to the indicated keratins was performed on HSE of the S.I. and colon of the indicated mouse lines. The blots to mouse K8/18/19 are included as internal controls.
We used high salt extraction, which provides a near quantitative recovery of the total keratin pool, to assess the relative content of individual keratins in the small intestine of the various mouse lines. As shown in Figure 2A, the K20 transgene protein expression levels were WT1 ≅ WT2 > M2 >> M1, which parallel the K20 transgene copy number (hK20 expression in the M1 line is difficult to discern by Coomassie staining). Notably, K20 overexpression results in down-regulation of K19 steady-state protein levels (Figure 2A, compare lane 3 with lane 4 or 5). The specific bands (for one-dimensional gels, Figure 2A) and spots (for two-dimensional gels, Figure 2B) were assigned to individual keratins by using two-dimensional gel analysis (Figure 2B) and immune blotting of the one- and two-dimensional gels with IF-specific antibodies (our unpublished data).
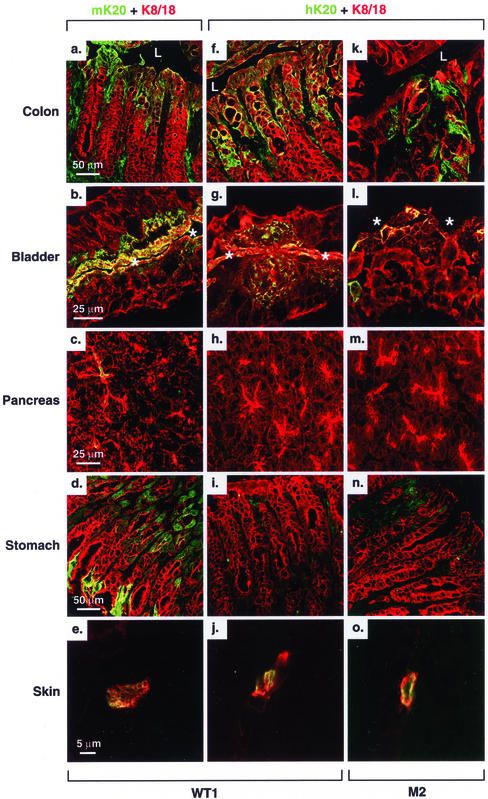
We then compared the tissue distribution of hK20 with endogenous mK20 by blotting with antibody's specific to mouse and human K20. Due to unavailability of antibodies that adequately recognize mouse K20 by immunoblotting, we generated a mAb (termed XQ1) that recognizes mouse but not human K20 (see MATERIALS AND METHODS). For this, we identified and sequenced a cDNA clone that corresponds to mK20 (Table 1). As shown in Figure 2C, hK20 maintained its expression in various segments of the small intestine (ileum, jejunum, and duodenum) and colon [right (ascending) and left (descending) colon]. The K20 transgene product also maintained its tissue/cell expression in the bladder (Figure 6, g and l) and in the scarcely found Merkel cells of the skin (Figure 6, j and o). The K20 transgene product was absent from the liver, as expected (Figure 2C, lane 7; confirmed by immune staining; our unpublished data). However, hK20 was also absent from mouse stomach and pancreas in contrast to its endogenous mouse K20 counterpart (Figures 2C for stomach and 6 for stomach and pancreas; pancreatic mK20 levels are too low to detect by Western blotting). This finding suggests that the regulatory elements that direct gastric and pancreatic expression of mK20 are likely to be outside of the 18-kb hK20 genomic construct that we used and/or that mK20 transcriptional machinery does not recognize the corresponding human sequences.
Figure 6.
Expression of mouse and human K20 in WT1 and M2 transgenic mouse organs. Tissues from colon, bladder, pancreas, stomach, and ear skin were fresh frozen, sectioned, and then double stained with the indicated antibodies (Ks20.8 for mK20 and ITKs20.10 for hK20, green; and 8592 for K8/18, red) as described in MATERIALS AND METHODS. Yellow-colored areas represent colocalization of K8/18 with mouse (a–e) or human (f–o) K20. Ks20.8 was used for staining because it recognizes mouse K20 by staining better than XQ1 (the latter provides a better signal for blotting purposes as used in Figure 2). Lumen, L (a, f, and k) or * (b, g, and l).
Effect of the R80H K20 Mutation on Keratin Filament Organization in Tissues
We examined keratin filament organization in tissues that overexpressed the R80H K20 mutant compared with similar tissues that overexpressed WT hK20. Mutant and WT hK20 integrate into the existing endogenous mouse keratin filament networks, which is detectable in all the tissues examined (Figure 6). There was some mosaicism in hK20 expression, particularly in the colon, in that not all epithelial tip cells uniformly expressed K20 (our unpublished data). This mosaic expression phenomenon was observed in other keratin-overexpressing transgenic mouse models such as those that overexpress mK19 (Bader and Franke, 1990). Tissues expressing WT K20 in the WT1 and WT2 lines had normal-looking filaments in all cases tested (Figure 7, a, b, e, f, i, and j; our unpublished data). However, in M2 mice, the keratin filaments collapsed into cytoplasmic dots and thick cables as noted in the ileum (Figure 7, g and h) and bladder (our unpublished data). Filament disruption was not seen in the ileum (Figure 7, k and l) or bladder (our unpublished data) of M1 mice likely due to lower hK20 expression level in this line, which suggests that the K20 R80H mutation behaves in a dominant-negative manner when overexpressed at moderate levels. Combined with the cell transfection experiments of Figure 4, our results show that the R80H mutation causes keratin filament disruption in vitro and in vivo. There was no obvious effect on tissue histology after testing all organs that expressed WT or R80H hK20 in WT1/2 and M1/2 transgenic mice (our unpublished data).
Figure 7.
Keratin filament organization of WT and R80H transgenic mice. Ileums of WT1 (a, b, e, and f), M2 (c, d, g, and h), WT2 (i and j), and M1 (k and l) were double stained with anti-hK20 and anti-mouse K8/18. e–h are high magnifications of a–d. Areas in squares show either normal filament (e and f) or dots (g and h). L, lumen.
Transgenic Mouse Cross-Breeding Demonstrates Reversible Rescue or Collapse of Keratin Filaments in the Small Intestine, Depending on the Dosage of WT versus Mutant K18 and K20
We tested the functional redundancy of K18 and K20 at the level of filament organization by breeding mice that overexpress WT hK18 (TG2 line) or that overexpress hK18 R89C (F22 and F30 lines), with the K20 mouse lines WT2 (which is similar to WT1) or M1 (which overexpresses low levels of K20). We chose for the initial breeding the M1 rather than the M2 line because M1 mice have normal-looking keratin filaments in the small intestine. As reported previously (Ku et al., 1995), WT K18-overexpressing mice (TG2) have normal-looking keratin filaments throughout their intestine (our unpublished data), whereas F22 (Figure 8A, e) and F30 (our unpublished data) have normal keratin filaments in the majority of epithelial cells but are found collapsed into cytoplasmic dots and thick cables in goblet cell of the small intestine (Figure 8A,e), likely related to the strong K18 staining in these cells (Figure 1e). Interestingly, keratin filament collapse occurs only in the F30 × M1 but not in the F22 × M1 ileums, nor in the ileums (or colons; our unpublished data) of the TG2 × WT2 or F30 × WT2 cross-breeds (Figure 9A). Analysis of the individual keratins in the small intestine of these cross-bred mice was also carried out. In TG2 and F30 ileums, the relative K18 levels were h > m and h ≅ m, respectively (Figure 9B, lanes 1 and 2). These levels were not significantly affected after cross-breeding with WT1 or M1 mice (Figure 9B, lanes 3–6). In the F30 × M1 mouse intestine, keratin filament disruption (Figure 9A) is likely caused by the addition of a very small amount of K20 that is barely perceptible by Coomassie staining.
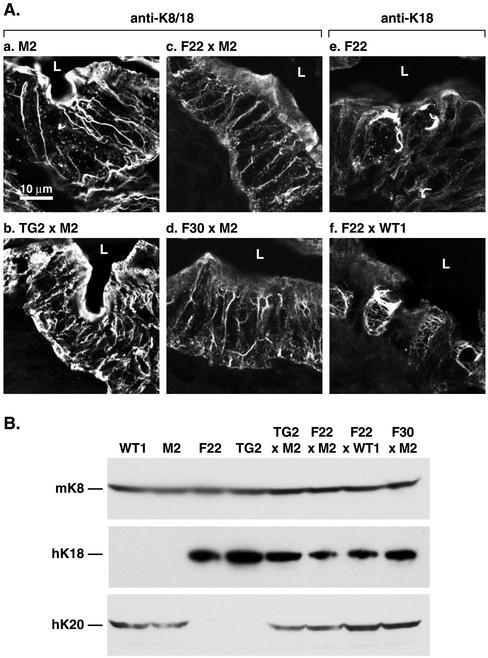
Figure 8.
Analysis of filament rescue in cross-bred K20- (WT1 and M2) and K18 (TG2, F22, and F30)-overexpressing transgenic mice. (A) Transgenic mice that overexpress WT (TG2) or R89C (F22 and F30) K18 were bred with mice that overexpress WT K20 (WT1) or R80H K20 (M2). Staining was done using anti-K8/18 or anti-K18 antibodies. (B) HSE of the small intestine were prepared from the indicated transgenic lines. Extracts were then subjected to Western blotting by using antibodies to mK8, hK18, and hK20.
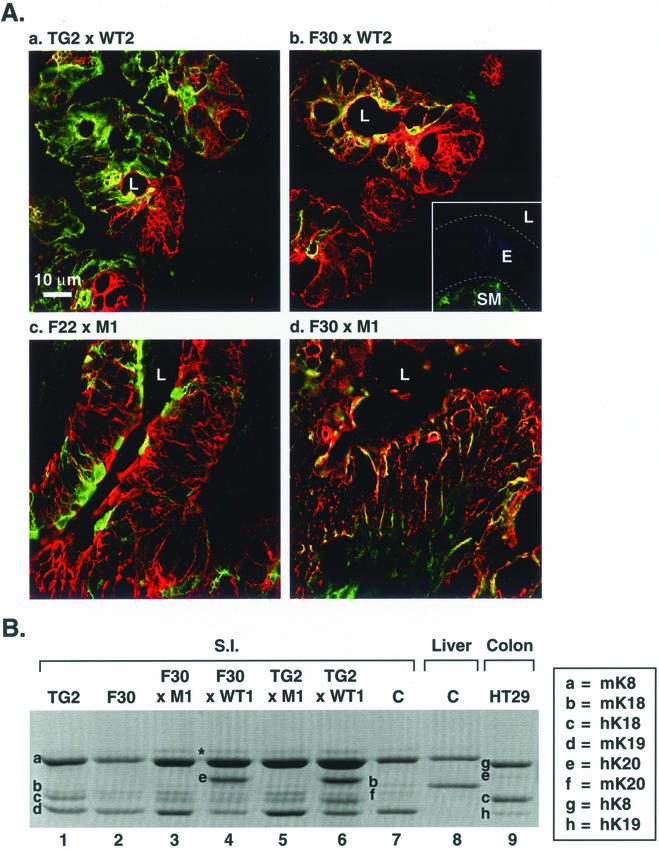
Figure 9.
(facing page). Analysis of filament disruption in cross-bred K20- (WT2 and M1) and K18 (TG2, F22, and F30)-overexpressing transgenic mice. (A) Transgenic mice that overexpress wild-type hK18 (TG2) were bred with WT2 and M1 mice. Also, mice that overexpress hK18 R89 (R89C) (F22 and F30) were bred with the K20 WT2 and M1 lines that overexpress wild-type or R80H K20, respectively. Ileums from the indicated cross-bred lines were then double stained with anti-K8/18 (red) and anti-hK20 (green) antibody. Note that filament collapse and formation of cable-like structures occurs only in the F30 × M1 ileum but not in the ileum of the other cross-breeds. Secondary antibody control (in the absence of primary antibody) is shown as an inset in b (L, lumen; E, epithelium; SM, smooth muscle). (B) HSE of small intestine or liver were obtained from the indicated transgenic and nontransgenic “C” mice or from human colonic HT29 cells, followed by SDS-PAGE, and then Coomassie Blue staining. The band indicated by an asterisk is seen occasionally and likely corresponds to vimentin.
The above-mentioned analysis was extended by mating M2 K20 mice with F22 or F30 mice to test for an additive K18 + K20 mutational effect and by TG2 × M2 and F22 × WT1 matings to test for rescue of the mutant K18 R89C or K20 R80H by wild-type K20 or K18 overexpression, respectively. When hK18 WT (TG2) and R89C (F22 or F30) mice are mated with M2 mice, the filament network in small intestinal epithelia of TG2 × M2 mice nearly normalize compared with M2 mice (compare Figure 8A, a and b). This indicates that wild-type hK18 compensates for mutant hK20 in filament organization and thus rescues the M2 phenotype. Similarly, the goblet cell phenotype of F22 × WT1 mice (Figure 8A, e) is also rescued when wild-type hK20 is overexpressed (Figure 8A, f). The filament disruption noted in F22 × M2 is similar to that seen in F30 × M2 (Figure 8A, c and d). Confirmation of the expression of hK18 and hK20 proteins in the various intercrosses was demonstrated by keratin-specific immune blotting as shown in Figure 8B. A schematic summary of the filament organization phenotype of these interbreedings is shown in Figure 10. Together, our results indicate that K20 and K18 are functionally redundant at the level of keratin filament assembly in the intestine.
Figure 10.
Effect of WT or mutant K18, and/or WT or mutant K20, overexpression on keratin filament organization in transgenic mouse small intestine. The schematic shows the impact on keratin filament organization upon overexpressing WT or R80H K20 (x-axis), or WT or R89C K18 (y-axis), compared with control nontransgenic FVB/n mice (shown in A, which also includes codes for the cell types shown in B–I). Keratin organization in epithelial cells of the small intestine is illustrated by the two typical patterns: 1) normal-looking filaments; and 2) a mixture of dots, cables, and short filaments.
DISCUSSION
Keratin Topography in the Small and Large Intestine
The findings described herein support and extend previous description of keratin distribution in mouse small and large intestine (Franke et al., 1979; O'Guin et al., 1987; Takemura et al., 1988), which is schematically summarized in Figure 3. The major points that are likely to be biologically relevant are: 1) K18 and K20 are differentiation-specific keratins in the small intestine. For example, K20 is primarily expressed in the more differentiated villus cells, whereas K18 is nearly absent in many of these cells (except for goblet cells) and is preferentially found in basal and midzonal regions of the epithelium. 2) K18 and K20 show significant differences in their distribution patterns in the small and large intestine. In that regard, K20 is restricted to a more pronounced epithelial tip distribution in the colon, whereas K18 is more abundant and less restricted in its colonic distribution, compared with the small intestine. In contrast, K8 and K19 are found nearly uniformly throughout the epithelium of the small intestine and colon. K20 is more abundant in the small versus the large intestine, which provides a likely explanation for the dominant negative effect of the R80H mutation in the ileum but not in the colon. 3) There seems to be mouse and human species differences in K20 expression. For example, K20 expression in mouse small intestine is most prominent in the ileum and, even so, is still relatively minor compared with K19. In contrast, K20 is a major keratin in human small intestine, particularly in the duodenum (Moll et al., 1990).
Targeting the Conserved Arginine of IF Proteins to Study Keratin and Other IF Protein Biology
In this study, Arg80 of K20 was targeted for mutation in a similar manner to the previous targeting of the same conserved K18 arginine (Arg89), which is located within the N-terminal aspect of helix 1A of the rod domain. This arginine is conserved in most IF proteins, including lamins, and across species, including snail IF proteins (Fuchs and Coulombe, 1992). Mutation of this conserved arginine occurs in several epidermal keratins and results in a severe phenotype (Coulombe et al., 1991; Fuchs and Weber, 1994). Its targeting to generate an animal model to study keratin function and potential disease association was first described in transgenic mice that overexpressed K18 R89C. Mutation of the conserved arginine causes keratin filament disassembly in cultured cells that express K18 (Ku et al., 1995) or K20 (Figure 4) and in tissues of transgenic mice that express K18 (Ku et al., 1995) or K20 (Figure 7). Collectively, the findings in this study that defined an in vivo role for K20 in keratin filament organization and the findings of the multiple studies that examined overexpression of K18 R89C in transgenic mice (reviewed by Omary et al., 2002) predict that the approach of overexpressing IF proteins with mutations that target this conserved arginine are likely to be fruitful.
Phenotype of Transgenic Mice That Overexpress Human K20 R80H
The major phenotype of the R80H K20 transgenic mice is a filament organization defect in enterocytes of the small intestine, which parallels the expression level of the mutant K20 protein. This defect occurs in the ileum of M2 but not M1 mice that, in turn, express lower K20 levels, and does not occur in the colon of M2 mice likely due to the higher relative level of K18 and the lower relative level of K20 in colonocytes. Disruption of keratin filaments did not result in a histological or disease-like phenotype based on several tests that we carried out. For example, the stool weight in WT1 and M2 mice was similar and both mouse strains were equally susceptible to rotavirus-induced infection and had similar virus shedding profiles (our unpublished data). The dichotomy between keratin filament disruption and the presence or lack of a physiological effect on the involved cell/tissue under basal or stress conditions seems to depend on the affected cell/tissue type. For example, mice that express K18 R89C have disrupted keratin filaments in hepatocytes and pancreatic acinar cells. Despite similar keratin filament disruption in both cell types, the liver becomes highly susceptible to injury, whereas the pancreas remains relatively resilient (Toivola et al., 2000a). Lack of an obvious physiological/histologic phenotype in M2 (or F30 × M2) mice may relate to: 1) the observation that keratin filament disruption did not involve the entire epithelium, 2) the presence of gene modifiers, or 3) the possibility that a given keratin may serve different functions depending on the cell/tissue location of its expression.
The 18-kb genomic construct we used to generate the WT1/2 and M1/2 transgenic lines faithfully maintained K20 expression in the small and large intestine, bladder, and Merkel cells. Regional distribution of the K20 transgene was also maintained (i.e., surface versus base of the epithelium). Two organs that did not maintain hK20 expression were the stomach and pancreas. Expression of K20 in gastric epithelia is well established (Moll et al., 1990), whereas expression in the pancreas depends on the species and the experimental conditions. For example, several studies report the presence of K20 in rat ductal cells (Moll et al., 1990; Bouwens, 1998), whereas ductal expression of mouse K20 is detectable at very low levels (Figure 6) but is induced in an autoimmune diabetes model (Anastasi et al. 1999) or after caerulein-induced pancreatitis (our unpublished observations). We did not observe pancreatic hK20 transgene induction after caerulein-induced pancreatitis (our unpublished data) or under basal conditions (Figure 6). Therefore, it is likely that the regulatory elements that control K20 expression in the stomach and pancreas are outside the 18-kb genomic region that we used in this study. This differs from K18 whereby a genomic sequence of 10-kb contained all the necessary elements for normal tissue specific expression in simple epithelia (Abe and Oshima, 1990). However, we cannot exclude the possibility that gastric/pancreatic regulatory factors may not recognize the human transgene.
Functional Redundancy of K18 and K20 at the Level of Keratin Filament Organization
The dominant negative filament organization phenotype noted in small intestinal enterocytes of the M2 transgenic line supports an in vivo role for K20 in keratin filament organization. This role is further substantiated by the intermixed cross-breeding of the transgenic mice that overexpress wild-type K18 or K20 or mutant K18 or K20 (summarized in Figure 10). Hence, wild-type K18 rescues mutant K20 and vice versa, and the effects of the K20 and K18 mutations are additive in the same cell in terms of their filament disruptive capacity. This provides in vivo evidence that K18 and K20 serve redundant functions in terms of keratin filament organization in the intestine. In addition, our findings provide an explanation for a K20-mediated sparing role of keratin filament organization in the majority of transgenic mouse enterocytes that overexpress K18 R89C (Ku et al., 1995), although K19 is likely to play a bigger “sparing” role given its abundance as the major type I keratin in the intestine and its distribution throughout the intestinal epithelium (Figures 1, 2, 3). Previous in vitro studies showed that K18, compared with K20, is a preferred partner for binding with K8 but K20 and K8 do associate and form filaments (Hofmann and Franke 1997) as confirmed in this study in transfected cells in culture (Figure 4) and by colocalization of filaments containing K20 and K18 in tissues (Figure 6). Availability of transgenic mice that overexpress wild-type and mutant K20 should provide useful in vivo models to study K20 function and regulation.
Acknowledgments
We thank Drs. Helene Baribault, Thomas Magin, and Andrea Quaroni for the generous gifts of antibodies; Kris Morrow for preparing the figures; Steve Avolicino (Histo-tec Laboratory, Hayward, CA) for histology staining; and Xiaomu Zheng and Phuoc Vo for excellent technical assistance. This work was supported by National Institutes of Health grant DK-52951 and by National Institutes of Health Digestive Disease Center grant DK-56339.
Abbreviations used: g, genomic; h, human; HSE, high salt extract(ion); IF, intermediate filament; K, keratin; m, mouse; M, mutant; mAb, monoclonal antibody; WT, wild-type.
References
- Abe, M., and Oshima, R.G. (1990). A single human keratin 18 gene is expressed in diverse epithelial cells of transgenic mice. J. Cell Biol. 111, 1197-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasi, E., Ponte, E., Gradini, R., Bulotta, A., Sale, P., Tiberti, C., Okamoto, H., Dotta, F., and Mario, U.D. (1999). Expression of Reg and cytokeratin 20 during ductal cell differentiation and proliferation in a mouse model of autoimmune diabetes. Eur. J. Endocrinol. 141, 644-652. [DOI] [PubMed] [Google Scholar]
- Bader, B.L., and Franke, W.W. (1990). Cell type-specific and efficient synthesis of human cytokeratin 19 in transgenic mice. Differentiation 45, 109-118. [DOI] [PubMed] [Google Scholar]
- Bouwens, L. (1998). Cytokeratins and cell differentiation in the pancreas. J. Pathol. 184, 234-239. [DOI] [PubMed] [Google Scholar]
- Calnek, D., and Quaroni, A. (1993). Differential localization by in situ hybridization of distinct keratin mRNA species during intestinal epithelial cell development and differentiation. Differentiation 53, 95-104. [DOI] [PubMed] [Google Scholar]
- Chandler, J.S., Calnek, D., and Quaroni, A. (1993). Identification and characterization of rat intestinal keratins. Molecular cloning of cDNAs encoding cytokeratin 8, 19, and a new 49-KDa type I cytokeratin (cytokeratin 21) expressed by differentiated intestinal epithelial cells. J. Biol. Chem. 266, 11932-11938. [PubMed] [Google Scholar]
- Chou, C.F., Riopel, C.L., Rott, L.S., and Omary, M.B. (1993). A significant soluble keratin fraction in “simple epithelial cells: lack of an apparent phosphorylation and glycosylation role in keratin solubility. J. Cell Sci. 105, 433-445. [DOI] [PubMed] [Google Scholar]
- Coulombe, P.A., Hutton, M.E., Letai, A., Hebert, A., Paller, A.S., and Fuchs, E. (1991). Point mutations in human14 genes of epidermolysis bullosa simplex patients: genetic and functional analyses. Cell 66, 1301-1311. [DOI] [PubMed] [Google Scholar]
- Coulombe, P.A., and Omary, M.B. (2002). “Hard” and “soft” principles defining the structure, function and regulation of keratin intermediate filaments. Curr. Opin. Cell Biol. 14, 110-122. [DOI] [PubMed] [Google Scholar]
- Fuchs, E., and Coulombe, P.A. (1992). Of mice and men: genetic skin diseases of keratin. Cell 69, 899-902. [DOI] [PubMed] [Google Scholar]
- Fuchs, E., and Weber, K. (1994). Intermediate filaments: structure, dynamics, function and disease. Annu. Rev. Genet. 63, 345-382. [DOI] [PubMed] [Google Scholar]
- Fuchs, E., and Cleveland, D.W. (1998). A structural scaffolding of intermediate filaments in health and disease. Science 279, 514-519. [DOI] [PubMed] [Google Scholar]
- Franke, W.W., Mahmood, N., Appelhans, B., Schmid, E., Freudenstein, C., Osborn, M., and Weber, K. (1979). The organization of cytokeratin filaments in the intestinal epithelium. Eur. J. Cell Biol. 19, 255-268. [PubMed] [Google Scholar]
- Harnden, P., Mahmood, N., and Southgate, J. (1999). Expression of cytokeratin 20 redefines urothelial papillomas of the bladder. The Lancet 353, 974-977. [DOI] [PubMed] [Google Scholar]
- Herrmann, H., and Aebi, U. (2000). Intermediate filaments and their associates: multi-talented structural elements specifying cytoarchitecture and cytodynamics. Curr. Opin. Cell Biol. 12, 79-90. [DOI] [PubMed] [Google Scholar]
- Hofmann, I., and Franke, W.W. (1997). Heterotypic interactions and filament assembly of type I and type II cytokeratins in vitro: viscometry and determination of relative affinities. Eur. J. Cell Biol. 72, 122-132. [PubMed] [Google Scholar]
- Ku, N.-O., Gish, R., Wright, T.L., and Omary, M.B. (2001). Keratin 8 mutations in patients with cryptogenic liver disease. N. Engl. J. Med. 344, 1580-1587. [DOI] [PubMed] [Google Scholar]
- Ku, N.-O., Liao, J., and Omary, M.B. (1998). Phosphorylation of human keratin 18 serine-33 regulates binding to 14–3-3 proteins. EMBO J. 17, 1892-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku, N.-O., Michie, S.A., Oshima, R.G., and Omary, M.B. (1995). Chronic hepatitis, hepatocyte fragility and increased soluble phosphoglycokeratins in transgenic mice expressing a keratin 18 conserved arginine mutant. J. Cell Biol. 131, 1303-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku, N.-O., Michie, S.A., Soetikno, R.M., Resurreccion, E.Z., Broome, R.L., Oshima, R.G., and Omary, M.B. (1996). Susceptibility to hepatotoxicity in transgenic mice that express a dominant-negative human keratin18 mutant. J. Clin. Investig. 98, 1034-1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku, N.-O., Wright, T.L., Terrault, N.A., Gish, R., and Omary, M.B. (1997). Mutation of human keratin 18 in association with cryptogenic cirrhosis. J. Clin. Investig. 99, 19-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ku, N.-O., Zhou, X., Toivola, D.M., and Omary, M.B. (1999). The cytoskeleton of digestive epithelia in health and disease. Am. J. Physiol. 277, G1108-G1137. [DOI] [PubMed] [Google Scholar]
- Leech, S.N., Kolar, A.J., Barrett, P.D., Sinclair, S.A., and Leonard, N. (2001). Merkel cell carcinoma can be distinguished from metastatic small cell carcinoma using antibodies to cytokeratin 20 and thyroid transcription factor 1. J. Clin. Pathol. 54, 727-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magin, T.M., Hesse, M., and Schroder, R. (2000). Novel insights into intermediate-filament function from studies of transgenic and knockout mice. Protoplasma 211, 140-150. [Google Scholar]
- Moll, R., Franke, W.W., Schiller, D.L., Geiger, B., and Krepler, R. (1982). The catalog of human cytokeratins: patterns of expression in normal epithelia, tumors and cultured cells. Cell 31, 11-24. [DOI] [PubMed] [Google Scholar]
- Moll, R., Lowe, A., Laufer, J., and Franke, W.W. (1992). Cytokeratin 20 in human carcinomas. A new histodiagnostic marker detected by monoclonal antibodies. Am. J. Pathol. 140, 427-447. [PMC free article] [PubMed] [Google Scholar]
- Moll, R., Schiller, D., and Franke, W.W. (1990). Identification of protein IT of the intestinal cytoskeleton as a novel type I cytokeratin with unusual properties and expression patterns. J. Cell Biol. 111, 567-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll, R., Zimbelmann, R., Golsdchmidt, M., Keith, M., Laufer, J., Kasper, M., Koch, P., and Franke, W.W. (1993). The human gene encoding cytokeratin 20 and its expression during fetal development and in gastrointestinal carcinomas. Differentiation 53, 75-93. [DOI] [PubMed] [Google Scholar]
- O'Guin, M., Galvin, S., Schermer, A., and Sun, T.-T. (1987). Pattern of keratin expression defines distinct pathways of epithelial development and differentiation. Curr. Top. Dev. Biol. 22, 97-125. [DOI] [PubMed] [Google Scholar]
- Omary, M.B., Ku, N.-O., and Toivola, D.M. (2002). Keratins: guardians of the liver. Hepatology 35, 251-257. [DOI] [PubMed] [Google Scholar]
- Oshima, R.G. (2002). Apoptosis and keratin intermediate filaments. Cell Death Differ. 9, 486-492. [DOI] [PubMed] [Google Scholar]
- Quaroni, A., Calnek, D., Quaroni, E., and Chandler, J.S. (1991). Keratin expression in rat intestinal crypt and villus cells. J. Biol. Chem. 266, 11923-11931. [PubMed] [Google Scholar]
- Takemura, R., Masaki, T., and Hirokawa, N. (1988). Developmental organization of the intestinal brush-border cytoskeleton. Cell Motil. Cytoskeketon 9, 299-311. [DOI] [PubMed] [Google Scholar]
- Toivola, D.M., Omary, M.B., Ku, N.-O., Peltola, O., Baribault, H., and Eriksson, J.E. (1998). Protein phosphatase inhibition in normal and keratin 8/18 assembly-incompetent mouse strains supports a functional role of keratin intermediate filaments in preserving hepatocyte integrity. Hepatology 28, 116-128. [DOI] [PubMed] [Google Scholar]
- Toivola, D.M., Ku, N.-O., Ghori, N., Lowe, A.W., Michie, S.A., and Omary, M.B. (2000a). Effects of keratin filament disruption on exocrine pancreas-stimulated secretion and susceptibility to injury. Exp. Cell Res. 255, 156-170. [DOI] [PubMed] [Google Scholar]
- Toivola, D.M., Baribault, H., Magin, T.M., Michie, S.A., and Omary, M.B. (2000b). Simple epithelial keratins are dispensable for cytoprotection in two pancreatitis models. Am. J. Physiol. 279, G1343-1354. [DOI] [PubMed] [Google Scholar]
- Wildi, S., Kleeff, J., Maruyama, H., Maurer, C.A., Friess, H., Buchler, M.W., Lander, A.D., and Korc, M. (1999). Characterization of cytokeratin 20 expression in pancreatic and colorectal cancer. Clin. Cancer Res. 10, 2840-2847. [PubMed] [Google Scholar]