Abstract
Improved methods of specimen preparation and dual-axis electron tomography have been used to study the structure and organization of basal bodies in the unicellular alga Chlamydomonas reinhardtii. Novel structures have been found in both wild type and strains with mutations that affect specific tubulin isoforms. Previous studies have shown that strains lacking δ-tubulin fail to assemble the C-tubule of the basal body. Tomographic reconstructions of basal bodies from the δ-tubulin deletion mutant uni3-1 have confirmed that basal bodies contain mostly doublet microtubules. Our methods now show that the stellate fibers, which are present only in the transition zone of wild-type cells, repeat within the core of uni3-1 basal bodies. The distal striated fiber is incomplete in this mutant, rootlet microtubules can be misplaced, and multiflagellate cells have been observed. A suppressor of uni3-1, designated tua2-6, contains a mutation in α-tubulin. tua2-6; uni3-1 cells build both flagella, yet they retain defects in basal body structure and in rootlet microtubule positioning. These data suggest that the presence of specific tubulin isoforms in Chlamydomonas directly affects the assembly and function of both basal bodies and basal body-associated structures.
INTRODUCTION
Microtubule organizing centers (MTOCs) are responsible for the temporal and spatial organization of cytoplasmic microtubules (MTs) and thus play a central role in many biological processes. Examples of MTOCs include the centrosome that organizes spindle MTs in organisms ranging from animals to yeasts, and the basal body that initiates the MTs of cilia and flagella. Despite much ongoing research on centrioles and basal bodies (reviewed in Kellogg et al., 1994; Preble et al., 2000; Doxsey, 2001), little is known about their assembly. The factors that regulate their duplication and control their function are also largely mysterious. A combined approach, by using mutants that lose control of these processes together with high-resolution three-dimensional (3-D) structural studies, may give a deeper understanding of the molecules that provide the information necessary for centriole and basal body assembly.
The biflagellate green alga Chlamydomonas reinhardtii has been a useful organism for the study of MTOCs because genetic and molecular analysis can be used to identify the roles of specific macromolecules in basal body function. Insights into basal body structure and function have already come from the study of strains with mutations in specific tubulin isoforms. For example, δ-tubulin, the fourth member of the tubulin superfamily, plays a critical role in assembling triplet MTs in Chlamydomonas (Dutcher and Trabuco, 1998) and Paramecium basal bodies (Garreau de Loubresse et al., 2001). In addition, ε-tubulin, the fifth member of the tubulin superfamily, plays a role in assembling doublet and triplet MTs in Chlamydomonas (Dutcher et al., 2002).
Much of what is known about basal body fine structure comes from studies using classic methods for electron microscopy (EM; Ringo, 1967; Johnson and Porter, 1968; Cavalier-Smith, 1974). Each basal body has a structural polarity. Its proximal region is formed from nine sets of angled, triplet MT blades, each consisting of a complete A-tubule (containing 13 protofilaments) and two incomplete tubules called B and C (each containing 11 protofilaments). The proximal region of the basal body also contains a ninefold symmetric “pinwheel” structure at its center. The triplet MTs continue into the distal region of the basal body and distinct transitional fibers radiate out from each triplet blade. The A- and B-tubules are continuous with the doublet MTs in the flagellum, and the C-tubule terminates at the distal end of the basal body. In Chlamydomonas, there is a specialized region, the “transition zone,” between the basal body and the flagellum. This zone has an important biological function, because its component proteins have been shown to affect MT severing (reviewed in Quarmby and Lohret, 1999). Also in this region centrin is included in an elaborate structure that looks like a nine-pointed star when viewed in cross section and an osmiophilic H in longitudinal view. Immediately distal to the transition zone the central pair MTs begins, forming the classic 9 + 2 arrangement of the flagellum proper.
Several additional structures associate with the mature basal bodies to form a complicated 3-D arrangement at the anterior end of the cell. These include the proximal striated fibers that connect the two mature basal bodies at their proximal base and centrin-containing fibers that form the nucleus-basal body connector, plus a distal striated fiber (Salisbury et al., 1988; Sanders and Salisbury, 1989, 1994). Two immature, or “probasal bodies”, lie adjacent to the mature basal bodies. Finally, four bundles of rootlet MTs form a cruciate array that radiate out from the basal bodies and bends toward the cell's posterior (LeDizet and Piperno, 1986; Holmes and Dutcher, 1989).
Recently, electron tomography has been shown to be a powerful method with which to study the 3-D fine structure of MTOCs (Moritz et al., 1995a,b; Bullitt et al., 1997; O'Toole et al., 1999). This method is conceptually similar to CT scans in medical imaging, and results in computer-generated reconstructions that can be sliced and imaged in any orientation (reviewed in Frank, 1992). Thus, electron tomography is an ideal method with which to explore complex biological structures, such as the basal body. In this article, we have used dual-axis tomography to study the fine structure of basal bodies and associated organelles in 3-D. The increased resolving power of this method has revealed novel structures in wild-type cells and several important structural alterations in mutant strains.
MATERIALS AND METHODS
Chlamydomonas Strains, Culture Conditions, and Genetic Analysis
C. reinhardtii strains used in this study include wild-type 137c mt+, uni3-1, uni3-1; tua2-6; and tua2-6 (Fromherz, Gomez-Ospina, Giddings, Dutcher; unpublished data). The genotypes of the mutant strains were verified by crossing to wild-type cells and examining the flagellar phenotypes in meiotic progeny. Cultures were grown at 25°C under constant illumination in Sager and Granick medium as modified in Preble et al., 2001.
Preparation of Cells for Electron Microscopy
We have developed an improved fixation protocol that uses high pressure freezing followed by freeze substitution (Preble et al., 2001). Briefly, aliquots of cells grown in suspension were spun at 500 × g, and the pellets were resuspended in 150 mM mannitol. The samples were spun again at 500 × g and the resulting loose cell pellet was then transferred to brass sample holders and rapidly frozen in a Balzers HPM010 high-pressure freezer (BAL-TEC; Technotrade International, Manchester, NH). The frozen cells were freeze-substituted for 3 d at -90°C in 0.5% glutaraldehyde and 0.1% tannic acid in acetone, rinsed in acetone followed by 2% OsO4 in acetone at -20°C for 1 d, and then warmed to 4°C, rinsed in acetone, and embedded in epon/araldite resin.
Serial thin (50–70 nm) or thick (250–400 nm) sections were cut using a Reichart (Leica, Wetzlar, Germany) Ultracut-E microtome, and the section ribbons were collected onto Formvar-coated copper slot grids. The sections were poststained in 2% uranyl acetate in 70% methanol followed by Reynold's lead citrate. For tomography, 15-nm colloidal gold particles (BBI International, Sigma, St. Louis, MO) were affixed to each surface to serve as fiducial markers for subsequent alignment (Ladinsky et al., 1994; O'Toole et al., 1999). Finally, the grids were carbon-coated to stabilize the grids under the electron beam.
Electron Microscopy
Serial thin sections were imaged in a Philips CM10 EM (FEI, Mahwah, NJ) operating at 80 kV. Approximately 15 data sets based on serial sections of the basal body through the transition zone were collected from cells of each strain to document a particular phenotype and to aid in the interpretation of tomographic data. Thick sections were first imaged in a Philips CM10 EM at 100 kV to identify cells with basal bodies in approximate cross section. The location of the cells was mapped by imaging the section at low magnification, and the map was used to locate the same cell in a high-voltage instrument where contrast is greatly reduced.
For tomography, the grids were placed in a Gatan tilt-rotate specimen holder (model 650; Gatan, Pleasanton, CA) and imaged in a JEM1000 high-voltage electron microscope operating at 750 kV. Images were captured digitally using a semiautomated data collection procedure developed in the Boulder 3-D laboratory by using software that incorporates Digital Micrograph (Gatan) to capture images on a 1024 × 1024 pixel charge-coupled device camera (Gatan) at a pixel size of 1.4 nm. Serial, tilted views were collected every 1.5° over a +60° range. Then the grid was rotated 90° and a second tilt series was acquired. In total, 43 dual-axis tomograms were reconstructed to examine the 3-D fine structure of basal bodies in wild-type and mutant strains.
Tomographic Reconstruction and Image Analysis
Dual-axis tomographic reconstruction was carried out using the IMOD software package as described previously (Kremer et al., 1996; Mastronarde, 1997; O'Toole et al., 1999). Briefly, the tilted views were aligned using the positions of the colloidal gold particles, and tomograms were calculated using an R-weighted back projection algorithm. The two tomograms were then aligned to each other and combined. Finally, dual-axis tomograms from serial sections were aligned and combined using the methods described by Ladinsky et al. (1994, 1999) and Marsh et al. (2001).
Tomographic reconstructions were displayed and analyzed using the IMOD viewing program (Kremer et al., 1996). MTs of the basal body and rootlet bundles were tracked and modeled, and a projection of the model was displayed to study the relationships of these organelles in 3-D.
Online Supplemental Material
The figures presented in this article are selected, single frames extracted from a complete tomographic volume or model. The movie supplements that correspond to each figure contain serial tomographic slices through the entire volume of the reconstruction. All movies were generated using the dmconvert program on a Silicon Graphics Octane computer and saved in QuickTime format by using jpeg compression.
RESULTS
Tomographic Reconstruction Reveals New Structures in the Wild-Type Basal Body
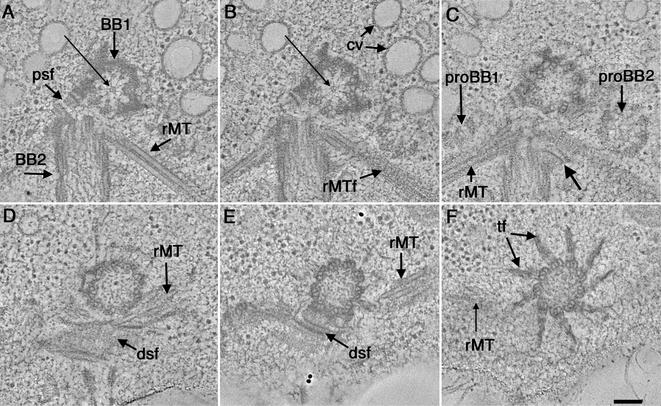
Examples of tomographic slices through a region containing wild-type basal bodies are shown in Figure 1, A–F. One basal body is shown in approximate cross section (Figure 1A, BB1) and the other in longitudinal view (Figure 1A, BB2). These six slices are spaced by 20–60 nm and display changes in basal body organization from the proximal (Figure 1A, BB1) to the distal regions of the structure (Figure 1F). The complete reconstructed volume of this basal body through the transition zone is shown in Video Sequence 1. This movie contains 216, 2.3-nm serial tomographic slices reconstructed from three serial 200-nm sections. When stepping through the serial tomographic slices, the details of the basal body and associated organelles can be appreciated in 3-D. The tomograms confirm the nine sets of angled, triplet MTs (Ringo, 1967; Cavalier-Smith, 1974), but additional features can be seen, due to the increased 3-D resolution of the tomograms.
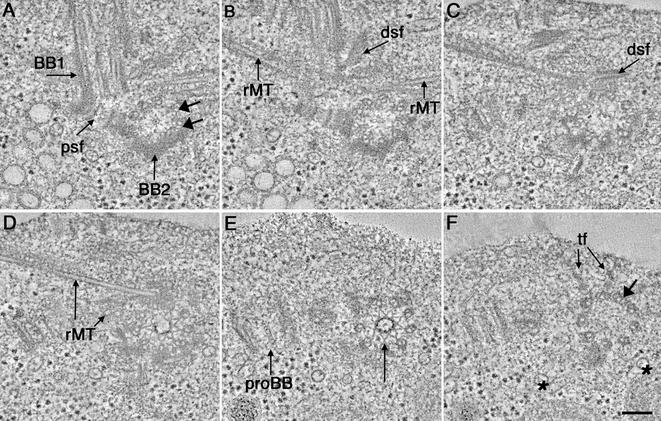
Figure 1.
Selected tomographic slices from the proximal to distal (A–F, respectively) region of a wild-type basal body complex. (A) One basal body is shown in cross section (BB1) and the other basal body in longitudinal view (BB2). The proximal base of BB1 consists of an amorphous, electron-dense ring and there is a ninefold symmetric pinwheel structure in its center (arrow). A two-membered rootlet microtubule bundle is seen (rMT). (B) The pinwheel structure of BB1 has a center formed from three rings (arrow). Other obvious features include the membrane compartments of the CV and the ordered arrangement of the striated fiber associated with the rootlet microtubule bundle (rMTf). (C) Two probasal bodies (proBB1 and proBB2) lie adjacent to the mature basal bodies. Other features include a four-membered rMT bundle as well as a fiber connecting BB2 to this rootlet bundle (arrow). (D and E) The dsf and rMTs are indicated. (F) Transitional fibers (f) radiate out from the triplets at the distal end of the basal body. Video sequence 1 shows a movie of 216 serial tomographic slices through this volume. Bar, 100 nm.
At the extreme proximal end of the basal body, there is an amorphous, electron-dense ring from which the triplet MTs emerge (Figure 1, A and B; BB1). There is also a proximal striated fiber (Figure 1A, psf) that connects the two mature basal bodies at their proximal ends. Nine sets of angled, triplet MTs, or “microtubule blades,” are linked to a central pinwheel structure (Figure 1, A and B; BB1, long arrow). The fine structure of the pinwheel, originally described by Ringo, 1967, can be further dissected into distinct structures, including nine electron-dense knobs that connect the distal ends of the pinwheel's spokes to the A-tubules of each triplet (Figure 1A, BB1), and a central hub formed from three rings (Figure 1, A and B, long arrow). When stepping through the serial tomographic slices, the structure and position of probasal bodies (proBB; Figure 1C) as well as the position of rootlet MT bundles (rMTs; Figure 1, A–F; Video Sequence 1) can be identified and followed. The order within the striated fibers that are associated with the rootlet MTs is well preserved (Figure 1B, rMTf). A fiber connecting the basal body to the rootlet MT can also be detected (Figure 1C, arrow). Probasal bodies are present in the proximal region; these are formed from nine triplet MTs. In this cell, a portion of one is seen in longitudinal view (Figure 1C, proBB1), and the other is seen in approximate cross section (Figure 1C, proBB2). These probasal bodies are much smaller than their mature counterparts. They are approximately 200 nm in width and 70–100 nm in length and can be tracked only through 40 tomographic slices (Video Sequence 1).
Specific triplets can be identified and tracked throughout the volume of these reconstructions. For example, the distal striated fiber, seen herein in transverse section (Figure 1, D and E, dsf), attaches to triplets 9,1,2 (Hoops and Witman, 1983). Thus, with markers such as the distal striated fiber, structural asymmetries that are present in the basal body can be studied in detail in 3-D. Immediately distal to the region where the distal striated fiber connects, transitional fibers can be seen radiating out from the triplet MTs (Figure 1F, tf). In this region, the triplet MT blades are not as sharply angled and form a smooth, circular arrangement. The transitional fibers are well preserved in these preparations and have a striated appearance that is not observed in chemically fixed cells.
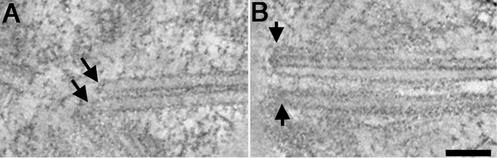
The MTs of the rootlet bundles (Figure 2A, arrows) and the MT ends at the base of the basal body (Figure 2B, arrows) can be imaged clearly in the tomographic slices. The IMOD imaging software contains a tool, the “slicer window,” that allows one to extract a slice cut at any orientation or position from the 3-D image data (Kremer et al., 1996; O'Toole et al., 1999). The tomographic slices were extracted so that the MTs could be imaged in longitudinal view, and information about their ends could be studied in detail. The MTs in the rootlet bundles are close and parallel to each other; their ends are anchored between the basal bodies. As seen in Figure 2A (arrows) these MT ends are distinctly capped. Similarly, the proximal ends of triplet MTs from the basal body are capped by dense, amorphous material (Figure 2B, arrows). Fibers attached to the basal body MT ends have also been observed (Figure 2B, top arrows).
Figure 2.
Microtubule ends are distinctly capped. (A) Arrows indicate capped microtubule ends from a two-membered rootlet bundle. (B) Triplet microtubule ends are capped at the proximal base of a basal body (arrows). Bar, 100 nm.
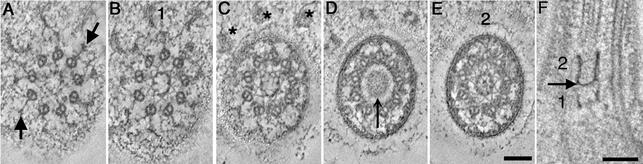
New structures have also been detected in tomographic slices through the transition zone (Figure 3; Video Sequence 1). This is the region where triplet MTs become doublets. At the proximal region of the transition zone, doublet MTs are bound to the flagellar membrane by Y-shaped connectors (Figure 3A, arrows). In Spermatozopsis similes, these structures include a 210-kDa protein (Lechtreck et al., 1999). Also in this region, the transitional fibers end in distinct knobs situated directly under the plasma membrane (Figure 3C, *; Video Sequence 1). As initially described by Ringo, 1967, the transition zone contains two distinct stellate fiber arrays that can readily be tracked through the tomographic slices. The first stellate fiber array consists of a nine-pointed star with its vertices centered on the doublet A-tubules and electrondense triangular points arranged in a circular hub at its center (Figure 3, B and C). A second stellate array is present at the distal end of the transition zone. This also consists of a nine-pointed star, but it has a much more elaborate central hub (Figure 3E). When stepping through serial tomographic slices, an amorphous disk is seen separating the two stellate arrays (Figure 3D, arrow). This disk spans only ∼10–15 nm and has never been detected in conventional thin sections. When viewed in longitudinal view, the two stellate arrays look like an osmiophilic H (Figure 3F; 1,2) made up of a distal and proximal cylinder with the amorphous disk showing as an electron-dense cross bar at the base of the distal cylinder (Figure 3F, arrow).
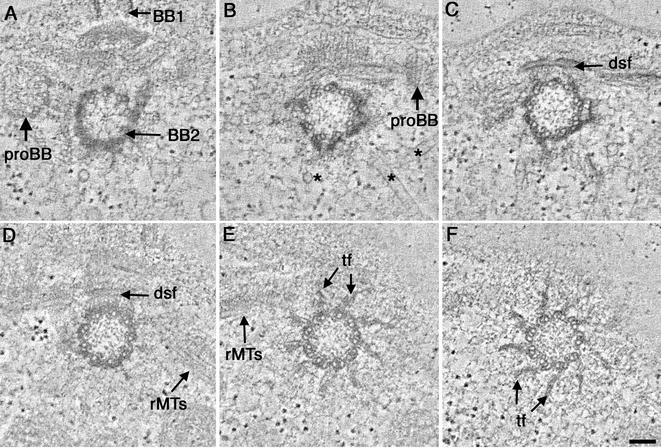
Figure 3.
Selected tomographic slices through the transition zone of a wild-type cell. (A) Proximal region of the transition zone contains doublet MTs and Y-shaped connectors (arrows). (B and C) First stellate fiber array (1) consists of a nine-pointed star containing a central hub formed from electron-dense triangular points. Distal tips of the transitional fibers form knobs that attach under the plasma membrane (C, *). (D) The first stellate fiber array is replaced by a central, amorphous disk (arrow). (E) A second stellate fiber array (2) at the distal end of the transition zone consists of a nine-pointed star with an elaborate center. (F) The two stellate fiber arrays (1, 2) look like an osmiophilic H in longitudinal view, separated by an electron-dense cross bar (arrow). Bar, 100 nm.
Transition Zone Structures Are Misplaced in the δ-Tubulin Deletion Mutant uni3-1
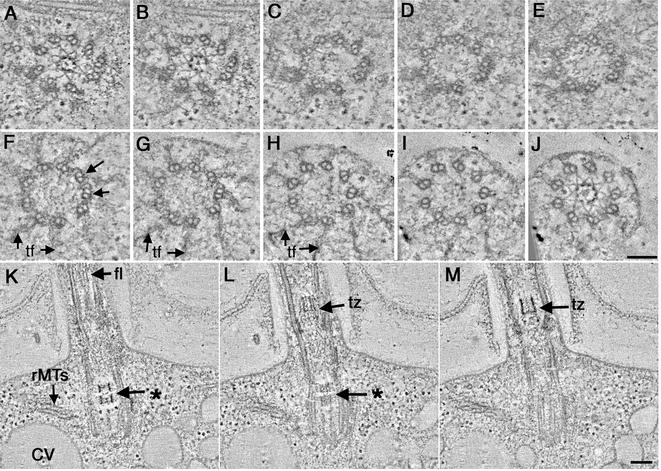
We have examined 13 tomograms and 16 sets of serial thin sections of basal bodies from uni3-1 to characterize its structural phenotype. Examination of tomographic slices confirmed that the uni3-1 basal body is formed mostly from doublet MTs (Figure 4; Video Sequence 2). In these cells, electron-dense material occupies the region where the C-tubule would normally be (Figure 4, A–E). In some basal bodies, triplet MTs were detected, but these have only been observed near the distal end of the basal body (Figure 4F, arrows). Even without the C-tubule, the doublets in the proximal region of the basal body look sharply angled, quite like the wild-type triplet blades. The pinwheel structure is present at the proximal end of the basal body (Video Sequence 2); it may be responsible for organizing the angled doublets in this region.
Figure 4.
Basal body defects in uni1-3 cells shown in cross-sectional (A–J; proximal to distal, respectively) and longitudinal view (K–M). A stellate fiber array is assembled in the proximal region of the basal body (A and B) as well as in the transition zone (J). The basal body is formed from mostly doublet MTs, yet some triplets are evident at its distal tip (F, arrows). In longitudinal view, the stellate fibers look like an osmiophilic H in the transition zone (M; tz). In this cell, a partial H is indicated in the basal body (K; *). rMTs (K) as well as membrane compartments of the CV are present. This abnormal basal body assembled a flagellum (K, fl). Bar, 100 nm.
Remarkably, in ∼40% of the uni3-1 cells examined, a stellate fiber array was detected in the proximal region of the basal body (Figure 4, A and B) as well as in the transition zone (Figure 4J), its exclusive position in wild-type cells. The stellate fibers that assemble within the uni3-1 basal body resemble the more proximal stellate fiber array that is present in the wild-type transition zone (compare Figure 4, A and B, with Figure 3, B and C). It comprises a nine-pointed star with a central hub formed from electron-dense triangular points. In all cells examined, the abnormally positioned transition zone material did not include the two stellate fiber arrays separated by an amorphous disk that is characteristic of the wild-type transition zone. Moreover, these abnormally placed stellate fibers were assembled exclusively in the proximal region of the uni3-1 basal body, and no such array was detected in the distal region of the basal body where the transitional fibers are found (Figure 4, F–H, TF; Video Sequence 2). However, as shown in Figure 4J and Video Sequence 2, the uni3-1 cells can assemble an apparently normal transition zone on these abnormal basal bodies.
As shown by others, the stellate fiber arrays that form the transition zone of wild-type cells look like an osmiophilic H in longitudinal view (Figure 3F; Ringo, 1967; Johnson and Porter, 1968; Cavalier-Smith, 1974). Tomographic reconstructions of uni3-1 cells in longitudinal view illustrate the presence of osmiophilic material, both in the proximal region of the basal body and in the transition zone (Figure 4, K–M; Video Sequence 3). Other features that normally surround the basal body are also evident, including rootlet MTs (Figure 4K, rMTs) and the membrane channels of the contractile vacuole (Figure 4K, CV). Selected tomographic slices through this cell show that the osmiophilic material within the proximal portion of the basal body does not form a complete H; the electron-dense cross bar is missing (Figure 4, K and L; *). In this cell, the osmiophilic density in the basal body only spans ∼80 nm, whereas the transition zone forms an osmiophilic H that is much longer (Figure 4, L and M; tz). Interestingly, this cell was able to build a flagellum, even with the abnormally placed transition zone material in the basal body (Figure 4, K–M, fl; Video Sequence 3).
The structures of a uni3-1 basal body that assembled a flagellum (Figure 5A; Video Sequence 4) and one that did not (Figure 5B; Video Sequence 5) were examined using electron tomography. The distal tip of the flagellum-forming basal body contained some triplet MTs (Figure 5A, arrows). This cell built a normal transition zone, consisting of doublet MTs and the two distinct stellate fiber arrays (Figure 5A; 1 and 2) separated by the amorphous disk (Figure 5A, arrow). The central pair MTs can be seen at the distal end of the transition zone (Figure 5A; cp). The adjacent serial thick sections confirmed the 9 + 2 arrangement of MTs in this flagellum (our unpublished data).
Figure 5.
Transition zones from uni3-1 cells that built a flagellum (A) and one that did not (B). (A) A subset of triplet MTs is indicated at the distal tip of the basal body (arrows). The transition zone is formed from doublet MTs, and two distinct stellate fiber arrays (1 and 2, respectively) that are separated by an amorphous disk (long arrow). The central pair of MTs is indicated at the distal tip of the transition zone (cp). (B) Triplet as well as singlet MTs is indicated at the distal tip of the basal body (arrows). Only the first stellate fiber array is assembled (1) and then the flagellar membrane closes. Bar, 100 nm.
A basal body that did not assemble a flagellum is shown in Figure 5B and Video Sequence 5. The first stellate fiber array is present and consists of a nine-pointed star and a central hub formed from electron-dense triangular points (Figure 5B, 1). Distal to the first stellate array, the MTs become disorganized and the flagellar membrane closes (Figure 5B; Video Sequence 5). Membrane blebs can be seen at positions beyond the region where the flagellar membrane has closed (Figure 5B; Video Sequence 5). Cells containing short, cone-shaped flagella were often seen in these preparations as reported previously (Dutcher and Trabuco, 1998). It is possible that these cells are more sensitive and have undergone flagellar autonomy.
The δ-Tubulin Deletion Mutant uni3-1 Contains Incomplete Fiber Systems
The distal striated fiber is a centrin-containing fiber system that is characterized by alternating dense and lightly stained filaments (Ringo, 1967; Cavalier-Smith, 1974; Salisbury et al., 1988). Selected tomographic slices through a wild-type cell show a distal striated fiber connecting the two mature basal bodies at their distal ends (Figure 6, dsf). The robust nature of this fiber system can be fully appreciated by stepping through the complete tomographic reconstruction of the wild-type cell in Video Sequence 6. The rootlet MTs are positioned immediately below this distal striated fiber (Figure 6A wild-type, rMTs; Video Sequence 6). In addition, the osmiophilic H shape of the transition zones is evident when stepping through tomographic slices (Figure 6A, wild type; *).
Figure 6.
The distal striated fiber connects the two wild-type basal bodies but is not assembled correctly in uni3-1 cells. (A) Selected tomographic slices through two wild-type basal bodies in longitudinal view. The dsf connects the two basal bodies at their distal ends. Bundles of rMTs are anchored in the dense plate beneath the distal striated fiber. The transition zone of each basal body looks like an osmiophilic H (*). (B) Selected tomographic slices through two uni3-1 basal bodies in longitudinal view are not connected at their distal ends but are connected at their proximal base by a pfs. Osmiophilic material assembles within the basal body as well as in the transition zone (*). RMTs look disorganized. Bar, 100 nm.
Analogous images of uni3-1 cells reveal an incomplete distal striated fiber (Figure 6B uni3-1, *; Video Sequence 7). Although some fibrous material can be detected, the robust fiber of wild-type cells is not assembled here. The classic H-shaped density of the transition zone can be seen (Figure 6B, uni3-1; * above left basal body), as well as osmiophilic material in the basal body core (Figure 6B, uni3-1; * on right basal body). The two mature basal bodies in this cell are, however, connected by a fiber at their proximal ends (Figure 6B, uni3-1, psf). The rootlet MTs that would normally be anchored directly below the distal striated fiber (Figure 6A wild-type, rMTs) seem misplaced in this uni3-1 cell (Figure 6B, uni3-1, rMTs; Video Sequence 7).
uni3-1 Cells Carrying the tua2 Suppressor Retain Basal Body Defects
Fromherz, Gomez-Ospina, Giddings, Dutcher (unpublished data) have identified extragenic suppressors of the uni3-1 strain that restore flagellar number in the absence of δ-tubulin. One of these loci maps to α2-tubulin. Tomographic analysis of 11 basal bodies from tua2-6; uni3-1 cells has revealed a number of subtle structural defects (Figure 7; Video Sequence 8). For example close examination of the basal bodies shown in Figure 7 reveals that triplet MTs can be detected in the proximal region of the basal body (Figure 7A; BB2, arrows) and again in the distal portion (Figure 7F, arrows), but the C-tubules are not continuous over the full length of the basal body. Otherwise, the basal bodies and their associated structures seem quite normal. For example, the two mature basal bodies (Figure 7A; BB1, BB2) are connected be a proximal striated fiber (Figure 7A, psf) and a probasal body is seen (Figure 7, D and E, arrow).
Figure 7.
Selected tomographic slices through the basal body region of a tua2-6; uni3-1 cell (A–F, proximal to distal, respectively). (A) One basal body is seen in longitudinal view (BB1) and the other in cross section (BB2). The two basal bodies are connected at their proximal bases by a psf. A subset of triplet microtubules present in BB2 is indicated (arrows). (B) rMTs as well as a portion of the dsf can be seen. (C) dsf. (D) Bundles of rMTs can be seen in cross and longitudinal view. (E) proBB and a stellate fiber array is assembled within BB2 (arrow). (F) Transitional fibers are indicated at the distal tip of BB2 (tf) and some triplet microtubles are present (arrow). Membranous tubes traverse the volume of the tomogram (*). Bar, 100 nm.
Tomographic reconstructions of the tua2-6; uni3-1 strain, reveal that fiber systems in this mutant are often incomplete or misplaced. In Figure 7, a stellate fiber array assembles in the proximal region of the basal body (Figure 7, D and E; arrow) but is excluded from the distal portion of the basal body where transitional fibers are formed (Figure 7F, tf). When stepping through serial tomographic slices we saw membranous tubes that were not present in wild-type or uni3-1 cells (Figure 7F, *; Video Sequence 8). These tubes are different in organization from the membrane system of the contractile vacuole, and they have been detected in all tua2-6; uni3-1 cells examined. The distal striated fiber in this cell does not seem to connect the distal ends of the two mature basal bodies (Figure 7, B and C; dsf). Serial tomographic slices show that the rootlet MT bundles are also not anchored properly in this cell (Video Sequence 8). Thus, although the δ-tubulin–lacking cells that carry this suppressor regain the ability to build both flagella, defects in basal body structure and position of associated organelles remain.
tua2-6 Cells Have Wild-Type Basal Body Morphology
Tomographic analysis of 11 basal bodies from cells carrying only this suppressor allele, designated tua2-6, show that these strains retain the ability to assemble triplet MTs in the basal body (Figure 8, A–F). Tomographic slices through a proximal region of the basal body show that normal features, such as the dense plate and pinwheel, can be identified (Figure 8A) as well as normal-looking probasal bodies (Figure 8, A and B; proBB). A distal striated fiber is also present in this cell, connecting the basal body in cross section to the adjacent longitudinal basal body (Figure 8, B and C; dsf). Membranous tubes similar to those detected in tua2-6; uni3-1 were also seen in the tua2-6 strain (Figure 8B).
Figure 8.
Selected tomographic slices through a tua2-6 basal body (A–F; proximal to distal, respectively). Probasal bodies (A and B; proBB) and membrane tubes (*) are present in the proximal region of the basal body. The distal striated fiber, shown in transverse section (dsf; C and D) connects the two basal bodies, and bundles of rMTs (D and E) can be detected. Transitional fibers radiate out from the triplet microtubule blades at the distal end of BB2 (E and F). Bar, 100 nm.
Video Sequence 9 is a movie of serial tomographic slices through the proximal region of the basal body in Figure 8, A–C, as well as the tomographic reconstruction of the adjacent serial section containing the distal portion of this cell's basal body. The latter includes the transition zone and a short segment of the flagellum from this tua2-6 cell. When stepping through the serial, tomographic slices it is evident that the stellate fibers are only detected in the transition zone of this strain (Video Sequence 9). Selected tomographic slices through a distal portion of the basal body from another tua2-6 cell are shown in Figure 8, D–F, and serial tomographic slices in Video Sequence 9. Bundles of rootlet MTs seem to be placed normally in this cell (Figure 6, D and E). The transitional fibers that radiate out from the triplet MT blades (Figure 7, E and F; Video Sequence 9) seem similar to those of wild-type cells.
3-D Relationships of Organelles with Basal Bodies in Wild-Type and Mutant Strains
In two of the strains used in this study (uni3-1 and tua2-6; uni3-1) the rootlet MTs were misplaced with respect to the basal bodies. To display the relationships of organelles in 3-D, the positions of MTs were tracked through the tomographic volumes and modeled. Figure 9 shows models of rootlet MT bundles and the MTs of only one basal body for reference from wild-type and mutant strains. The other mature basal body, as well as the probasal bodies, were left out of the model for simplicity. The four bundles of rootlet MTs are arranged in a cruciate array in wild-type cells, with two bundles containing a 3-over-1 MT arrangement (Figure 9A, purple) and two bundles containing two MTs (Figure 9A, light blue). The nine sets of angled triplet MTs that are a hallmark of the wild-type basal body are also seen (Figure 9A, green).
Figure 9.

Distribution of rootlet microtubules in wild-type and mutant strains. (A) In wild-type cells, four bundles of rootlet microtubules form a cruciate array. The triplet microtubules of one basal body (green) were modeled for reference. Four-membered rootlets display a 3-over-1 microtubule arrangement (pink) and two membered rootlets contain two roughly parallel microtubles. (B) Examples of rootlet microtubule bundles from two different uni3-1 cells. The basal bodies are formed from mostly doublet microtubules (green) although some triplets can be detected. The rootlet microtuble bundles look disorganized. Some four-membered bundles (pink) are present, but often they do not form a 3-over-1 arrangement. Two–membered bundles were observed (blue) as well as microbules that did not bundle (yellow). (C) tua2-6; uni3-1 cells contained both four-membered (pink) and two membered (blue) but their arrangement was not completely normal. The basal body (green) contains both doublet and triplet microtubles. (D) Cells that are simply tua2-6 contain basal bodies with triplet microtubules (green) and four-membered (pink) and two-membered (blue) rootlet microtubule bundles with a normal, cruciate arrangement.
Models from two uni3-1 cells are shown in Figure 9B. Basal bodies were formed from mostly doublet MTs (Figure 9B, green). The basal body doublets were angled correctly yet some doublets seemed abnormally spaced in the basal body (Figure 9B, top). In some cells, the rootlet bundles looked grossly disorganized. In these cells, four-membered rootlets could be identified (Figure 9B, purple), but they were often not organized in a 3-over-1 MT pattern. MTs were also detected in the region of the basal body complex that were not associated with any type of bundle (Figure 9B, yellow). The positioning of rootlet bundles was quite variable, with some cells containing more organized bundles (our unpublished data).
Cells carrying uni3-1 and its suppressor tua2-6 retained structural defects in their basal body complex (Figure 9C). Doublet and triplet MTs were present within the same basal body (Figure 9C, green). The C-tubule of the triplets was not continuous through the length of the basal body. In this cell, the rootlet MTs formed normal bundles with two fourmembered bundles (Figure 9C, purple) and two two-membered bundles (Figure 9C, light blue), yet their position relative to the basal body was abnormal.
Cells carrying the tua2-6 allele in a wild-type UNI3 background looked wild-type in MT organization (Figure 9D). Basal bodies contained nine sets of angled, triplet MTs (Figure 9D, green). The rootlet MT bundles were organized in a cruciate array as in wild-type cells. The four-membered rootlets were organized in the normal, 3-over-1 arrangement (Figure 9D, purple). The MTs of two-membered rootlets shown in Figure 9D (light blue) continued out the volume of the reconstruction and were not abnormally shortened.
DISCUSSION
The Value of Tomography for Study of Complex Cellular Substructures in 3-D
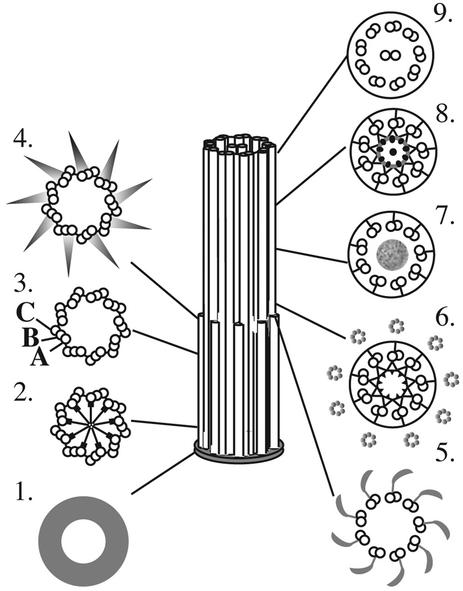
The tomographic analyses described herein complement and extend what has been learned about basal body structure from conventional electron microscopy. The increased 3-D resolving power of this method, combined with improved methods for specimen preparation that are based on rapid freezing, has allowed us to identify new structures in the wild-type basal body (Figure 10). Novel details of the pinwheel structure from the proximal basal body region (Figure 10; 2), the striated morphology of the transitional fibers (Figure 10; 4), as well as the identification of the amorphous disk in the transition zone (Figure 10; 7) are a few of the examples that were described. The rootlet MT bundles that are associated with the basal body complex were also tracked, and the projected 3-D models have revealed structural phenotypes that were not detected in a single thin section.
Figure 10.
Structural organization of the wild-type basal body, transition zone, and flagellum as modified from Preble et al. (2001). Cross sections (1–9) are displayed from proximal to distal, respectively. Tomographic analysis has revealed new features in the central hub of the pinwheel (2) and a striated appearance of the transitional fibers (4). New detail in the transition zone include an amorphous disk (7) that separates the two stellate fiber arrays (6 and 8).
Tomography allows the investigator to select the orientation of slices cut from the image data to obtain the most informative views of specific organelles (Kremer et al., 1996; O'Toole et al., 1999). Using this technique, we imaged the ends of MTs in rootlet bundles adjacent to the basal bodies, as well as the triplet MT ends at the proximal base of the basal body. The rootlet MT bundles are anchored in the dense plate, directly under the distal striated fiber (Ringo, 1967), and our tomographic slices showed these ends to be distinctly capped. The triplet MT ends at the proximal end of the basal body were similar. All these MT ends are strikingly similar to those found adjacent to the spindle pole body in budding yeast, which are characterized by a distinct cap with fibrous material connecting them to the spindle pole body proper (O'Toole et al., 1999). Similarly, tomographic analysis of Drosophila centrosomes, in combination with immunolocalization, has identified γ-tubulin in a complex with other proteins, as a cap at the MT minus ends (Moritz et al., 1995a, 2000). In Chlamydomonas, γ-tubulin has been localized to the proximal region of the basal body (Silflow et al., 1999), so it seems likely that a cap-like structure composed of γ-tubulin and other proteins is a conserved feature of MT minus ends.
The basal body of Chlamydomonas has rotational as well as proximal/distal asymmetry. It also displays positional asymmetry of organelles relative to the basal body complex (Holmes and Dutcher, 1989). Tomography is an ideal method for studying positional asymmetry because one can step through serial tomographic slices and understand the complexity of the volume reconstructed. The technique can also be of value to study the phenotypes of strains that carry mutations that disrupt cell asymmetry. It would however, be less useful for analysis of global cellular asymmetries, due to the large number of serial thick sections that would be required.
Presence of Specific Tubulin Isoforms Affects Basal Body Structure
After its discovery in Chlamydomonas, δ-tubulin has been identified in a number of organisms, including ciliates, trypanosomes, rodents, and humans (reviewed in Schiebel, 2000; Oakely, 2000). Although antibodies to δ-tubulin localize to the basal bodies in Chlamydomonas (Dutcher, 2001) different localization patterns have been reported in animal cells. Although Chang and Stearns (2000) found δ-tubulin at the centrosomes of H20S human sarcoma cells, Smrkza et al. (2000) reported centrosome localization in only mitotic C2 myoblast cells. They also found high levels of δ-tubulin expression in mouse testis where p localizes to the centriolar vaults and the manchette of the mouse spermatid. Furthermore, a clear homolog of δ-tubulin is absent in Saccharomyces cerevisiae, Caenorhabditis elegans, Drosophila, and Arabidopsis thaliana (Dutcher, 2001). These organisms, with the exception of Drosophila, do not assemble basal bodies/centrioles with triplet MTs. The δ-tubulin gene may therefore be present only in those organisms where the assembly of triplet MTs is necessary.
Our tomograms confirmed the observation made previously that basal bodies in cells with a δ-tubulin deletion contain mostly doublet MTs; the C-tubule is missing. However, the tomograms have also revealed that a subset of triplets can be found at the extreme distal end of uni3-1 basal bodies that were competent to build a flagellum. Tomograms and thin section analysis showed that the C-tubule was also assembled in part of the basal bodies in tua2-6; uni3-1 cells. Here, too, the C-tubule was not continuous over the entire basal body but was most commonly seen at its extreme distal end.
The partial triplet MTs detected in the mutant strains raises questions about basal body assembly; perhaps the distal region of the basal body is assembled or modified before probasal body elongation. Such distal localization has been observed with other basal body proteins. For example p210 is a component of the Y-shaped connectors in the green alga S. similis. This protein localizes to the distal end of the basal body, but also to probasal bodies of interphase cells (Lechtreck et al., 1999). More recently the distal basal body protein Vfl1p has been detected in the probasal bodies of Chlamydomonas interphase cells (Silflow et al., 2001). Modification of the distal end of basal bodies in Chlamydomonas, such as assembly of the C-tubule, may also occur first, which would result in triplets being seen at the extreme distal end of the basal body in uni3-1 and tua2-6; uni3-1 strains.
Another possibility is that C-tubules may assemble in the absence of δ-tubulin but be unstable and disassemble. In Paramecium, basal bodies first assemble a ring of nine singlet MTs followed by the assembly of the B- and C-tubule, respectively (Dippel, 1968). Assembly of MTs in the Chlamydomonas basal body has been studied using strains that carry mutations that affect basal body templating. The bld2-1 strain contains incomplete basal bodies (Goodenough and St. Clair, 1975) and results in cytokinesis defects (Ehler et al., 1995). Goodenough and St. Clair (1975) noted that the bld2-1 cells had primarily rings of nine singlet MTs; rarely, they observed doublet and triplet MTs that seemed to be fraying or disassembling. bld2-1 basal bodies in the presence of the extragenic rgn1-1 suppressor contain rings of MTs in various stages of assembly (Preble et al., 2001). It is likely that the assembly of basal bodies in Chlamydomonas is similar to that of Paramecium and that the loss of the C-tubule seen in strains carrying a δ-tubulin deletion is a consequence of instability in the C-tubule. The subset of triplets that is retained at the distal end of some uni3-1 and tua2-6; uni3-1 cells may be stabilized by the transitional fibers or by MT capping proteins at the distal end. Thus, δ-tubulin may be needed for maintenance of the C-tubule rather than its initiation. Immunoelectron microscopy of these strains with antibodies to δ-tubulin may help to resolve these issues.
Structural Consequences of Basal Bodies Defects
It is likely that the absence of triplet MTs in the basal bodies of strains carrying the δ-tubulin deletion gives rise to down-stream effects that produce the structural phenotypes observed in this study. In wild-type basal bodies, the centrincontaining stellate fibers are assembled only in the transition zone, where doublets MTs are present. Our study confirms a structural polarity to the wild-type transition zone that consists of two stellate fiber arrays with distinct morphologies (Ringo, 1967). These two arrays were separated physically by an amorphous disk, a structure that had not previously been identified. In the uni3-1 cells and tua2-6; uni3-1 cells, transition zone material assembled within the core of the basal bodies in a large percentage of the cells examined. Furthermore, when viewed in cross section, this array resembled the more proximal transition zone star that assembles in wild-type cells. Where does the material come from? Strains carrying a missense mutation in VFL2 lack assembled centrin fibers, but unassembled centrin is localized to the lumen of the basal body (Taillon et al., 1992). Perhaps unassembled centrin is normally present in the lumen of the Chlamydomonas basal body, and the presence of doublet, rather than triplet, MTs in the basal body is a signal to assemble the stellate fibers. Altered transition zones have also been detected in other uniflagellate strains of Chlamydomonas, including uni1-1 and uni1-2, but the ectopic material was placed distal to the transition zone in these cells (Huang et al. 1982), providing further evidence that the transition zone material only assembles in regions where doublet MTs are present.
The aberrant stellate fiber array that assembled in uni3-1 and uni3-1; tua2-6 cells was observed in the proximal region of the basal body and was excluded from the more distal region of the basal body that contains transitional fibers. Although the length of the aberrant transition zone material within the basal body varied from cell to cell, its placement within the core of the basal body had a distinct polarity. Recently, the VFL1 gene product has been localized to a subset of triplet MTs at the distal lumen of the basal body (Silflow et al., 2001). Perhaps the presence of distinct proteins such as Vfl1p may interfere with the abnormal assembly of stellate fibers in this region of uni3-1 or tua2-6; uni3-1 basal bodies.
In wild-type basal bodies, the distal striated fiber normally assembles on triplets 9, 2, and 1 in a region directly proximal to that containing the transitional fibers (Hoops and Witman, 1983). Our images show that this fiber normally connects to the A-, B-, and C-tubules of the triplet. Perhaps the absence of the C-tubule in this region of uni3-1 basal bodies prevents the assembly of the distal striated fiber. Disorganized or mislocalized distal striated fibers have also been observed in bld2-1 cells, which contain incomplete basal bodies (Preble et al., 2001). Basal bodies from these strains contain singlet, rather than triplet MTs so mislocalized distal striated fibers may again be a structural consequence of the lack of complete triplet MTs.
The 3-D relationships between rootlet MT bundles and the basal body complex revealed useful information about positional defects in the mutants studied. For example, it was not obvious in thin sections or individual tomographic slices that rootlet MTs had become unorganized. When the MTs were tracked and their 3-D model displayed, the abnormalities became obvious. Previously, it was shown that the uni3-1 strain had a large fraction of cells with cleavage furrow defects (Dutcher and Trabuco, 1998; Fromherz, Gomez-Ospina, Giddings, Dutcher; unpublished data). Because the rootlet MTs are responsible for positioning the cleavage furrow (Ehler et al., 1995), it is not surprising that some uni3-1 cells display cleavage abnormalities.
Defects in rootlet MT positioning and cleavage furrow placement have also been described for strains in which the distal striated fiber is not present or is assembled incorrectly. In the variable flagellar mutants, vfl1, vfl2, and vfl3 the distal striated fiber is either absent or only partially assembled (Wright et al., 1983, 1985; Adams et al., 1985). The dense plate that normally is present beneath the distal striated fiber is also absent. Because the rootlet MTs are anchored in the dense plate, it follows that defects in distal striated fiber assembly lead to defects in dense plate assembly and disorganization of rootlet MTs. The disorganization of rootlet MTs then gives rise to the cleavage furrow defects in these strains. Thus, the structural defects described in this study can be explained by a similar mechanism in which the distal striated fiber cannot assemble properly on the abnormal basal body doublet MTs, leading to defects in the dense plate assembly and rootlet MT anchoring, and ultimately to cleavage furrow defects.
Basal Body Maturation as a Mechanism to Overcome Structural Defects
Structural and biochemical differences between old and new basal bodies/centrioles have been observed in many organisms. In animal cells, for example, the mother centriole contains distal appendages, initiates the primary cilium, and elaborates the pericentriolar material that initiates and anchors cytoplasmic MTs (reviewed in Doxsey, 2001). Antibodies that recognize cenexin (Lange and Gull, 1995) show exclusive association with the mother centriole. Observations of living cells by using green fluorescent protein-centrin have documented differences in centriole behavior during the cell cycle, with the mother centriole retaining a central position in the cell, whereas the daughter centriole is motile through the cytoplasm (Piel et al., 2000). Just before centriole replication, the movements and characteristics of the daughter become indistinguishable from those of the mother centriole, which suggests a maturation of the daughter centriole throughout interphase (Piel et al., 2000). Thus, there is precedent for maturation of the centriole during the cell cycle.
In Chlamydomonas, the uni1 strain forms only a single flagellum, and this flagellum is assembled on the mother basal body rather than the daughter. This flagellum, which is opposite to the eyespot, is the older, more mature basal body (Huang et al., 1982; Holmes and Dutcher, 1989). Previous studies have established that uni3-1 cells can contain zero, one, or two flagella and that flagellar number is dependent on the mitotic history of the cell (Dutcher and Trabuco, 1998). The flagellum that assembles in uni3-1 cells also assembles on the older basal body. It is possible that as the basal body in these cells matures, various proteins are slowly recruited to its different regions.
The suppressor, tua2-6 in combination with the uni3-1 cells may restore some features of the basal body as a result a modification to α-tubulin (Fromherz, Gomez-Ospina, Giddings, Dutcher; unpublished data). Basal bodies in these tua2-6; uni3-1 cells are competent to build both flagella and may bypass or speed the cell cycle maturation necessary for uni3-1 cells. In summary, the data presented show that the presence or absence of specific tubulin isoforms directly affects the 3-D structural organization and function of the basal body complex in Chlamydomonas.
Supplementary Material
Acknowledgments
We thank David Mastronarde for development of the automated image capture software and for helpful advice for tomographic reconstruction. We thank Andrew Staehelin for use of the highpressure freezer and Mary Morphew for useful suggestions for specimen preparation. This work was supported by grant RR-00592 to J.R. McIntosh from the National Center for Research Resources of the National Institutes of Health.
The online version of this article contains video material for some figures. Online version is available at www.molbiolcell.org.
References
- Adams, G.M.W., Wright, R.L., and Jarvik, J.W. (1985). Defective temporal and spatial control of flagellar assembly in a mutant of Chlamydomonas reinhardtii with variable flagellar number. J. Cell Biol. 100, 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullitt, E., Rout, M.P., Kilmartin, J.V., and Akey, C.W. (1997). The yeast spindle pole body is assemble around a central crystal of Spc42p. Cell 89, 1077-1086. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith, T. (1974). Basal bodies and flagellar development during the vegetative cell cycle and the sexual cycle of Chlamydomonas reinhardtii. J. Cell Sci. 16, 529-556. [DOI] [PubMed] [Google Scholar]
- Chang, P., and Stearns, T. (2000). δ-Tubulin and ε-tubulin: two new human centrosomal tubulins reveal new aspects of centrosome structure and function. Nat. Cell Biol. 2, 30-35. [DOI] [PubMed] [Google Scholar]
- Dippel, R.V. (1968). The development of basal bodies in Paramecium. Proc. Natl. Acad. Sci. USA 61, 461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey, S. (2001). Re-evaluating centrosome function. Nat. Rev. Mol. Cell. Biol. 2, 688-698. [DOI] [PubMed] [Google Scholar]
- Dutcher, S.K. (2001). The tubulin fraternity: alpha to eta. Curr. Opin. Cell Biol. 13, 49-54. [DOI] [PubMed] [Google Scholar]
- Dutcher, S.K., Morrissette, N.S., Preble, A.M., Rackley, C., and Stanga, J. (2002). Epsilon tubulin is an essential component of the centriole. Mol. Biol. Cell 13, 3859-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher, S.K., and Trabuco, E.C. (1998). The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes δ-tubulin, a new member of the tubulin superfamily. Mol. Biol. Cell 9, 1293-1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehler, L.L., Holmes, J.A., and Dutcher, S.K. (1995). Loss of spatial control of the mitotic spindle apparatus in a Chlamydomonas reinhardtii mutant strain lacking basal bodies. Genetics 141, 945-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank, J. (1992). Introduction: principles of electron tomography. In: Electron Tomography: Three Dimensional Imaging with the Transmission Electron Microscope, ed. J. Frank, New York: Plenum Press, 1-13.
- Garreau de Loubresse, N., Ruiz, F., Beisson, J., and Klotz, C. (2001). Role of delta-tubulin and the C-tubule in assembly of Paramecium basal bodies. BMC Cell Biol. 2, 4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough, U. W., and St. Clair, H. S. (1975). BALD-2: a mutation affecting the formation of doublet and triplet sets of MTs in Chlamydomonas reinhardtii. J. Cell Biol. 66: 480-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes, J.A., and Dutcher, S.K. (1989). Cellular asymmetry in Chlamydomonas reinhardtii. J. Cell Sci. 94, 273-285. [DOI] [PubMed] [Google Scholar]
- Hoops, H.J., and Witman, G.B. (1983). Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonas flagella. J. Cell Biol. 97, 902-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, B., Ramanis, Z., Dutcher, S., and Luck, D.L. (1982). Uniflagellar mutants of Chlamydomonas: evidence for the role of basal bodies in transmission of positional information. Cell 29, 745-753. [DOI] [PubMed] [Google Scholar]
- Johnson, U.G., and Porter, K.R. (1968). Fine structure of cell division in Chlamydomonas reinhardtii. Basal bodies and microtubules. J. Cell Biol. 38, 403-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg, D.R., Moritz, M., and Alberts, B.A. (1994). The centrosome and cellular organization. Annu. Rev. Biochem. 63, 639-674. [DOI] [PubMed] [Google Scholar]
- Kremer, J.R., Mastronarde, D.N., and McIntosh, J.R. (1996). Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116, 71-76. [DOI] [PubMed] [Google Scholar]
- Ladinsky, M.S., Kremer, J.R., Furcinitti, P.S., McIntosh, J.R., and Howell, K.E. (1994). HVEM tomography of the trans-Golgi network: structural insights and identification of a lace-like vesicle coat. J. Cell Biol. 127, 29-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladinsky, M.S., Mastronarde, D.N., McIntosh, J.R., Howell, K.E., and Staehelin, L.A. (1999). Golgi structure in three dimensions: functional insights from the normal rat kidney cell. J. Cell Biol. 144, 1135-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange, B.M., and Gull, K. (1995). Molecular marker for centriole maturation in the mammalian cell cycle. J. Cell Biol. 130, 919-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechtreck, K-F., Teltenkötter, A., and Grunow, A. (1999). A 210 kDa protein is located in a membrane-microtubule linker at the distal end of mature and nascent basal bodies. J. Cell Sci. 112, 1633-1644. [DOI] [PubMed] [Google Scholar]
- LeDizet, M., and Piperno, G. (1986). Cytoplasmic microtubules containing acetylated alpha-tubulin in Chlamydomonas reinhardtii: spatial arrangement and properties. J. Cell Biol. 103, 13-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh, B.J., Mastronarde, D.N., Buttle, K.F., Howell, K.E., and McIntosh, J.R. (2001). Organellar relationships in the Golgi region of the pancreatic beta cell line, HIT-T15, visualized by high resolution electron tomography. Proc. Natl. Acad. Sci. USA 98, 2399-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde, D.N. (1997). Dual-axis tomography: an approach with alignment methods that preserve resolution. J. Struct. Biol. 120, 343-352. [DOI] [PubMed] [Google Scholar]
- Moritz, M., Braunfeld, M.B., Fung, J.C., Sedat, J.W., Alberts, B.M., and Agard, D.A. (1995a). Three-dimensional structural characterization of centrosomes from early Drosophila embryos. J. Cell Biol. 130, 1149-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moritz, M., Braunfeld, M.B., Guenebaut, V., Heuser, J., and Agard, D.A. (2000). Structure of the γ-tubulin ring complex: a template for microtubule nucleation. Nat. Cell Biol. 2, 365-370. [DOI] [PubMed] [Google Scholar]
- Moritz, M., Braunfeld, M.B., Sedat, J.W., Alberts, B., and Agard, D.A. (1995b). Microtuble nucleation by γ-tubulin-containing rings in the centrosome. Nature 378, 638-640. [DOI] [PubMed] [Google Scholar]
- Oakley, B.R. (2000). An abundance of tubulins. Trends Cell. Biol. 10, 537-542. [DOI] [PubMed] [Google Scholar]
- O'Toole, E.T., Winey, M., and McIntosh, J.R. (1999). High voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell 10, 2017-2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel, M., Meyer, P., Khodjakov, A., Reider, C.L., and Bornens, M. (2000). The respective contributions of mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 149, 317-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preble, A.M., Giddings, T.H., and Dutcher, S.K. (2000). Basal bodies and centrioles: their function and structure. Curr. Top. Dev. Biol. 49, 207-233. [DOI] [PubMed] [Google Scholar]
- Preble, A.M., Giddings, T.H., and Dutcher, S.K. (2001). Extragenic bypass supressors of mutations in the essential gene BLD2 promote assembly of basal bodies with abnormal microtubules in Chlamydomonas reinhardtii. Genetics 157, 163-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarmby, L.M., and Lohret, T.A. (1999). Microtubule severing. Cell Motil. Cytoskeleton 43, 1-9. [DOI] [PubMed] [Google Scholar]
- Ringo, D.L. (1967). Flagellar motion and fine structure of the flagellar apparatus in Chlamydomonas. J. Cell Biol. 33, 543-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury, J.L., Baron, A.T., and Sanders, M.A. (1988). The centrinbased cytoskeleton of Chlamydomonas reinhardtii: distribution in interphase and mitotic cells. J. Cell Biol. 107, 635-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, M.A., and Salisbury, J.L. (1989). Centrin-mediated microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J. Cell Biol. 108, 1751-1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders, M.A., and Salisbury, J.L. (1994). Centrin plays an essential role in microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J. Cell Biol. 124, 795-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiebel E. (2000). Two new tubulins differ in a split decision. Nat. Cell Biol. 2, E3-E4. [DOI] [PubMed] [Google Scholar]
- Silflow, C.D., Liu, B., LaVoie, M., Richardson, E.A., and Palevitz, B.A. (1999). γ-Tubulin in Chlamydomonas: characterization of the gene and localization of the gene product in cells. Cell Motil. Cytoskeleton 42, 285-297. [DOI] [PubMed] [Google Scholar]
- Silflow, C. D., LaVoie, M., Tam L-W., Tousey, S., Sanders, M., Wu, W., Borodovsky, M., and Lefebvre, P.A. (2001). The Vfl1 protein in Chlamydomonas localizes in a rotationally asymmetric pattern at the distal ends of basal bodies. J. Cell Biol. 153, 63-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smrzka, O.W., Delgehyr, N., and Bornens, M. (2000). Tissue-specific expression and subcellular localization of mammalian δ-tubulin. Curr. Biol. 10, 413-416. [DOI] [PubMed] [Google Scholar]
- Taillon, B.E., Adler, S.A., Suhan, J.P., and Jarvik, J.W. (1992). Mutational analysis of centrin: an EF-hand protein associated with three distinct contractile fibers in the basal body apparatus of Chlamydomonas. J. Cell Biol. 119, 1613-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, R.L., Chojnacki, B., and Jarvik, J.W. (1983). Abnormal basal-body number, location, and orientation in a striated fiber-defective mutant of Chlamydomonas reinhardtii. J. Cell Biol. 96, 1697-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, R.L., Salisbury, J., and Jarvik, J.W. (1985). A nucleus-basal body connector in Chlamydomonas reinhardtii that may function in basal body localization or segregation. J. Cell Biol. 101, 1903-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.