Abstract
Introduction of double-stranded RNA (dsRNA) can elicit a gene-specific RNA interference response in a variety of organisms and cell types. In many cases, this response has a systemic character in that silencing of gene expression is observed in cells distal from the site of dsRNA delivery. The molecular mechanisms underlying the mobile nature of RNA silencing are unknown. For example, although cellular entry of dsRNA is possible, cellular exit of dsRNA from normal animal cells has not been directly observed. We provide evidence that transgenic strains of Caenorhabditis elegans transcribing dsRNA from a tissue-specific promoter do not exhibit comprehensive systemic RNA interference phenotypes. In these same animals, modifications of environmental conditions can result in more robust systemic RNA silencing. Additionally, we find that genetic mutations can influence the systemic character of RNA silencing in C. elegans and can separate mechanisms underlying systemic RNA silencing into tissue-specific components. These data suggest that trafficking of RNA silencing signals in C. elegans is regulated by specific physiological and genetic factors.
INTRODUCTION
Double-stranded RNA (dsRNA) has the capability to render genes nonfunctional in a sequence-specific manner. When introduced into cells, dsRNA can activate mechanisms that target the degradation of cognate cytoplasmic mRNAs (Montgomery et al., 1998; Tuschl et al., 1999; Klink and Wolniak, 2000) and thus can effectively silence full gene expression at the posttranscriptional level. This genetic interference effect (termed RNA interference, RNAi) is temporary and the effect is dosage dependent. RNAi has been observed in many cell types from divergent eukaryotes, including protozoa, fungi, plants, invertebrates, and mammals (Hannon, 2002). Additionally, RNAi is part of a larger network of “RNA silencing” mechanisms (Baulcombe, 2002; Wassenegger, 2002).
RNAi phenotypes have generally been elicited in Caenorhabditis elegans and other nonvertebrate species by using dsRNA molecules that are rather long (>100 base pairs). Once inside cells, long dsRNA molecules are cleaved into double-stranded small interfering RNAs (siRNAs) that are 21–25 base pairs in length by an enzyme with RNaseIII-like activity (Dicer) (Hamilton and Baulcombe, 1999; Parrish et al., 2000; Yang et al., 2000; Bernstein et al., 2001; Grishok et al., 2001; Ketting et al., 2001; Knight and Bass, 2001). Cleavage into siRNAs is an early step in the RNAi silencing mechanism (Elbashir et al., 2001b; Parrish and Fire, 2001; Klahre et al., 2002), and the rde-4 gene product in C. elegans likely influences Dicer-dependent cleavage of dsRNAs as evidenced by observations 1) that rde-4 mutant animals injected with long dsRNAs do not accumulate siRNAs and do not exhibit an RNA silencing response; 2) that RDE-4 is found in tight physical association with DICER; and 3) that the RNA silencing defects in rde-4 mutant animals can be overcome by injection of precleaved siRNAs into mutant animals (Tabara et al., 1999; Parrish and Fire, 2001; Tabara et al., 2002). Although RNAi can be triggered by long dsRNAs or by siRNAs in C. elegans, the key to effective RNAi responses in mammalian systems has been the use of siRNAs (Elbashir et al., 2001a; Caplen et al., 2001). siRNAs are long enough to mediate sequence-specific mRNA cleavage, yet are short enough to avoid activation of nonsequence specific dsRNA responses (such as interferon responses) that exist in mammalian systems.
dsRNAs that are delivered into extracellular spaces can elicit systemic RNA silencing in diverse organisms, including C. elegans, planaria, Coleoptera, cnidaria, and plants (Palauqui et al., 1997; Fire et al., 1998; Lohmann et al., 1999; Sanchez Alvarado and Newmark, 1999; Bucher et al., 2002), as evidenced by observations of RNA silencing in cells that are far removed from the initial site of dsRNA delivery. In C. elegans, four methods are available for delivery of dsRNA into the organism: 1) injection of dsRNA into any site of the animal (Fire et al., 1998; Grishok et al., 2000), 2) feeding animals with bacteria engineered to express dsRNA (Timmons et al., 2001), 3) soaking animals in dsRNA (Tabara et al., 1998), and 4) in vivo transcription of dsRNA from transgene promoters (Tabara et al., 1999; Tavernarakis et al., 2000). Injection, feeding, and soaking can result in RNAi in all cells of the treated animal and its progeny, an indication that the RNAi signal is mobile and can be taken up by different tissues. The mobile behavior of the RNA silencing signal could reflect a combination of different transport mechanisms, including cellular uptake of dsRNA from the coelomic fluid, exit of dsRNA from cells, direct intercellular trafficking of dsRNA between coupled cells, and/or partitioning of the dsRNA pool upon cell division. Because RNAi and related mechanisms are thought to be an organismal response to challenge from viral and transposon parasites (reviewed in Plasterk, 2002), we reasoned that the systemic character of the response might depend rather specifically on physiological conditions.
It is conceivable that systemic RNAi in C. elegans might involve a rather simple and broadly active mechanism of dsRNA uptake by individual cells. C. elegans does not have an active circulatory system; instead, dsRNA may gain direct access to cells via the coelomic fluid. (Similarly, in other organisms, dsRNA may gain access to cells via the circulatory system, culture fluid, etc.) In injection, needle-mediated tissue disruption undoubtedly facilitates access to the coelomic fluid. In delivery by feeding and soaking, dsRNA may be distributed to cells from the gut in the same manner as nutrients. It is also conceivable that dsRNA residing in “infected” cells could undergo successive rounds of cellular exit and re-entry into adjacent “uninfected” cells, culminating in a systemically affected animal. The latter assumes that dsRNA can exit as well as enter cells, and raises questions about the cellular autonomy of dsRNA effects. Although there is considerable evidence for an ability of animal cells to import dsRNA, there is little data that supports the notion of robust cellular exit of dsRNA in animal systems. This question is particularly intriguing given that transport of an RNA silencing signal between host and graft has been demonstrated in plants (Palauqui et al., 1997), whereas there remains a lack of conclusive evidence for movement of RNA molecules between cells in multicellular animals. Addressing the ability of dsRNA to exit cells is important not only in elucidating the full mechanism of RNAi but also in understanding how dsRNA can elicit biologically significant systemic responses (e.g., a systemic antiviral response in an organism after localized infection).
In an effort to more fully understand the nature of systemic RNA silencing in a complex, multi-tissue animal, we are assessing the ability of in vivo-delivered dsRNA molecules to elicit systemic RNA silencing. We have introduced transgenes into C. elegans that express dsRNA under the direction of tissue-specific promoters in one type of cell. We note that although transgene-mediated delivery of dsRNA is effective in eliciting tissue-specific RNA silencing, robust systemic RNA silencing was not observed. Unexpectedly, we have found that exogenous delivery of unrelated dsRNA molecules to these same transgenic strains can elicit a detectable systemic RNA silencing phenotype. We have also observed that animals defective for fed-1 or fed-2 are unable to mount a robust systemic silencing response to ingested dsRNAs, yet these mutants can display systemic silencing in response to tissue-specific transcription of dsRNAs from transgenes. While both sid-1 mutants (Winston et al., 2002) and fed mutants fail to respond to ingested dsRNA, sid-1 mutants, but not fed mutants, fail to exhibit systemic silencing in response to silencing signals transcribed within cells. These observations demonstrate a capability for dsRNA export by cells, highlight the complexity of the systemic silencing mechanisms in multicellular animals and raise the possibilities of multiple and/or tissue-specific mechanisms for cellular uptake and export of RNA silencing signals.
MATERIALS AND METHODS
DNA Constructs
For gfp hairpin RNA Production in C. elegans. Three promoters were used for tissue-specific expression in C. elegans: myo-3 drives expression in body wall muscle in late embryos and larvae (Figure 1a) (Fire et al., 1998), vit-2 in the gut during vitellogenesis (MacMorris et al., 1994), and unc-119 in neurons (Maduro and Pilgrim, 1995). These promoters are well characterized, generate abundant transcript, are particularly tissue specific, and activate transcription relatively late in development. DNA upstream of myo-3 (2300 bp) was obtained from plasmid L2534 by using standard cloning techniques; 250 bp of vit-2 promoter was amplified from genomic DNA by using oligonucleotides LT095 (tgctctagagatccaactgtattacttgaa) and LT096 (cggggtaccggctgaaccgtgattggactg); and 1200 bp of DNA upstream of unc-119 was obtained from plasmid pBY103 (generously provided by M. Maduro and D. Pilgrim, University of Alberta, Calgary, AB, Canada). The promoters were fused to two copies of green fluorescent protein DNA in inverted repeat orientation separated by 900 base pairs of nonrelated spacer sequence (Timmons et al., 2001). RNA transcribed from this sequence has the capacity to fold back into a hairpin double-stranded RNA. This same inverted repeat sequence has been used in the feeding protocol and was functional in eliciting RNAi (our unpublished data). The myo-3 promoter was fused to the gfp hairpin DNA sequence to generate pLT98, the vit-2 promoter sequence was used similarly to generate pLT156, and the unc-119 promoter for pLT164. We similarly configured two additional versions of gfp hairpin DNA under the transcriptional regulation of these worm promoters in hopes of better cytoplasmic accumulation of dsRNA: 1) the original version does not have introns in either gfp repeat and has a 900-base pair spacer between inverted repeats; as described 2) a second version replaces the spacer sequence with intronic sequences that should splice out when expressed in the worm, leaving behind a perfect hairpin RNA with no spacer (Smith et al., 2000); and 3) a third configuration contains four introns in the first gfp repeat sequence and also an intronic spacer sequence. The results obtained using intron-containing gfp hairpin RNAs were similar to the results obtained in Figure 1 by using configurations without introns (our unpublished data).
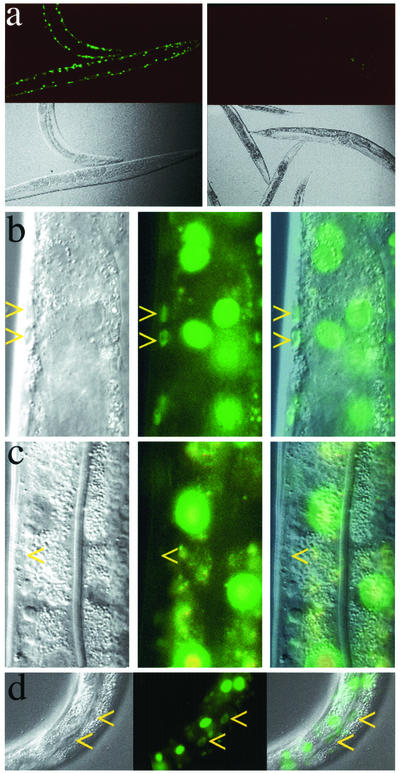
Figure 1.
Transgenes expressing gfp hairpins are effective triggers for RNAi. (a) GFP fluorescence (top two panels) and Nomarski (bottom two panels) images of worms harboring an integrated myo-3::GFP transgene expressing GFP in muscle. (The GFP reporters used in all these experiments were tagged with a nuclear localization signal.) The animals depicted on the right harbor an additional myo-3::gfp hairpin transgene array capable of expressing dsgfp RNA in muscle. The images on the left depict the normal accumulation pattern of GFP expressed from the reporter transgene. The images on the right demonstrate that the myo-3::gfp hairpin construct can deliver enough dsgfp RNA to mediate effective silencing of GFP in muscle cells. (b) Nomarski (left), GFP fluorescence (middle), and composite (right) images of an animal with a rpL28::GFP transgene expressing GFP in all cells. Arrows point to muscle nuclei with GFP fluorescence. Larger fluorescent nuclei are polyploid gut nuclei. (c) Nomarski (left), GFP fluorescence (middle), and composite (right) images of an animal with two transgenes: the myo-3::gfp hairpin transgene as in (a, right) and the rpL28::GFP transgene reporter used in b. Arrow points to muscle nucleus that fails to express GFP. Nuclei nearby are not affected. (d) Nomarski (left), GFP fluorescence (middle), and composite (right) images of an animal with a let-858::GFP transgene expressing GFP in all cells and a vit-2::gfp hairpin transgene expressing dsgfp RNA in the gut. Arrows point to nuclei that show reduced GFP accumulation in gut. Two explanations are possible for the lack of interference in other gut cells: the extrachromosomal vit-2::gfp hairpin transgene array may have been mitotically segregated out of the unaffected cells; and this promoter has previously been demonstrated to drive expression in a dynamic pattern (MacMorris et al., 1994; our unpublished observations).
For dsRNA Production. The nonsequence related dsRNA was derived from a plasmid (pLT190) containing 1 kb of sequence corresponding to the bacterial tetracycline resistance gene (Peden, 1983). This sequence has no homology to gfp nor to C. elegans genes. dsRNA was transcribed from pLT190 by using an in vitro transcription kit (Ambion, Austin, TX). A 400-base pair fragment of the rde-1 gene was subcloned into a plasmid with opposable T7 promoter sites (L4440) and was used as a template for in vitro transcription (Ambion). Doubly transgenic animals were soaked in this solution, and progeny animals were monitored for loss of fluorescence. Plasmid pPD128.110 (Timmons and Fire, 1998) was used to transcribe dsgfp RNA used in soaking experiments.
Soaking Conditions
For Induced Export. Animals were soaked in several different control solutions, including water, M9 media, injection buffer (Stinchcomb et al., 1985), M9 media/50 mM NaCl, 0.5 mg/ml dsDNA oligonucleotides in water, and 0.5 mg/ml plasmid DNA in water (Tabara et al., 1998). Animals were also soaked in dstetA RNA in injection buffer or water (Mello et al., 1991) 4 h overnight at 15°C in Eppendorf tubes. Concentrations of dstetA RNA were 0.5–2 μg/μl. Animals were allowed a 4-h recovery period before examination. Recovered animals were incubated at 15°C, 20°C, and 25°C to test for temperature effects of induced systemic silencing. Temperature did not influence the degree to which systemic silencing could be induced by exogenous dsRNA delivery, nor did daily heat pulses of 37°C for 30 min (our unpublished data). rde-1(ne300) and sid-1(qt2) mutant animals containing the trigger and target transgenes were soaked in 2 mg/ml concentrations of dstetA RNA as described, allowed to recover, and 50 treated animals harboring both transgenes were scored for systemic silencing on the day of recovery and on two subsequent days. Systemic silencing effects were not noted in any of the soaked animals harboring these mutations. We also monitored treated animals for phenotypes associated with RNAi silencing defects such as germline desilencing of transgenes (green fluorescent protein [GFP] expression in the germline) and increased frequency of nondisjunction (presence of males) (Kelly et al., 1997). None of these phenotypes was observed. Control soaking conditions (without dsRNA) did not elicit systemic silencing.
For RNAi. Animals were soaked in injection buffer or water by using dsRNA concentrations of 1–2 mg/ml at 15°C overnight. GFP expression levels were monitored in recovered animals and their progeny.
C. elegans Strains
Two C. elegans strains harboring chromosomally integrated transgenes (PD7325 and PD8160) were used. Both these strains stably and reproducibly express GFP in the nuclei of all somatic cells. (The GFP harbors a nuclear localization signal.) The transgene in PD7325 (dpy-20(e1282); ccIn7325[BK48+pMH86]) expresses gfp from a let-858 promoter (Kelly et al., 1997). Strain PD8160 (provided by J. Fleenor, Carnegie Institution) harbors a ccIn8160 transgene that drives GFP from a ribosomal protein L28 promoter (Consortium, 1998). Strain PD4251 contains an integrated array of myo-3::GFP and stably expresses GFP in muscle nuclei. The ccIn8160 insertion, gfp hairpin transgene array, and sid-1(qt2) mutant were brought together into the same strain (YY304) by standard genetic manipulation. The ccIn8160 insertion, gfp hairpin transgene array, and each fed mutant were brought together to produce strains YY216 and YY209. Other strains used in these experiments include: sid-1(qt2); fed-1(ne309)III, and fed-2(ne319)IV. None of the fed loci correspond to sid-1 or to the sid-1-related locus that encodes the ZK721.1 protein. In some cases we have used the following protocol to minimize contaminations of our stocks: ampicillin (100 μg/ml), tetracycline (10 μg/ml), and kanamycin (10 μg/ml) was added to freshly thawed stocks that were recovered onto normal growth media seeded with wild-type OP50 bacteria. After recovery from freezing, animals were moved to fresh plates, and the population was allowed to increase over the course of a few days. The animals were then collected and lysed in a solution of 10% bleach/1 N NaOH. The embryos that survived this treatment were washed with water and plated. The resulting L1-L3 larvae were then soaked in solutions of antibiotics in M9 media overnight and replated.
Generation of Additional C. elegans Stocks
The gfp hairpin plasmids described above were injected into wild-type N2 worms along with the dominant rol-6 transformation marker (Mello et al., 1991), and heritable transgenic lines were established. The gfp hairpin transgenes were maintained as extrachromosomal arrays. These are lost at some frequency during meiotic and mitotic cell divisions (Stinchcomb et al., 1985). Doubly transgenic animals were generated by crossing worms harboring a gfp hairpin array with worms harboring a GFP-expressing array using standard genetic manipulations. Six different myo-3::gfp hairpin lines were generated. All were analyzed in an rpL28::GFP background, and three of these were analyzed in a let-858::GFP background. Four vit-2::gfp hairpin lines were generated. All were analyzed in a let-858::GFP background; three were crossed into rpL28::GFP. One unc-119::gfp hairpin line was established for each GFP reporter. The GFP fluorescence in ventral nerve cord and nerve ring (the major neuropils) was not significantly reduced in these lines. Additionally, GFP fluorescence in neurons was not reduced in unc-119::gfp reporter lines harboring the unc-119::gfp hairpin. These observations are in agreement with previous reports of a more limited capacity for RNA silencing of some genes in C. elegans neurons (Timmons et al., 2001; Simmer et al., 2002).
RESULTS
Tissue-specific Expression of a gfp hairpin Does Not Elicit Systemic RNA Silencing in All Animals
We wondered whether RNAi could be restricted to particular tissues in C. elegans if the dsRNA trigger were presented to a subset of cells within the animal (Figures 1 and 2). We used GFP as an RNAi target so that RNAi could be observed in individual cells by monitoring for loss of GFP fluorescence. The generality of cell-specific RNAi in C. elegans was tested by using several well-described promoters to drive expression of the gfp hairpin in distinct tissues. The promoters are not only tissue restricted but also transcribe to relatively high levels and initiate transcription relatively late in development. A myo-3 promoter was used to drive expression of a gfp hairpin in muscle cells, a vit-2 promoter for intestinal cells, and an unc-119 promoter for neuronal cells. Multiple C. elegans lines were obtained for each gfp hairpin construct injected (see MATERIALS AND METHODS).
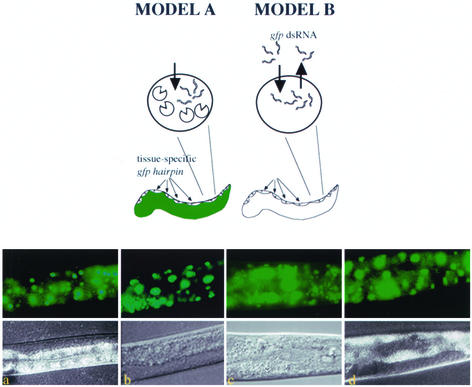
Figure 2.
Assessments of dsRNA cell autonomy. The models depict the experimental design to address the autonomous nature of RNA silencing signals and predicts the RNAi phenotypes of transgenic animals if in vivo-transcribed RNA silencing signals remain cell autonomous (left) or are capable of cellular exit (right). A transgene expressing a gfp hairpin in a tissue-specific manner was introduced into animals expressing a GFP reporter in all cells by standard genetic crosses. Model A predicts that animals would not exhibit systemic silencing for GFP if dsRNA cannot exit the tissue of origin. Cell autonomous RNAi (white) is exhibited as tissue-specific loss of GFP. Model B predicts that animals would exhibit systemic silencing for GFP if RNA silencing signals can exit the cells of origin. (a) The GFP expression pattern in an animal harboring a chromosomally integrated let-858::GFP transgene. (b) A transgene with myo-3::gfp hairpin sequences that delivers dsgfp RNA in muscle (Figure 1, a and c) was crossed into the strain represented in a. (c) A transgene with vit-2::gfp hairpin sequence that delivers dsgfp RNA in gut (Figure 1d) was crossed into animals harboring let-858::GFP. (d) A transgene with unc-119::gfp hairpin sequences that delivers dsgfp RNA in neurons (see MATERIALS AND METHODS) was crossed into animals harboring let-858::GFP. Images were captured using a 40× objective so the overall GFP fluorescence of animals could be compared. RNA silencing was not generally observed in non-hairpin–expressing cells as evidenced by uniform GFP fluorescence in somatic cells.
An RNAi phenotype (loss of GFP fluorescence) was observed in tissues where transcription of the gfp hairpin was expected, demonstrating the effectiveness of the constructs in delivering dsRNA to cells (Figure 1). For example, the GFP fluorescence level in muscle cells of animals harboring a myo-3::GFP reporter was reduced, as expected, when a myo-3::gfp hairpin transgene was crossed into the strain (Figure 1a, compare left and right panels). The loss of fluorescence observed in muscle cells was dependent on an intact RNAi mechanism: when doubly transgenic animals (accumulating GFP in muscle nuclei and gfp hairpin in muscle) were injected with dsRNA corresponding to the rde-1 gene or were crossed into an rde-1 mutant background, no loss of fluorescence was observed in muscle (our unpublished data). Similarly, the gfp hairpin was effective in gut cells: the fluorescence intensity was reduced in gut nuclei of doubly transgenic animals (accumulating GFP in nuclei of all cells and gfp hairpin in gut; Figure 1d), although with fewer animals exhibiting effects in comparison with animals silenced from a myo-3::gfp hairpin transgene. The gfp hairpin driven from an unc-119 promoter was not effective in eliciting RNAi in neurons; this tissue has previously been noted to be refractory to dsRNA for some genes in wild-type C. elegans (Timmons et al., 2001; Simmer et al., 2002).
We then explored the possibility that RNA silencing signals might move bidirectionally across cell membranes (Figure 2). Clearly, dsRNA can be taken up by C. elegans cells when delivered by injection, feeding, or soaking; however, the ability of animal cells to export RNA silencing signals has not been fully investigated. We tested for this capacity by observing whether gfp hairpins expressed in specific tissues could elicit ectopic silencing phenotypes (in cells that do not transcribe the gfp hairpin). Doubly transgenic strains were generated that express GFP in all cells and gfp hairpin RNA in a tissue-restricted manner, and all cells were monitored for RNAi (loss of GFP fluorescence) (Figure 2). With respect to the two GFP-expressing transgenes used as RNAi targets in our experiments (rpL28::GFP and let-858::GFP; see MATERIALS AND METHODS) each was maintained as a chromosomal insertion and GFP fluorescence in all cells was consistent and uniform in the absence of the second gfp hairpin transgene.
The RNA silencing phenotypes observed in doubly transgenic animals remained restricted to mostly those cells previously demonstrated to respond to the tissue-specific promoter (Figure 2). Robust systemic RNA silencing was not observed for any of the combinations of GFP-expressing and gfp hairpin-expressing transgenes. (Figure 2; our unpublished data). Specifically, the animals we observed exhibited interference with GFP expression in those tissues that expressed the interference trigger (the gfp hairpin), but were not substantially silenced for GFP in other tissues.
Systemic RNA Silencing from a gfp hairpin Can Be Induced
We considered several explanations for lack of systemic RNA silencing in our doubly transgenic animals: cells may lack any mechanism for dsRNA exit, our experimental conditions may be unfavorable for observation of dsRNA export, or a generalized alarm signal may be needed to trigger dsRNA export. In the latter two cases, we might expect that our doubly transgenic animals could be induced to elicit systemic RNA silencing. In particular, we reasoned that exposure to high concentrations of dsRNA might affect the mobility of an intracellular dsRNA by titrating out mechanisms that confine dsRNA within cells, by titrating mechanisms that convert dsRNAs to unexportable forms, by titrating mechanisms that eliminate the dsRNA character of the trigger, or by specifically activating dsRNA export systems. The complexity of the injected material can also affect the RNA silencing response in many systems, including C. elegans. RNAi exhibits premature saturation when several dsRNA sequences are introduced to the organism simultaneously, resulting in attenuated RNAi phenotypes for each of the dsRNAs (Gonczy et al., 2000; Parrish et al., 2000). The experiments below derive from a hypothesis that the introduction of a second species of dsRNA to doubly transgenic animals might stimulate saturation in the cell where the gfp hairpin was transcribed, and that gfp hairpin molecules not engaged in RNA silencing mechanisms might freely exit that cell. Thus, one possible outcome from exposure of these animals to external dsRNA was that cells might be induced to export an RNAi silencing signal.
In our initial tests of saturation-induced systemic RNA silencing, doubly transgenic animals (expressing a gfp hairpin in a tissue-specific manner and a GFP reporter in all cells) were injected with a solution of dstetA RNA. The bacterial tetA region used in our experiments does not have homology to worm sequences or to GFP. All combinations of doubly transgenic strains were tested by injection, and we observed reduced levels of GFP fluorescence in some of the injected animals and in their progeny (our unpublished data). We considered that the reduced fluorescence might be a result of tissue disruption from the injection needle. We next used the soaking method to deliver dstetA RNA to doubly transgenic animals (Figure 3). Again, we noted a reduced fluorescence in the treated animals and in their progeny.
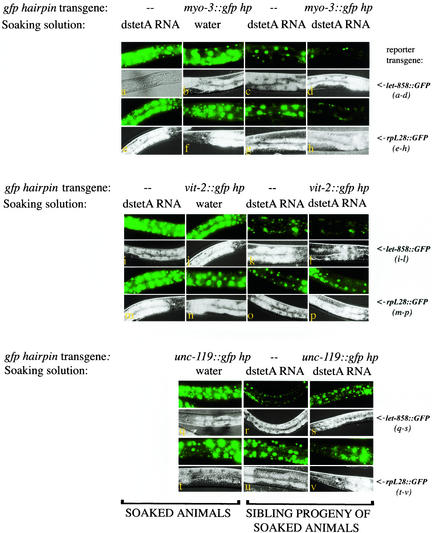
Figure 3.
Systemic RNAi is inducible. Exposing cells to unrelated dsRNAs might facilitate intracellular release of an RNA silencing signal derived from an in vivo transcribed gfp hairpin. Pairs of images are representative of each experiment; GFP fluorescence images are above, Nomarski below. Two different integrated transgenes expressing GFP in all cells were used in these experiments: a let-858::GFP transgene (a, b, c, d, i, j, k, l, q, r, and s) or a rpL28::GFP transgene (e, f, g, h, m, n, o, p, t, u, and v); GFP is the RNAi target in these experiments. Similar results were observed for each reporter. A second transgene driving tissue-specific expression of a gfp hairpin dsRNA was present in some of the strains used. Animals were soaked in high concentrations of dstetA RNA or in water, and GFP fluorescence was monitored in the soaked animals (a, b, e, f, i, j, m, n, q, and t) or in the resulting progeny (c, d, g, h, k, l, o, p, r, s, u, and v). (The soaked animals were allowed a recovery period of 12–18 h before imaging. The accumulation of GFP in these control animals was not severely affected under these harsh conditions.) Animals in b, d, f, and h harbored a second transgene expressing a gfp hairpin dsRNA from a myo-3 promoter in addition to the GFP-expressing transgene. Animals in j, l, n, and p have a vit-2::gfp hairpin transgene. Animals in q, s, t, and v have an unc-119::gfp hairpin transgene. Soaking GFP-expressing animals in dstetA RNA does not affect GFP fluorescence (a, e, i, and m). Soaking doubly transgenic animals in control solutions does not elicit reduced GFP fluorescence (b, f, j, n, q, and t). Two progeny classes arise from doubly transgenic animals: both classes are homozygous for the GFP reporter transgene; one class inherits the gfp hairpin transgene (d, h, l, p, s, and v); the second class does not (c, g, k, o, r, and u) (Stinchcomb et al., 1985). (Animals in c, g, k, o, r, and u were recovered after photography and allowed to produce progeny. The gfp hairpin transgene was not observed in the progeny of these animals.) When doubly transgenic animals are soaked in dstetA RNA, both classes of progeny exhibit some loss of GFP fluorescence. The effects in singly transgenic progeny of soaked animals (c, g, k, o, r, and u; Table 1) may result from induced maternal deposition of gfp hairpin into these animals while resident as a germline/embryo in the treated hermaphrodite, implying exit of gfp hairpin RNAs from the maternal somatic cells. Loss of GFP expression is not observed in tissues that have been previously reported to be intractable for RNAi (neurons and some pharyngeal cells).
Exposure of animals to extracellular nonspecific dsRNA by soaking (or injecting) led to an RNA silencing response in increased numbers of cells in diverse non-neuronal tissues of treated animals and their progeny (Figure 3). This effect was most striking when the gfp hairpin was driven in muscle or gut (Figure 3 and Table 1). This effect was dependent on four elements: an intracellular RNAi trigger, extracellular (nonspecific) dsRNA, a functional RNAi system, and a functional systemic transport system, as evidenced by the following results. 1) Soaking singly transgenic let-858::GFP or rpL28::GFP reporter strains (lacking the intrinsically expressed gfp hairpin) in solutions containing unrelated dsRNA (dstetA RNA or dsunc-22 RNA solutions) had no effect on GFP fluorescence levels (Figure 3). 2) Soaking doubly transgenic animals (GFP reporter + gfp hairpin) in several different control solutions, including water, M9 media, M9 media/50 mM NaCl, 0.5 mg/ml dsDNA oligonucleotides in water, 0.5 mg/ml plasmid DNA in water, and 0.1 mg/ml ssRNA in M9 did not elicit systemic loss of GFP fluorescence (Figure 3; our unpublished data). 3) Nonspecific dsRNA was ineffectual in eliciting systemic silencing in a strain lacking an essential component of the RNAi machinery (RDE-1) (our unpublished data). 4) Nonspecific dsRNA was ineffectual in eliciting systemic silencing in a strain lacking a functional sid-1 gene: doubly transgenic animals that harbored the sid-1(qt2) mutation did not exhibit systemic silencing when soaked in dstetA RNA solutions (our unpublished data). Sid-1 encodes a transmembrane protein that is required for efficient RNAi and has been implicated in systemic RNA silencing (Winston et al., 2002). The dependence on a functional Sid-1 protein demonstrates that this inducible system relies on endogenous mechanisms for dissemination of RNA silencing signals.
Table 1.
Analysis of RNAi phenotypes of soaked progeny
|
Transgenes in progeny (animals assayed)
|
||||||
|---|---|---|---|---|---|---|
|
let-858::GFP
|
let-858::GFP gfp hairpin
|
|||||
|
No. of progeny showing systemic RNAi
|
||||||
| Transgenes in parent (soaked animals) | Strong | Weak | None | Strong | Weak | None |
| Control soaking | ||||||
| let-858::GFP; myo-3::gfp hairpin | 0 | 0 | 50 | 0 | 1 | 49 |
| let-858::GFP; vit-2::gfp hairpin | 0 | 2 | 48 | 0 | 0 | 50 |
| let-858::GFP; unc-119::gfp hairpin | 0 | 0 | 50 | 0 | 0 | 50 |
| let-858::GFP | 0 | 0 | 30 | |||
| dsRNA soaking | ||||||
| let-858::GFP; myo-3::gfp hairpin | 0 | 4 | 46 | 18 | 10 | 22 |
| let-858::GFP; vit-2::gfp hairpin | 0 | 4 | 46 | 11 | 12 | 27 |
| let-858::GFP; unc-119::gfp hairpin | 0 | 1 | 49 | 2 | 9 | 41 |
| let-858::GFP | 0 | 0 | 30 | |||
We monitored RNA silencing phenotypes in the progeny of treated, doubly transgenic animals. The doubly transgenic animals give rise to two classes of progeny: doubly transgenic progeny (with GFP- and gfp hairpin-expressing transgenes) and singly transgenic progeny (with a GFP-expressing transgene only, but lacking the hairpin transgene). Extrachromosomal arrays, such as the transgene expressing the gfp hairpin, are generally lost at some frequency during meiosis (and occasionally during mitosis) in worms (Stinchcomb et al., 1985; Mello et al., 1991). Because we monitored RNA silencing in progeny of treated animals, our results provide direct evidence of some deposition of mobile silencing signals to the germline. This is demonstrated in the class of singly transgenic progeny that carried only the GFP reporter transgene, but which were the progeny of animals also expressing the gfp hairpin (Figure 3 and Table 1). Because this class of progeny no longer has a transcriptional supply of gfp hairpin, the RNA silencing phenotypes observed were likely derived from maternal deposition of gfp hairpin. These progeny animals showed a decrease in the level of GFP fluorescence, depending (as described above) on the presence in the parent of the gfp hairpin transgene and exogenous dsRNA. (We verified that these animals were lacking transgene sequences and were not mosaics by recovering affected animals after photomicroscopy and by monitoring the progeny for the absence of the transformation marker phenotype.) By using polymerase chain reaction-based assays to assess gfp hairpin transgene inheritance patterns, we had previously demonstrated that myo-3::gfp hairpin lines do produce a few animals (<10% of progeny) that do not express the transformation marker (roller) phenotype but are in fact mosaics. These mosaic animals generally gave rise to some progeny with the roller phenotype (our unpublished data). The silencing phenotypes in singly transgenic progeny were not as expressive nor as penetrant as observed in doubly transgenic progeny, and silencing was not observed in subsequent generations.) These experiments suggest an induced release of a mobile silencing trigger from cells in the parent, followed by entry of the silencing trigger into the germline, and subsequent, albeit limited, RNA silencing in progeny.
We noted promoter-dependent differences in the extent of systemic silencing elicited by the gfp hairpin in response to exogenous dsRNA. Animals expressing a gfp hairpin in muscle from the myo-3 promoter exhibited the most extensive silencing in affected animals. Animals expressing a gfp hairpin in gut from the vit-2 promoter and in neurons from the unc-119 promoter exhibited a comparatively weaker systemic silencing. Although the induced systemic silencing phenotype from the vit-2::gfp hairpin is striking in some cells of injected animals, the penetrance of induced systemic silencing within the progeny of treated animals is lower in comparison to the myo-3::gfp hairpin system. Most likely this difference is a reflection of the dynamic nature of the vit-2 promoter transgene (MacMorris et al., 1994) and that the vit-2 promoter drives expression of the gfp hairpin in fewer cells than the myo-3 promoter. The promoter-specific differences with respect to inducible RNA silencing may also be attributed to differential transcriptional abilities of the promoters, to differences in frequency of mosaicism of the gfp hairpin transgene, or may reflect genuine cell type-specific differences in dsRNA exit machinery.
Systemic Silencing from RNA Silencing “Triggers” Synthesized within Cells Is Regulated by Multiple Cellular Factors
Our observations suggesting cellular entry and exit of dsRNA in C. elegans led us to apply genetic analyses in an effort to identify components of the import-export system. The different dsRNA delivery methods allowed us to anticipate and define potentially distinct dsRNA uptake routes. Defects for any of these uptake pathways would lead to predictable, tissue-specific patterns of RNAi insensitivity. For example, animals exposed to dsRNA by using ingestion-based delivery protocols might use an environmental uptake pathway to take up dsRNA from the environment via the gut. If so, the ingestion-based delivery methods would allow us to isolate mutants that are defective in this form of uptake.
We obtained mutants defective in environmental uptake from the same genetic screen that generated RNAi-defective (rde) animals (Tabara et al., 1999). dsRNA was delivered to animals by allowing them to feed on bacteria overexpressing dsRNA, and mutants were isolated based on their failure to exhibit RNAi. Unlike the rde mutant animals, the mutants we selected still exhibit silencing phenotypes when dsRNA is injected into them (Figure 4A, d). At least two different mechanistic defects could lead to this method of delivery-dependent RNAi phenotype: a defect in dsRNA uptake or a hypomorphic defect in the RNAi mechanism. Mutations at the fed-1 locus give rise to phenotypes that seem to fit the former description.
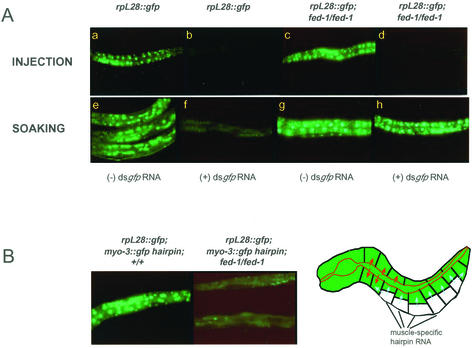
Figure 4.
fed-1(ne309) mutant animals defective in responding to ingested dsRNAs, but are not defective in systemic RNA silencing. (A) Animals were homozygous for a chromosomally integrated rpL28::GFP transgene expressing GFP in all cells: Both wild-type animals (a, b, e, and f) and fed-1 animals (c, d, g, and h) harbor this transgene. Animals were injected with (b and d) or soaked in (f and h) dsgfp RNA. RNAi (loss of fluorescence) was observed in wild-type using both methods of delivery (b and f). All cells of fed-1 animals exhibit RNAi when the animal is injected (d), but not when the animals are soaked (h) in dsgfp RNA. (The animals depicted were progeny of animals that had been injected into the gut; similar results are obtained from germline injections.) (B) fed-1 is not defective in dissemination of RNA silencing signals. An additional transgene (myo-3::gfp hairpin) was crossed into the animals in Figure 3A. Left, the gfp hairpin expressed in muscle does not elicit robust RNA silencing in cells other than muscle in a wild-type animal. Right, the gfp hairpin expressed in muscle exhibits systemic RNA silencing in a fed-1(ne309) background. Diagram, the phenotypic analyses suggest that fed-1 mutant is defective in uptake of dsRNA from gut sources (red arrows) has an intact RNA silencing mechanism in most cells and is not inhibited (possibly hyperactive) in dissemination of cell-intrinsic RNA silencing signals from somatic cells (blue arrows).
fed-1(ne309) mutants are defective in their response to dsRNA introduced from the environment, as evidenced by the failure of even high doses of dsRNA delivered by soaking to induce RNAi in any cell of the animal (Figure 4A, h). However, with respect to the relative ability to respond to injected dsRNA, the cells of fed-1 mutants, including gut cells, are comparable to the cells of wild-type animals. Most fed-1 cells, including gut, exhibit RNAi when dsRNA is delivered by injection (Figure 4A, d); in contrast, no cells of fed-1 mutants, including gut cells, exhibit RNAi when dsRNA is delivered by ingestion. The developing germline of fed-1 mutants also has the capacity for dsRNA uptake: the progeny of fed-1 mutants that were injected into the gut with dsRNA exhibit RNA silencing (Figure 4A, d).
The capacity to disseminate RNA silencing signals throughout the animal is not defective in fed-1 mutants: a myo-3::gfp hairpin dsRNA expressed within muscle cells is capable of eliciting systemic RNAi in this mutant (Figure 4B). Interestingly, systemic silencing seems more robust in this mutant compared with wild-type. The expressivity of the systemic RNA silencing phenotype is not equal for all affected mutants within the population. (This was also observed in the progeny of soaked wild-type animals.) Some doubly transgenic fed-1 mutants have little GFP fluorescence (as in Figure 4B), most have some GFP fluorescence (fluorescence in roughly 30–60% of nonneuronal cells), and a few animals express GFP in most cells (>60% of nonneuronal cells). Again, as was observed for systemically affected (soaked) wild-type animals, expressivity of systemic silencing is not a heritable attribute: When fed-1(ne309) doubly transgenic animals were reared individually, each animal gave rise to doubly transgenic progeny exhibiting the described distribution of GFP intensities, irrespective of whether the parent animal exhibited a strong degree of systemic silencing or a weak amount of systemic silencing: 5–25% of the singly transgenic progeny derived from cloned doubly transgenic animals exhibited a loss of GFP fluorescence in 20–40% of nonneuronal cells, suggesting that silencing signals in a fed-1 mutant can be distributed to progeny, most likely via germline import of GFP signals from somatic cells of the expressing parent. The silenced state of these singly transgenic animals was not transferred to subsequent generations: silenced singly transgenic fed-1 animals gave rise to fully unaffected progeny (our unpublished data). [Singly transgenic animals that exhibited silencing were carefully assayed for array mosaicism.] We have developed polymerase chain reaction-based assays to detect the presence of the gfp hairpin transgene and have observed that nonroller animals in this stock that harbor the transgene (mosaics) generally gave rise to some progeny that exhibit the roller phenotype (100/100; our unpublished data). Most (>90%) of the fed-1 singly transgenic animals that exhibited systemic silencing and that were the progeny of doubly transgenic animals did not give rise to roller progeny, confirming that the gfp hairpin array in silenced animals was lost during parental meiosis and not during embryonic mitosis.
We note similar phenotypic responses to dsRNA in a second and distinct mutant that we have termed fed-2. fed-2 mutants also fail to respond robustly to dsunc-22 RNA or dspos-1 RNA when delivered by feeding, but do respond to dsunc-22 RNA and dspos-1 RNA when delivered by injection, and do exhibit systemic RNA silencing phenotypes from a cell-intrinsic gfp hairpin (our unpublished data). Unlike wild-type animals (Figure 3), RNA silencing triggered by gfp hairpin expression in fed mutants is not influenced by ingestion of exogenous dsRNAs. The collective phenotypic observations imply that fed mutants are defective in mechanisms that allow dissemination of RNA silencing signals from dsRNAs imported by gut cells.
Although our data provides evidence of machinery for cellular export of RNA silencing signals, the nature of the exported signal is not apparent. dsRNAs undergo processing intracellularly into ∼22 nucleotide dsRNA fragments (siRNAs) that are sufficient to mediate silencing (Hamilton and Baulcombe, 1999; Parrish et al., 2000; reviewed in Tuschl, 2001), and siRNAs are capable of entering cells and eliciting RNAi phenotypes. In vivo-transcribed dsRNAs also undergo processing into siRNAs. Thus, a systemic gfp silencing signal might be exported in the form of long dsRNA, siRNAs, or an as-yet-unidentified molecule. (In plants, two distinct populations of siRNAs have been observed in tissues undergoing RNA silencing, and one class correlates with systemic RNA silencing; Hamilton et al., 2002.) Long dsRNAs (∼100 base pairs) injected into worms can enter cells and elicit RNAi in treated animals and progeny, but the ability of long, intact molecules to act systemically has not been fully investigated. Systemic phenotypes might arise as a consequence of organismal trafficking of long dsRNAs through the circulatory system and uptake by each cell. Alternatively, systemic phenotypes might result from a relay mechanism where signaling molecules (such as siRNAs) are generated within cells and are transferred to nearby cells.
We can address the ability of long dsRNA to act systemically by monitoring for RNA silencing in the progeny of animals treated with long dsRNA. dsRNA injected into the body cavity of wild-type animals can elicit RNAi in F1 progeny (Fire et al., 1998), indicating that the oocyte/germline has the capacity to take up dsRNA. Because wild-type animals process dsRNAs into siRNAs, both size species may travel to and be taken up by the developing germline cells. (Previously, siRNAs have been demonstrated to elicit RNA silencing in progeny of injected worms; Parrish and Fire, 2001; Simmer et al., 2002). In contrast, homozygous rde-4 mutant animals injected with dsRNA do not accumulate siRNAs (Parrish and Fire, 2001; Tabara et al., 2002), are defective for RNA silencing, and produce homozygous rde-4 progeny that are also defective in RNA silencing. RNA silencing is not defective in rde-4/+ heterozygous animals and this provides a means to examine the ability of large dsRNAs to act systemically: Large dsRNA molecules were introduced into rde-4 homozygotes by injection into the gut (or germline); the treated animals were crossed to wild-type males; and the resulting rde-4/+ heterozygous progeny were monitored for RNAi phenotypes (Figure 5). RNA silencing was observed in the heterozygous progeny of rde-4 homozygotes injected with dsRNA into the gut or germline. Given an inability to process large dsRNAs into siRNAs in the rde-4/rde-4 animals, these results suggest that unprocessed dsRNAs can elicit systemic RNA silencing.
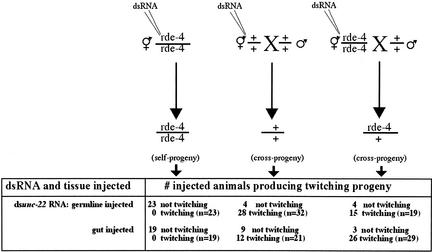
Figure 5.
Processing into siRNAs is not a requirement for import into C. elegans germ cells. Wild-type (center) and rde-4(ne299) animals (left and right) were injected with dsunc-22 RNA. The injected animals were placed individually on culture plates. Where indicated (center and right), wild-type males were also added to plates. The males harbored a PD8160 transgene (expressing GFP), thus allowing identification of cross-progeny, and only plates with cross-progeny were scored in these instances. The progeny were scored for the presence of an unc-22 phenotype (twitching). In plates where twitching was observed, 10–80% of progeny animals exhibited a strong twitching phenotype. The variation in numbers of affected progeny may reflect differences in the volume of dsRNA injected into each parent animal. (The n represents the number of plates scored and also represents the numbers of animals surviving injection or the numbers of surviving animals that produced cross-progeny expressing GFP.)
DISCUSSION
DsRNA has the capacity to act noncell autonomously, resulting in a systemically affected organism. dsRNA introduced from exogenous sources can spread to distant tissues in creatures as diverse as plants and C. elegans. Systemic responses are noted in C. elegans after injection of dsRNA into any site of the animal and also from ingestion of dsRNAs. These methods of delivery also result in affected offspring, an indication that RNA silencing signals are mobilized to the germline. Similarly, exogenous RNA silencing signals delivered locally to plants (using Agro-infiltration, viral delivery, or particle bombardment of individual leaves) culminates in a systemically affected organism (Mlotshwa et al., 2002). A different test of systemic silencing consists of intracellular (transgene-driven) dsRNA expression within a subset of cells, followed by assays for interference in the neighboring cells that lack transgene. In plant systems, this type of protocol has been shown to produce a remarkable degree of spreading of a silencing signal from transgene-expressing host to a transgene-free graft (Boerjan et al., 1994; Palauqui et al., 1997; Palauqui and Balzergue, 1999; Fagard et al., 2000). In contrast, experiments in animal systems have revealed some degree of cell autonomy: localized synthesis of an interfering RNA in a subset of tissues of Drosophila (Kennerdell and Carthew, 2000) has been shown to produce interference in a similarly localized region of the animal. Plants contain a number of systems for intercellular exchange of macromolecular components (plasmodesmata, phloem, etc.). Extracellular mRNAs of defined sequence have been proposed to traffic long distances through these structures, exiting from one cell and functioning in a more distant cell (Ruiz-Medrano et al., 2001). In contrast, specific RNAs are not generally noted outside cells in animal systems.
To distinguish between unidirectional uptake of dsRNA and bidirectional movement of dsRNA across cell boundaries, we used C. elegans strains harboring two different transgenes: one transgene produced GFP in all cells, and a second transgene produced a double-stranded gfp hairpin RNA in a subset of cells. The use of GFP as an RNAi target allowed us to monitor effects on gene expression (loss of GFP fluorescence) at the response level of individual cells. We failed to note systemic silencing from any of the three tissue-specific promoters used to drive gfp hairpin expression, and this contradicted our anticipated results. Because only a few molecules of dsRNA per cell are required for an RNAi response in C. elegans (Fire et al., 1998; Kennerdell and Carthew, 1998), we considered that systemic silencing phenotypes might reflect “leaky promoter” activity in nonspecific tissues. We would thus have been presented a need to discern the mechanism responsible for systemic loss of GFP fluorescence (leaky promoter versus cellular exit of dsRNA).
These initial results (Figures 1 and 2) raised some intriguing questions as to the robustness of dsRNA exit mechanisms in C. elegans cells and suggested that cellular export of RNA silencing molecules is not a significant aspect of the systemic RNAi response, at least for systemic silencing signals that are derived from intracellular sources. (Similarly, dsRNAs transcribed within cells do not elicit systemic RNA silencing in Drosophila; Giordano et al., 2002.) Indeed, analyses of the fed mutants suggest that at least some cells in this animal have active mechanisms to prevent systemization of RNA silencing signals that are derived from the nucleus. However, systemization of RNA silencing signals has been observed previously using similar transgene configurations in C. elegans. In one instance, the myo-2 promoter was used to drive expression of a gfp hairpin, and loss of fluorescence was observed outside the normal expression range of this promoter (pharyngeal muscle) (Winston et al., 2002). Because the ectopic RNA silencing in this system was dependent upon a functional sid-1 gene, spreading of a silencing signal (as opposed to errant transcription) is indeed implicated. (Sid-1 encodes a protein with eleven putative transmembrane-spanning regions, and sid-1 mutants ineffectively respond to dsRNA delivered exogenously; thus, sid-1 is more likely to play a role in the uptake of RNA silencing signals or in their further dissemination rather than regulation of transgene expression.) However, the ectopic silencing induced by expression of myo-2::gfp hairpin was relatively weak (silencing was not observed in all cells of the animal) and was temperature dependent. Our contrasting observations of tissue-specific silencing may be a reflection of the relative robustness of the promoters used in the different experiments. Although the myo-3, vit-2, and unc-119 regions we used are considered strong transcriptional activators based on the fluorescence intensity of GFP that accumulates when under their regulation, the myo-2 and snb-1 promoters are exceptionally strong (snb-1 promoters have also been noted to elicit RNAi outside the nervous system (Honigberg, personal communication). Thus, an ability to effect tissue-specific RNAi in C. elegans by using transgene-delivered dsRNA may depend upon the choice of promoter used to drive dsRNA expression.
After treatment with exogenous, unrelated dsRNA we observed release of an RNA silencing signal from cells (Figure 3 and Table 1). Although exposure to high concentrations of dsRNA of defined sequence is probably not a normal event in the life of native C. elegans, the effects we have observed demonstrate that dsRNA derived from the environment can influence, as well as trigger, RNA-silencing mechanisms in this organism. The systemic silencing response we observed with exogenous nonspecific dsRNA in C. elegans was in all cases a partial and limited response. One intriguing possibility is that the immediate physiological inducer of systemic silencing may not be dsRNA but rather some other molecule produced during the experiments. In this respect, it is important to recall lessons from studies of metabolism in which key regulators can be by-products rather than central intermediates in a biochemical pathway (Jobe and Bourgeois, 1972).
Given the important role of RNAi in protecting cells from viruses, it might be expected that growth conditions that are indicative of a hostile or pathogen-rich environment might also induce similar systemic RNAi responses. We have noted that certain contaminated growth media produce a systemic response similar to that observed in the presence of exogenous dsRNA and in fed-1 and fed-2 mutants (our unpublished data). Several interesting possibilities exist that might explain systemic responses in such an environment: a dsRNA virus/bacteriophage present in the media might recapitulate the experimental conditions in Figure 3 by providing an abundant source of dsRNA or other triggering molecule; some environments might provide conditions under which the C. elegans RNAi system is “primed” to handle intracellular dsRNA; alternatively, some environments might allow systemic silencing by physically interfering with the integrity of C. elegans cells by providing molecules that perforate cells, for example. The nature of the contaminating microorganisms in the growth media has not been fully characterized. These experiments reinforce the suggestion that systemic RNAi may be part of a general mechanism for sensing and responding to environmental pathogens.
Our results suggest that multiple mechanisms are used in the systemic uptake and exit of dsRNA molecules in intact animals and these mechanisms can be revealed by physiological and genetic aberrations. For example, the constitutive systemic silencing observed in fed-1 and fed-2 mutants suggests an interesting interplay between cellular dsRNA uptake and exit mechanisms in this organism. The normal activities performed by these genes might include inhibiting the manufacture of a mobile silencing signal, or inhibiting the cellular exit or uptake of a mobile silencing signal; or preventing organismal or cellular purge, sequestration, or degradation of a silencing signal. The precise mechanism that is disrupted by these mutations is not known. The phenotypes of the fed mutants contrast sharply to that of sid-1 mutants (Winston et al., 2002): sid-1 facilitates mobilization of silencing signals and defects in sid-1 lead to a failure to respond to dsRNA delivered by feeding or soaking, a failure to exhibit systemic RNAi from transgene-derived dsRNAs expressed in somatic tissue and a reduced level of RNAi response in progeny of injected mutant animals. Like sid-1 mutants, the fed mutants are defective in responding to ingested dsRNAs; however, these mutants respond much more robustly to injected dsRNA than sid-1, leading to silencing in the progeny of fed animals (our unpublished data). One of our working models for the phenotypes exhibited by fed mutants (spreading of in vivo-derived signals and lack of spreading of signals derived by ingestion) is that these different phenotypic behaviors reflect a deficiency in a tissue-specific mechanism active in gut cells.
In C. elegans, the most readily observed responses to dsRNA are sequence-specific gene silencing effects (Montgomery et al., 1998). In contrast, mammalian cells exhibit a variety of prominent responses to dsRNA that are not sequence-specific in nature (Kaufman, 1999; Williams, 1999; Majde, 2000; Barber, 2001; Levy and Garcia-Sastre, 2001). The innate immune responses to dsRNA in mammalian systems involve both intracellular and extracellular detection of dsRNA (Alexopoulou et al., 2001). These reflect in part a conservative, efficient, and evolutionarily constrained effort to eliminate viruses or invading genomes. In some cases, the mammalian response to extracellular dsRNA is thought to result from circulating virus or the death and lysis of viral-infected cells. Our observations of inducible export of dsRNA in C. elegans raise the possibility that these protective responses that act systemically in higher animals may also be mediated by deliberate cellular dsRNA export mechanisms.
Acknowledgments
We thank Bill Kelly, Steve Kostas, Jamie Fleenor, S. Parrish, J. Yanowitz, M. McMorris, T. Blumenthal, M. Maduro, D. Pilgrim, N. Kaplan, and R. Morgan for reagents and helpful comments. Some nematode strains were obtained from the Caenorhabditis Genetics Center, funded by the National Institutes of Health National Center for Research Resources. This work was funded in part by funds from National Institutes of Health (GM-37706 [to A.F.], GM-58800 [to C.M.], HD08353 [to L.T.]), and from the COBRE Program of the National Center for Research Resources.
References
- Alexopoulou, L., Holt, A.C., Medzhitov, R., and Flavell, R.A. (2001). Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413, 732-738. [DOI] [PubMed] [Google Scholar]
- Barber, G.N. (2001). Host defense, viruses and apoptosis. Cell Death Differ. 8, 113-126. [DOI] [PubMed] [Google Scholar]
- Baulcombe, D. (2002). RNA silencing. Curr. Biol. 12, R82-R84. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Caudy, A.A., Hammond, S.M., and Hannon, G.J. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363-366. [DOI] [PubMed] [Google Scholar]
- Boerjan, W., Bauw, G., Van Montagu, M., and Inze, D. (1994). Distinct phenotypes generated by overexpression and suppression of S-adenosyl-l-methionine synthetase reveal developmental patterns of gene silencing in tobacco. Plant Cell 6, 1401-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher, G., Scholten, J., and Klingler, M. (2002). Parental RNAi in Tribolium (Coleoptera). Curr. Biol. 12, R85-R86. [DOI] [PubMed] [Google Scholar]
- Caplen, N.J., Parrish, S., Imani, F., Fire, A., and Morgan, R.A. (2001). Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc. Natl. Acad. Sci. USA 98, 9742-9747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium, The. (1998). Genome sequence of the nematode C. elegans: a platform for investigating biology. Science 282, 2012-2018. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001a). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494-498. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., and Tuschl, T. (2001b). RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 15, 188-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagard, M., Boutet, S., Morel, J.B., Bellini, C., and Vaucheret, H. (2000). AGO1, QDE-2, and RDE-1 are related proteins required for post-transcriptional gene silencing in plants, quelling in fungi, and RNA interference in animals. Proc. Natl. Acad. Sci. USA 97, 11650-11654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806-811. [DOI] [PubMed] [Google Scholar]
- Giordano, E., Rendina, R., Peluso, I., and Furia, M. (2002). RNAi triggered by symmetrically transcribed transgenes in Drosophila melanogaster. Genetics 160, 637-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonczy, P., et al. (2000). Functional genomic analysis of cell division in C. elegans using RNAi of genes on chromosome III. Nature 408, 331-336. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Pasquinelli, A.E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D.L., Fire, A., Ruvkun, G., and Mello, C.C. (2001). Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106, 23-34. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Tabara, H., and Mello, C.C. (2000). Genetic requirements for inheritance of RNAi in C. elegans. Science 287, 2494-2497. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J., and Baulcombe, D.C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286, 950-952. [DOI] [PubMed] [Google Scholar]
- Hamilton, A., Voinnet, O., Chappell, L., and Baulcombe, D. (2002). Two classes of short interfering RNA in RNA silencing. EMBO J. 21, 4671-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon, G.J. (2002). RNA interference. Nature 418, 244-251. [DOI] [PubMed] [Google Scholar]
- Jobe, A., and Bourgeois, S. (1972). lac Repressor-operator interaction VI. The natural inducer of the lac operon. J. Mol. Biol. 69, 397-408. [DOI] [PubMed] [Google Scholar]
- Kaufman, R.J. (1999). Double-stranded RNA-activated protein kinase mediates virus-induced apoptosis: a new role for an old actor. Proc. Natl. Acad. Sci. USA 96, 11693-11695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly, W.G., Xu, S., Montgomery, M.K., and Fire, A. (1997). Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics 146, 227-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell, J.R., and Carthew, R.W. (1998). Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95, 1017-1026. [DOI] [PubMed] [Google Scholar]
- Kennerdell, J.R., and Carthew, R.W. (2000). Heritable gene silencing in Drosophila using double-stranded RNA. Nat. Biotechnol. 18, 896-898. [DOI] [PubMed] [Google Scholar]
- Ketting, R.F., Fischer, S.E., Bernstein, E., Sijen, T., Hannon, G.J., and Plasterk, R.H. (2001). Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev. 15, 2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klahre, U., Crete, P., Leuenberger, S.A., Iglesias, V.A., and Meins, F., Jr. (2002). High molecular weight RNAs and small interfering RNAs induce systemic posttranscriptional gene silencing in plants. Proc. Natl. Acad. Sci. USA 99, 11981-11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klink, V.P., and Wolniak, S.M. (2000). The efficacy of RNAi in the study of the plant cytoskeleton. J Plant Growth Regul. 19, 371-384. [DOI] [PubMed] [Google Scholar]
- Knight, S.W., and Bass, B.L. (2001). A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293, 2269-2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, D.E., and Garcia-Sastre, A. (2001). The virus battles: IFN induction of the antiviral state and mechanisms of viral evasion. Cytokine Growth Factor Rev. 12, 143-156. [DOI] [PubMed] [Google Scholar]
- Lohmann, J.U., Endl, I., and Bosch, T.C. (1999). Silencing of developmental genes in hydra. Dev. Biol. 214, 211-214. [DOI] [PubMed] [Google Scholar]
- MacMorris, M., Spieth, J., Madej, C., Lea, K., and Blumenthal, T. (1994). Analysis of the VPE sequences in the Caenorhabditis elegans vit-2 promoter with extrachromosomal tandem array-containing transgenic strains. Mol. Cell. Biol. 14, 484-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro, M., and Pilgrim, D. (1995). Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics 141, 977-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majde, J.A. (2000). Viral double-stranded RNA, cytokines, and the flu. J. Interferon Cytokine Res. 20, 259-272. [DOI] [PubMed] [Google Scholar]
- Mello, C.C., Kramer, J.M., Stinchcomb, D., and Ambros, V. (1991). Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959-3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlotshwa, S., Voinnet, O., Mette, M.F., Matzke, M., Vaucheret, H., Ding, S.W., Pruss, G., and Vance, V.B. (2002). RNA silencing and the mobile silencing signal. Plant Cell Suppl. 14, S289-S301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery, M.K., Xu, S., and Fire, A. (1998). RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA 95, 15502-15507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palauqui, J.C., and Balzergue, S. (1999). Activation of systemic acquired silencing by localised introduction of DNA. Curr. Biol. 9, 59-66. [DOI] [PubMed] [Google Scholar]
- Palauqui, J.C., Elmayan, T., Pollien, J.M., and Vaucheret, H. (1997). Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16, 4738-4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrish, S., Fleenor, J., Xu, S., Mello, C., and Fire, A. (2000). Functional anatomy of a dsRNA trigger: differential requirement for the two trigger strands in RNA interference. Mol. Cell 6, 1077-1087. [DOI] [PubMed] [Google Scholar]
- Parrish, S., and Fire, A. (2001). Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. RNA 7, 1397-1402. [PMC free article] [PubMed] [Google Scholar]
- Peden, K.W. (1983). Revised sequence of the tetracycline-resistance gene of pBR322. Gene 22, 277-280. [DOI] [PubMed] [Google Scholar]
- Plasterk, R.H. (2002). RNA silencing: the genome's immune system. Science 296, 1263-1265. [DOI] [PubMed] [Google Scholar]
- Ruiz-Medrano, R., Xoconostle-Cazares, B., and Lucas, W.J. (2001). The phloem as a conduit for inter-organ communication. Curr. Opin. Plant Biol. 4, 202-209. [DOI] [PubMed] [Google Scholar]
- Sanchez Alvarado, A., and Newmark, P.A. (1999). Double-stranded RNA specifically disrupts gene expression during planarian regeneration. Proc. Natl. Acad. Sci. USA 96, 5049-5054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmer, F., Tijsterman, M., Parrish, S., Koushika, S.P., Nonet, M.L., Fire, A., Ahringer, J., and Plasterk, R.H. (2002). Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12, 1317-1319. [DOI] [PubMed] [Google Scholar]
- Smith, N.A., Singh, S.P., Wang, M.B., Stoutjesdijk, P.A., Green, A.G., and Waterhouse, P.M. (2000). Total silencing by intron-spliced hairpin RNAs. Nature 407, 319-320. [DOI] [PubMed] [Google Scholar]
- Stinchcomb, D.T., Shaw, J.E., Carr, S.H., and Hirsh, D. (1985). Extrachromosomal DNA transformation of Caenorhabditis elegans. Mol. Cell. Biol. 5, 3484-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara, H., Grishok, A., and Mello, C.C. (1998). RNAi in C. elegans: soaking in the genome sequence. Science 282, 430-431. [DOI] [PubMed] [Google Scholar]
- Tabara, H., Sarkissian, M., Kelly, W.G., Fleenor, J., Grishok, A., Timmons, L., Fire, A., and Mello, C.C. (1999). The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99, 123-132. [DOI] [PubMed] [Google Scholar]
- Tabara, H., Yigit, E., Siomi, H., and Mello, C.C. (2002). The dsRNA binding protein RDE-4 interacts with RDE-1, DCR-1, and a DExH-Box helicase to direct RNAi in C. elegans. Cell 109, 861-871. [DOI] [PubMed] [Google Scholar]
- Tavernarakis, N., Wang, S.L., Dorovkov, M., Ryazanov, A., and Driscoll, M. (2000). Heritable and inducible genetic interference by double-stranded RNA encoded by transgenes. Nat. Genet. 24, 180-183. [DOI] [PubMed] [Google Scholar]
- Timmons, L., Court, D.L., and Fire, A. (2001). Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103-112. [DOI] [PubMed] [Google Scholar]
- Timmons, L., and Fire, A. (1998). Specific interference by ingested dsRNA. Nature 395, 854. [DOI] [PubMed] [Google Scholar]
- Tuschl, T. (2001). RNA interference and small interfering RNAs. Chembiochem. 2, 239-245. [DOI] [PubMed] [Google Scholar]
- Tuschl, T., Zamore, P.D., Lehmann, R., Bartel, D.P., and Sharp, P.A. (1999). Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev. 13, 3191-3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wassenegger, M. (2002). Gene silencing. Int. Rev. Cytol. 219, 61-113. [DOI] [PubMed] [Google Scholar]
- Williams, B.R. (1999). PKR; a sentinel kinase for cellular stress. Oncogene 18, 6112-6120. [DOI] [PubMed] [Google Scholar]
- Winston, W.M., Molodowitch, C., and Hunter, C.P. (2002). Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295, 2456-2459. [DOI] [PubMed] [Google Scholar]
- Yang, D., Lu, H., and Erickson, J.W. (2000). Evidence that processed small dsRNAs may mediate sequence-specific mRNA degradation during RNAi in Drosophila embryos. Curr. Biol. 10, 1191-1200. [DOI] [PubMed] [Google Scholar]