Abstract
The yeast amphiphysin homologue Rvs167p plays a role in regulation of the actin cytoskeleton, endocytosis, and sporulation. Rvs167p is a phosphoprotein in vegetatively growing cells and shows increased phosphorylation upon treatment with mating pheromone. Previous work has shown that Rvs167p can be phosphorylated in vitro by the cyclin-dependent kinase Pho85p complexed with its cyclin Pcl2p. Using chymotryptic phosphopeptide mapping, we have identified the sites on which Rvs167p is phosphorylated in vitro by Pcl2p-Pho85p. We have shown that these same sites are phosphorylated in vivo during vegetative growth and that phosphorylation at two of these sites is Pcl-Pho85p dependent. In cells treated with mating pheromone, the MAP kinase Fus3p is needed for full phosphorylation of Rvs167p. Functional genomics and genetics experiments revealed that mutation of other actin cytoskeleton genes compromises growth of a strain in which phosphorylation of Rvs167p is blocked by mutation. Phosphorylation of Rvs167p inhibits its interaction in vitro with Las17p, an activator of the Arp2/3 complex, as well as with a novel protein, Ymr192p. Our results suggest that phosphorylation of Rvs167p by a cyclin-dependent kinase and by a MAP kinase is an important mechanism for regulating protein complexes involved in actin cytoskeleton function.
INTRODUCTION
The Saccharomyces cerevisiae proteins Rvs167p and Rvs161p are closely related members of the amphiphysin family of proteins, which includes mammalian amphiphysins I and II, and plant, Drosophila, nematode, and fission yeast isoforms. Amphiphysin family members contain an amino-terminal BAR domain, which is defined by homology to a protein family that includes BIN-1, Amphiphysin, and Rvs., and may contain a carboxy-terminal SH3-domain (Sivadon et al., 1997). Members of the amphiphysin family of proteins play roles in endocytosis and actin cytoskeleton function in a variety of organisms. For example, amphiphysin I and II are concentrated in nerve terminals of mature neurons and are believed to act as adaptor proteins in clathrin-mediated endocytosis.
The S. cerevisiae genes RVS167 and RVS161 were first identified in a screen for mutants that exhibited reduced viability upon starvation (Bauer et al., 1993). Deletion of RVS167 or RVS161 causes a phenotype consistent with a role for the Rvs proteins in cortical actin cytoskeleton organization, endocytosis, and membrane dynamics. The mutants exhibit loss of viability and unusual cell morphology in poor growth medium or salt-containing medium, delocalized actin distribution under suboptimal growth conditions, an abnormal (random) budding pattern in diploids, and defects in endocytosis and sporulation (Bauer et al., 1993). Ultrastructural studies have revealed that rvs167 mutants accumulate late secretory vesicles at sites of membrane and cell wall construction (Breton et al., 2001). Consistent with a role for Rvs167p in the actin cytoskeleton, Rvs167p colocalizes with cortical actin patches during vegetative growth and is concentrated at the shmoo tip during mating (Balguerie et al., 1999).
Like its mammalian homologues, Rvs167p is a multidomain protein, capable of interacting with a number of actin cytoskeleton proteins. Rvs167p forms a heterodimer with Rvs161p through their respective BAR domains (Navarro et al., 1997; Sivadon et al., 1997; Colwill et al., 1999). In addition, Rvs167p, like the amphiphysins, has a Src homology 3 (SH3) domain, a protein module well defined for binding prolinerich sequences (Pawson and Scott, 1997). The middle portions of Rvs167p and the amphiphysins are not conserved. In Rvs167p, the central portion of the molecule consists of a region rich in glycine, proline, and alanine (the GPA region), most of which is predicted to form no secondary structure elements and is notable because it contains no charged amino acids over a stretch of 107 residues.
Consistent with the multidomain structure of Rvs167p, two-hybrid and phage display screens have identified a large number of putative Rvs167p-interacting proteins, many with previously characterized roles in the actin cytoskeleton. These include Abp1p, Acf2p, Act1p, Las17p, Sla1p, Sla2p, and Srv2p (Amberg et al., 1995; Colwill et al., 1999; Madania et al., 1999; Bon et al., 2000; Drees et al., 2001; Tong et al., 2002). For most of these proteins, however, there has been no demonstration that their interaction with Rvs167p is direct, and the functional significance of the interaction is not clear. In addition, a number of synthetic lethal or negative growth synergistic interactions have been identified between RVS167 and other actin cytoskeleton genes, including SLA1, SLA2, SAC6, SRV2 (Lila and Drubin, 1997), YPT51/VPS21 (Singer-Krüger and Ferro-Novick, 1997), MYO1, MYO2, and certain alleles of ACT1 (Breton and Aigle, 1998), and SLT2 and KRE6 (Breton et al., 2001). Taken together, these data make clear that Rvs167p plays an important but apparently redundant role in organizing the actin cytoskeleton, possibly by acting as an adaptor to bring other proteins together at specific places or times in the yeast life cycle. However, the precise molecular role of Rvs167p remains to be defined.
A fundamental question in cytoskeleton biology is how the components of the actin cytoskeleton are regulated to coordinate rapid remodeling of the cytoskeleton. One likely mechanism is protein phosphorylation, which has been implicated in regulation of the actin patch-associated proteins Pan1p, Sla1p, Ent1p, and Clc1p in S. cerevisiae (Chu et al., 1999; Zeng and Cai, 1999; Watson et al., 2001; Zeng et al., 2001). Rvs167p may also be regulated by phosphorylation. Rvs167p is a phosphoprotein in log-phase cells and appears as an increased number of phosphoforms in cells arrested in G1 phase by treatment with mating pheromone (Lee et al., 1998). In addition, Rvs167p could be phosphorylated in vitro by the cyclin-dependent kinase (Cdk), Pcl2p-Pho85p, which had been immunoprecipitated from yeast (Lee et al., 1998). Pho85p is a multifunctional Cdk with roles in phosphate and glycogen metabolism, targeted proteolysis, cell cycle progression, and cell polarity (for reviews, see Carroll and O'Shea, 2002; Moffat et al., 2000). Consistent with the multifunctional nature of Pho85p, 10 genes encoding Pho85p cyclins, Pcls, have been identified and placed into subfamilies based on sequence similarity within the cyclin-box region (Measday et al., 1997). The Pcl1,2 subfamily includes three Pcls that are specifically expressed in the G1 phase of the cell cycle (Pcl1p, Pcl2p, and Pcl9p), as well as two that are not known to be regulated in the cell cycle (Pcl5p and Clg1p). Lee et al. (1998) showed that the phenotype of rvs167Δ strains has some similarities with the phenotypes of strains deleted for genes encoding Pho85p or members of the Pcl1,2 group of cyclins. Interestingly, the mammalian homologue of Pho85p, Cdk5, has been shown to phosphorylate the mammalian homologue of Rvs167p, amphiphysin I, in vitro (Floyd et al., 2001), suggesting that this regulation may be conserved.
In this study we have examined the possible regulation of Rvs167p by phosphorylation both by Pcl-Pho85p and by the MAP kinase Fus3p, which is involved in the response to mating pheromone.
MATERIALS AND METHODS
Media and Yeast Strain Manipulations
Yeast strains are listed in Table 1. Some strains were obtained from the yeast gene-deletion mutant collection constructed by the deletion consortium (Winzeler et al., 1999). Other strains were made using standard yeast genetic techniques (Sherman, 1991). Strain BY1344 is a Ura- derivative of strain BY714 (Lee et al., 1998), isolated by plating BY714 on SD medium containing 1 mg/ml 5-fluoroorotic acid (Boeke et al., 1984). Complete deletions of the indicated open reading frames (ORFs) were constructed by integrative transformation (Longtine et al., 1998). Standard yeast media were used (Sherman, 1991). To make salt-containing minimal medium, NaCl was added to synthetic dextrose medium (SD) to the final concentration (wt/vol) indicated. Phosphate-depleted medium (YPGal-Pi) was prepared as described (Warner, 1991). Where indicated, α-factor (from Louisiana State University Health Sciences Center Core Laboratory, New Orleans, LA) was added to log-phase cultures in YPD to a final concentration of 5 μM, and cells were grown for 1.5 h (2 h for the slow-growing strains BY391 and BY1076) before harvesting.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| BY263a | MATatrp1Δ63 ura3-52 lys2-801 ade2-107 his3Δ200 leu2-Δ1 | Measday et al. (1994) |
| BY391a | BY263 pho85Δ::LEU2 | Measday et al. (1997) |
| BY508a | BY263 rvs167Δ::TRP1 | Lee et al. (1998) |
| BY1078a | BY263 ark1Δ::His5 | See text |
| BY1076a | BY263 prk1Δ::TRP1 | See text |
| BY1326a | BY263 MATα rvs167Δ::TRP1 sla1Δ::kan | See text |
| BY1342a | BY263 slt2Δ::kan | Baetz et al. (2001) |
| BY1344a | BY263 pcl1Δ::LEU2 pcl2Δ::LYS2 pcl5Δ::TRP1 clg1Δ::URA3 pcl9Δ::HIS3 | See text |
| BY1404b | MATα ura3Δ0 leu2Δ0 his3Δ1 lys2Δ0 mfa1Δ::MFA-pr-HIS3 can1Δ0 rvs167Δ::Nat | See text; derivative of Y2454 (Tong et al., 2001) |
| BY1456b | MATaura3 leu2 his3 MFA1-pr-HIS3 rvs167Δ::Nat end3Δ::kan | Cross of consortium end3Δ::kan strain × BY1404 |
| BY1523 | MATaura3 leu2 his3 rvs167Δ::Nat end3Δ::kan sla1Δ::kan | Cross of BY1456 × BY1326 |
| BY1596b | MATafus3Δ::kan | Winzeler et al. (1999) |
| BY1615b | MATahog1Δ::kan | Winzeler et al. (1999) |
| EG123 | MATahis4 leu2 trp1 ura3 can1 gal2 suc2Δ | I. Herskowitz |
| YMP2 | EG123 fus3Δ::URA3 | A. Neiman (Peter et al., 1993) |
| YMP3 | EG123 kss1Δ::URA3 | A. Neiman (Peter et al., 1993) |
| YMP4 | EG123 fus3Δ::URA3 kss1Δ::URA3 | A. Neiman (Peter et al., 1993) |
Except as noted, strains are isogenic to the parent strain, BY263, an S288C derivative
Strains from the deletion consortium are isogenic to the parent strain, BY4741 (MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0), which is also derived from S288C (Brachmann et al., 1998)
Insect Cell Culture and Protein Purification
Pho85p tagged N-terminally with GST + 6× His and Pcl2p tagged N-terminally with 6× His were made by cloning the ORFs into the Baculovirus Transfer Vectors pAcGHLT and pAcHLT-c, respectively (PharMingen, San Diego, CA; Huang et al., 1999a). Rvs167p was tagged N-terminally with GST + 6× His in pAcGHLT. To produce active Fus3p, Fus3pK42R, or Kss1p kinase from insect cells, cells were coinfected with viruses expressing the MAP kinase module of STE5, STE11-4, STE7, and FUS3-myc, FUS3K42R-myc, or KSS1-myc (Breitkreutz et al., 2001). Sf9 insect cells were coinfected with the recombinant viruses and were harvested after 48 h. Insect cells were grown in Grace's insect medium at 27°C, infected, and lysed using standard procedures (Ausubel et al., 1994; PharMingen). To prepare Pcl2p-Pho85p, cells were coinfected with viruses expressing His-Pcl2p and His-GST-Pho85p, and then Cdk complexes were purified over glutathione Sepharose (Amersham, Piscataway, NJ) as previously described (Colwill et al., 1999). The reconstituted MAP kinase modules were purified as described by coimmunoprecipitation with anti-myc antibodies (Breitkreutz et al., 2001). Other GST-tagged proteins were purified over glutathione Sepharose using standard procedures. His-tagged proteins were purified using Ni-NTA agarose (Qiagen, Valencia, CA) using a native purification procedure recommended by the manufacturer.
Plasmids
Plasmids are described in Table 2. Site-directed mutagenesis was done using either standard two-step PCR mutagenesis techniques (Ho et al., 1989; Warner, 1991) or the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Primers were designed to amplify the region encoding each putative Pho85 phosphorylation site and surrounding area. The sequences of primers used in this article are available upon request. The integrity of all PCR products was confirmed by sequencing.
Table 2.
Plasmids used in this study
| Plasmid | Description | Source |
|---|---|---|
| pGST-GPA-SH3 | Codons 282-482 of RVS167 were subcloned in pGEX-1 (Pharmacia, Piscataway, NJ) to give RVS167 GPA-SH3 with an N-terminal GST tag. | Lee et al. (1998) |
| pGAL-RVS167 | RVS167 under the control of the GAL promoter in a 2-μm (high copy) URA3 plasmid. | Colwill et al. (1999) |
| pGPA-SH3-His | Codons 282-482 of RVS167 were amplified by PCR and subcloned into pET-21d+ (Novagen, Madison, WI) to give RVS167 GPA-SH3 with a C-terminal 6-His tag under the control of the T7 promoter. | This study |
| pBST-BEE1 | An NcoI-SalI fragment containing the LAS17 ORF was isolated from pAS1-WAS1 (Colwill et al., 1999) and ligated into pBST-KS+ (Stratagene, La Jolla, CA) to give full-length LAS17 under the control of the T7 promoter. | This study |
| pGEM+YMR192w | A 2.5-kb fragment containing the YMR192w ORF plus 36 bp upstream and 231 bp downstream was amplified using PCR and cloned into pGEM-T Easy (Promega, Madison, WI) to give YMR192w under the control of the T7 promoter. | This study |
| p413MET-RVS167 | A 1.6-kb BglII fragment containing the RVS167 ORF was subcloned into p413MET (Ronicke et al., 1997). | Colwill et al. (1999) |
| p413MET-RVS167-4A | p413MET-RVS167 derivative containing RVS167 with S299A, S321A T323A, and S379A substitutions | This study |
| p416MET-RVS167 | A 1.6-kb BglII fragment containing the RVS167 ORF from pBA1015 (Colwill et al., 1999) was subcloned into p416MET (Ronicke et al., 1997). | This study |
| p416MET-RVS167-3A | p416MET-RVS167 derivative containing RVS167 with S299A, S321A, and S379A substitutions | This study |
| p416MET-RVS167-4A | p416MET-RVS167 derivative containing RVS167 with S299A, S321A T323A, and S379A substitutions. | This study |
In Vitro Kinase Assays and Quantitative Phosphorylation
Approximately 10 pmol GST-GPA-SH3 was phosphorylated by 3 pmol Pho85p and 1 pmol Pcl2p in a 30 μl reaction with 100 mM Tris.Cl, pH 7.5, 10 mM MgCl2, 1 mM DTT, 50 μM ATP, and 50 μCi [γ-32P]ATP (Mandel, Guelph, Ontario, Canada). The reaction was incubated at 30°C for 30 min. Proteins were separated on 7.5% polyacrylamide gels, the gels were stained with Coomassie Blue and dried, the radioactive proteins were detected by autoradiography, and the gel slice containing the protein of interest was cut out and processed. To achieve quantitative phosphorylation of Rvs167p, reactions were carried out as described above with the following exceptions: ATP was added to a final concentration of 5 mM and reactions were incubated at 25°C for 16 h, and then an additional one half volume of kinase was added and the reaction was incubated at 25°C for an additional 6 h. Reactions were done with unlabeled ATP and, in parallel, “spiked” with [γ-32P]ATP so that incorporation of PO4 could be monitored. Quantification by phosphorimager showed that approximately 3 mol of phosphate were incorporated per mole of Rvs167p. “Mock” phosphorylation reactions were identical to quantitative phosphorylation reactions except that kinase was left out.
The reconstituted MAP kinase modules were used to phosphorylate ∼1 pmol GST-Rvs167p or 20 pmol GPA-SH3-His in a 30-μl reaction using [γ-32P]ATP as described by Breitkreutz et al. (2001). Labeled proteins were visualized by phosphorimager.
In Vivo Labeling of Rvs167p and Phosphopeptide Mapping
The galactose-inducible expression vector pGAL-RVS167 (Colwill et al., 1999) was transformed into strains BY263 and BY1344, and overexpression of RVS167 was confirmed by Western blot analysis (Lee et al., 1998). Transformants were grown to midlog phase in SD-Ura and then in YPD for about two generations and then were transferred to YPGal-Pi and allowed to double once. Because overexpression of RVS167 is toxic (Colwill et al., 1999), this doubling took ∼10–15 h. The cells were pelleted and concentrated 20-fold to give a final volume of 2 ml in YPGal-Pi, 32P-orthophosphate was added to a final concentration of 1 mCi/ml, and cells were incubated with shaking for 1 h. Cells were pelleted, washed with 50 mM NaF, and flash frozen. 32P-labeled Rvs167p was immunoprecipitated from cells (Lee et al., 1998), run on a 7.5% polyacrylamide gel, and detected by autoradiography.
The band corresponding to 32P-Rvs167p was cut out of the dried gel. The gel slice was rehydrated in water for 1 h, and peptides were isolated using an in-gel digestion procedure as described (Figeys et al., 2001), except that chymotrypsin (Roche Diagnostics, Indianapolis, IN) was used instead of trypsin because the GPA region contains a stretch of 107 amino acids with no charged residues. Phosphopeptide mapping was done using standard procedures (Boyle et al., 1991; van der Geer and Hunter, 1994). Chymotryptic peptides (200–2000 cpm total) were spotted onto 20 cm × 20 cm thin-layer glassbacked cellulose (TLC) plates (Merck, Darmstadt, Germany) at a spot 3 cm from the bottom and 7 cm from the cathode side. The plates were run in pH 4.72 electrophoresis buffer on an HTLE-7000 electrophoresis apparatus at 1000 V for 3 h and air-dried. Chromatography in the perpendicular direction (in phosphochromatography buffer) was done for 12–14 h, and the plate was exposed to a phosphorimager screen.
Western and Far Western Assays
Preparation of cell extracts and Western blot hybridization were done as described (Lee et al., 1998). Far Western hybridization was done as described (Guichet et al., 1997) using 35S-labeled Las17p or Ymr192p that had been synthesized using a coupled T7 polymerasereticulocyte lysate system (Promega, Madison, WI) primed with plasmid pBST-BEE1 or pGEM+YMR192w. To ensure that the in vitro translation reaction products hybridized with the Western blot contained no phosphatase activity, which could have removed phosphates from the phosphorylated Rvs167p, 32P-labeled Rvs167p was run on a gel and transferred to duplicate nitrocellulose membranes. One filter was hybridized with mock in vitro translated protein (primed with no DNA), and the other was stored at -20°C. After an overnight hybridization and washing, the 32P-Rvs167p on the two filters was quantified using phosphorimager analysis. This control experiment showed that no counts were lost from the Rvs167p protein on the filter during incubation with the in vitro translation mixture.
SGA Analysis and Complementation of Synthetic Lethality
Synthetic genetic array (SGA) analysis was done as described (Tong et al., 2001) with the modifications outlined below. A strain in which the RVS167 ORF had been replaced with the gene encoding nourseothricin resistance Nat (Goldstein and McCusker, 1999) and that contained plasmid p416MET-RVS167-3A, which encodes Rvs167p lacking the three phosphorylated serines (Table 1), was crossed to an ordered array of ∼4000 viable gene-deletion mutants (xxxΔ). The levels of Rvs167p in cells containing this plasmid under semirepressing conditions (0.17 mM methionine) are similar to endogenous levels of Rvs167p (see RESULTS). Diploids were sporulated, and double-mutant haploids (rvs167Δ xxxΔ) carrying p416MET-RVS167-3A were selected and then tested for growth on SD + 4% NaCl at 34°C, conditions under which some rvs167 double mutants have been shown to be inviable (Lila and Drubin, 1997). Strains that had apparent synthetic defects with RVS167-3A were retested by mating in parallel to an rvs167Δ strain containing vector, p416MET-RVS167-3A, or plasmid p416MET-RVS167, which encodes wild-type Rvs167p. This screen for genetic backgrounds that require phosphorylation of Rvs167p was not exhaustive because 1) an early (and thus incomplete) set of the gene-deletion mutants was used and 2) deletion strains that themselves grow slowly are beyond the sensitivity of this assay (Tong et al., 2001).
Complementation of the sla1Δ rvs167Δ strain was assayed on SD-His containing 4.1% NaCl at 34°C. Complementation of the end3Δ sla1Δ rvs167Δ strain was assayed on SD-Ura containing 3.2% NaCl at 30°C. The degree of complementation by RVS167-4A was transformant-dependent and highly variable in penetrance (our unpublished results), possibly due to variability in levels of expression of RVS167 from the plasmid.
RESULTS
Identification of In Vitro Pcl2p-Pho85p Phosphorylation Sites on Rvs167p
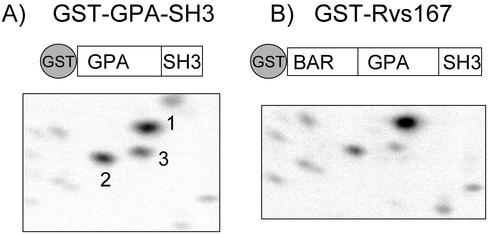
Previous work has shown that a truncated version of Rvs167p containing the GPA and SH3 domains can be phosphorylated by the Pcl2p-Pho85p cyclin-dependent kinase in vitro (Lee et al., 1998). To establish Rvs167p as a bona fide in vivo substrate of Pho85p, we sought to identify the Pcl2p-Pho85p phosphorylation sites. To this end, we first analyzed the phosphopeptide map of bacterially expressed GST-GPA-SH3 that had been phosphorylated in vitro using Pcl2p-Pho85p purified from insect cells (Figure 1A). This identified three major phosphopeptides (numbered 1, 2, and 3 in Figure 1A), suggesting that the GPA-SH3 region is phosphorylated at three sites. We next asked whether the same sites were phosphorylated in the context of the full-length protein by comparing this map with that of GST-Rvs167p purified from insect cells and phosphorylated in vitro with Pcl2p-Pho85p. We saw the same three major spots, as well as a similar pattern of minor spots (Figure 1B). This result suggests that the amino-terminal BAR domain is not needed for normal Pcl2p-Pho85p-dependent phosphorylation and is consistent with previous findings that Pcl2p does not interact with the BAR domain in a two-hybrid assay (Colwill et al., 1999). Thus, we did our subsequent phosphopeptide mapping on GST-GPA-SH3 mutant proteins purified from Escherichia coli.
Figure 1.
Phosphopeptide maps of full-length and truncated Rvs167p phosphorylated in vitro. Pcl2p-Pho85p purified from insect cells was used to phosphorylate purified proteins. (A) GST-GPA-SH3 purified from bacteria. (B) GST-Rvs167p purified from insect cells. Phosphorylated proteins were digested with chymotrypsin and analyzed as described in MATERIALS AND METHODS. Above the phosphopeptide maps are schematic representation of the Rvs167 proteins showing domains defined by homology to other proteins. BAR: Bin1/Amphiphysin/Rvs167 domain; GPA: region rich in glycine, proline, and alanine; SH3: Src Homology 3 domain.
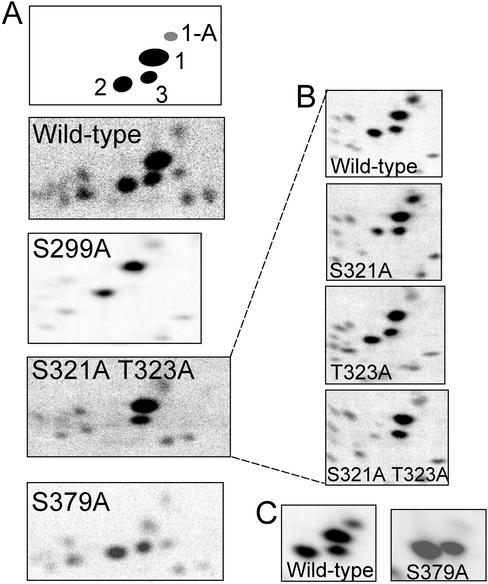
Like all Cdks, Pho85p is a proline-directed kinase: its target site for phosphorylation is a serine or threonine followed by a proline (O'Neill et al., 1996). To identify phosphorylation sites on Rvs167p, we constructed a series of mutants encoding single serine or threonine to alanine substitutions at each of the eight S/TP sites in the GPA and SH3 regions of Rvs167p and expressed these genes in E. coli. We then purified the mutant proteins, phosphorylated the proteins in vitro using Pcl2p-Pho85p, and repeated the phosphopeptide mapping. Comparison of wild-type GST-GPA-SH3 with mutants T407A, T412A, and T454A showed that none of the three major phosphopeptides was missing in the mutants (our unpublished results). In the phosphopeptide map of mutant S390A, some of the faint phosphopeptides were missing, suggesting that S390 may be a minor site for phosphorylation (our unpublished results). In contrast, the phosphopeptide map of mutant S299A lacked spot 3 (Figure 2A), indicating that S299 is phosphorylated by Pcl2p-Pho85p in vitro. In addition, mutant S379A gave a phosphopeptide map in which spot 1 was barely detectable (Figure 2A). This suggests that S379 is phosphorylated by Pcl2p-Pho85p but that the peptide containing S379 may comigrate with another (unidentified) minor peptide.
Figure 2.
Phosphopeptide analysis of wild-type and mutant GST-GPA-SH3 phosphorylated by Pcl2p-Pho85p in vitro. Chymotryptic phosphopeptides were prepared and analyzed as described in MATERIALS AND METHODS. (A) Phosphopeptide maps showing a single spot missing from each mutant map. A schematic diagram showing the major chymotryptic phosphopeptides seen in Rvs167p is shown at the top. Spot 1-A is a phosphopeptide whose presence varies from map to map; it is produced by chymotryptic cleavage at a secondary site in the protein. (B) Phosphopeptide maps of wild-type protein and various combinations of mutants S321A and T323A. (C) Phosphopeptide maps of wild-type protein and mutant S379A analyzed in parallel, showing that spot 1-A and spot 1 are absent in the S379A mutant map.
By predicted mobility (Boyle et al., 1991), spot 2 was predicted to contain a phosphopeptide (DPATATSPTPTGY) with both a serine and a threonine that were followed by a proline: S321 and T323 (italics). Mutant T323A gave the same phosphopeptide map as did wild-type protein (Figure 2B). In contrast, the phosphopeptide map of mutant S321A gave three spots, but spot 2 was less intense and had increased mobility in the chromatography dimension compared with the same spot on the wild-type map (Figure 2B). This suggested that in the mutant protein, an alternative residue was being phosphorylated. This would be consistent with phosphorylation of T323 in the absence of S321, based on predicted mobility calculations (Boyle et al., 1991). Indeed, in the phosphopeptide map of the S321A and T323A double mutant, spot 2 disappeared (Figure 2, A and B).
Because we used chymotrypsin, which is less specific than the more commonly used trypsin (Antal et al. 2001), we sometimes saw spots that were products of cleavage at secondary sites. The phosphopeptide containing S379, in particular, gave two spots in some of our maps (Figure 2A: spots 1 and 1-A and Figure 2C). The peptide containing S379 has two leucine residues, which are minor chymotrypsin digestion sites, and therefore a significant amount of partially digested peptide might be expected. We found that the intensity of peptide 1-A seemed to vary depending on the batch of chymotrypsin (compare phosphopeptide maps of wild-type Rvs167p in Figure 2, A and C).
In summary, we conclude that Pcl2p-Pho85p phosphorylates Rvs167p in vitro at three major sites: S299, S379, and S321, and at T323 if S321 is not present.
Phosphorylation of Rvs167p by Pcl-Pho85p In Vivo
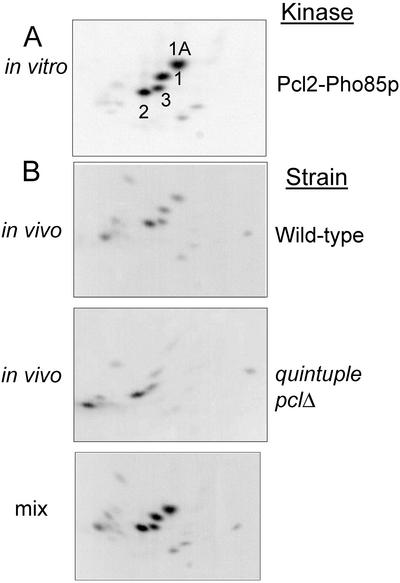
To determine whether Rvs167p is phosphorylated at the same sites in vivo as it is in vitro, we labeled wild-type cells with 32P-orthophosphate and did phosphopeptide mapping on immunoprecipitated 32P-Rvs167p. Even though overexpression of RVS167 is toxic and thus leads to a slowed doubling time (Colwill et al., 1999), we could only recover sufficient phosphorylated Rvs167p to use in phosphopeptide mapping experiments if we overexpressed RVS167 under the control of the GAL promoter on a high-copy plasmid.
Rvs167p isolated from wild-type cells gave the same phosphopeptide map as did Rvs167p GPA-SH3 phosphorylated in vitro with Pcl2p-Pho85p (Figure 3, A and B). When in vitro–and in vivo–labeled proteins were analyzed after mixing, all the peptides colocalized, indicating that Rvs167p is phosphorylated on S299, S321, and S379 in vivo (Figure 3B). In repeated in vivo–labeling experiments we have noticed that phosphorylation of S299 appears variable (our unpublished results).
Figure 3.
Comparative phosphopeptide analysis of Rvs167p phosphorylated in vitro and in vivo. (A) GST-GPA-SH3 was phosphorylated in vitro by Pcl2p-Pho85p (cf. Figures 1 and 2). (B) Rvs167p was immunoprecipitated from 32P-labeled wild-type cells and from 32P-labeled Δpcl1 Δpcl2 Δpcl5 Δpcl9 Δclg1 cells (see MATERIALS AND METHODS for details), and phosphopeptide analysis was performed as with the in vitro–labeled protein. In addition, equal numbers of counts from in vitro and in vivo–labeled proteins were mixed to give a phosphopeptide map (labeled mix).
To determine whether phosphorylation of Rvs167p was dependent on Pcl-Pho85p, we performed phosphopeptide mapping on Rvs167p from a yeast strain deleted for the genes encoding the five cyclins of the Pcl1,2 subfamily: PCL1, PCL2, PCL5, PCL9, and CLG1 (Measday et al., 1997). We chose to use this strain rather than one deleted for PHO85 itself because pho85Δ strains are slow growing and difficult to label in vivo, particularly because overexpression of RVS167 is toxic (Colwill et al., 1999). In protein from the mutant strain, spots 1 and 1-A, representing phosphorylation at S379, and spot 3, representing phosphorylation at S299, were reduced in intensity (Figure 3B). Spot 2 (S321) was not reduced in intensity. We conclude that Pcl-Pho85p is required in log-phase cells for phosphorylation of S379 and S299. However, because Pcl2p-Pho85p can phosphorylate Rvs167p at S321 in vitro, it is also possible that both Pcl-Pho85p and another kinase can phosphorylate Rvs167p at this site in vivo in a redundant manner.
FUS3 Is Needed for Phosphorylation of Rvs167p
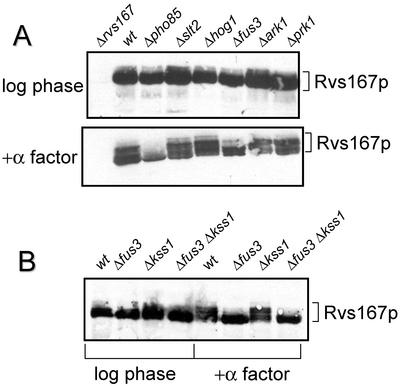
The results with the quintuple PCL delete strain showed that another kinase must be able to phosphorylate Rvs167p in vivo. We were therefore interested in identifying that kinase. Previous work showed that phosphoforms of Rvs167p could be detected by Western blot in extracts from log-phase cells, but multiple phosphoforms of Rvs167p were more clearly visible in cells treated with α-factor (Lee et al., 1998). We therefore used Western blots of both log-phase and α-factor–treated cells to detect Rvs167p phosphoforms in various strains in which a single gene encoding a kinase had been deleted.
As observed previously (Lee et al., 1998), the presence of phosphoforms of Rvs167p was reduced in both log-phase and α-factor–treated cells in a strain deleted for PHO85 in comparison to wild-type (Figure 4A). In contrast, Rvs167p from strains deleted for SLT2, HOG1, ARK1, or PRK1 had levels of phosphorylation similar to those seen in wild-type cells under both conditions (Figure 4A). However, the strain deleted for FUS3 also had reduced phosphorylation of Rvs167p. The reduction in Rvs167p phosphoforms was barely detectable in log-phase cells but was conspicuous in cells treated with α-factor (Figure 4A).
Figure 4.
Western blot analysis of Rvs167p phosphorylation in yeast strains deleted for genes encoding various kinases. Lysates were made from strains grown to log phase or treated with α-factor for 1.5 h (2 h for Δpho85 and Δprk1 strains). Lysates were analyzed as described in MATERIALS AND METHODS using anti-Rvs167p antiserum. (A) All strains were of the S288C strain background. (B) All strains were of the EG123 strain background.
Fus3p is a MAP kinase involved in the pheromone response and is partially functionally redundant with the homologous MAP kinase Kss1p (for review see Breitkreutz and Tyers, 2002; Gustin et al., 1998). We examined the dependence of Rvs167p phosphorylation on FUS3 and KSS1 in the EG123 strain background (Peter et al., 1993). In wild-type EG123 cells, we saw multiple Rvs167p phosphoforms in cells treated with α-factor (Figure 4B). In EG123 cells deleted for FUS3, there was a reduction in the number of phosphoforms of Rvs167p (Figure 4B), similar to that seen in the S288C strain background (Figure 4A). Rvs167p phosphoforms looked similar to those seen in wild-type cells in a strain deleted for KSS1, whereas in cells deleted for both FUS3 and KSS1, Rvs167p phosphoforms resembled those in fus3Δ cells. Thus FUS3, but not KSS1, is needed for efficient phosphorylation of Rvs167p in vivo.
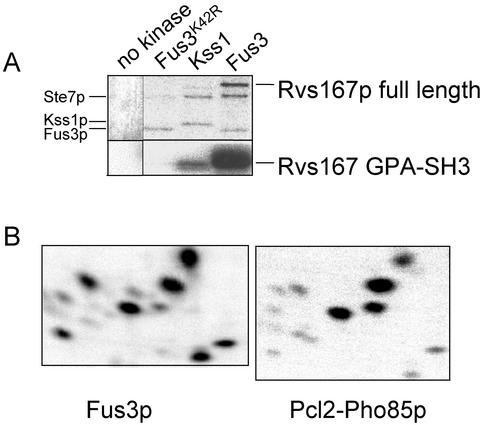
We next asked whether Fus3p was able to phosphorylate Rvs167p directly in vitro, using a purified MAP kinase module reconstituted in insect cells from recombinant components (Breitkreutz et al., 2001). Insect cells were coinfected with various combinations of baculoviruses that expressed STE11-4 (a constitutively active form of STE11), STE5, STE7, and FUS3-myc, KSS1-myc, or FUS3K42R-myc. MAP kinase complexes were then immunopurified from the cell lysates. Both the Fus3p-containing complex and the Kss1p-containing complex have been shown to phosphorylate Ste12p, a known substrate for both kinases (Breitkreutz et al., 2001). In our assay, Fus3p, but neither a catalytically inactive Fus3p (Fus3K42R) nor Kss1p, was able to phosphorylate full-length GST-Rvs167p (Figure 5A, top panel). We also tested whether Fus3p could phosphorylate Rvs167p GPA-SH3-His, which lacks the entire BAR domain. The GPA-SH3 fragment was phosphorylated efficiently by Fus3p, but not by Fus3-K42R or Kss1p (Figure 5A, bottom panel), indicating that, in vitro, the BAR domain of Rvs167p is not needed for phosphorylation by Fus3p. Phosphopeptide mapping of Rvs167p GPA-SH3 phosphorylated by Fus3p in vitro revealed that the same sites were phosphorylated as with Pcl2p-Pho85p, although not all with the same intensity (Figure 5B).
Figure 5.
Phosphorylation of GST-Rvs167p and GPA-SH3-His by Fus3p kinase. (A) Phosphorimage showing proteins phosphorylated in vitro using an immunoprecipitated MAP kinase module, reconstituted in insect cells, that contained Fus3K42R-myc, Kss1-myc, or Fus3-myc (see MATERIALS AND METHODS). (B) Comparative phosphopeptide analysis of Rvs167 GPA-SH3-His phosphorylated in vitro by a purified MAP kinase module containing Fus3p (left panel) or by Pcl2p-Pho85p (right panel, cf. Figures 1, 2, and 3).
In summary, the MAP kinase Fus3p is able to phosphorylate Rvs167p in vitro and is required for full phosphorylation of Rvs167p in response to α-factor in vivo.
Role of Phosphorylation In Vivo
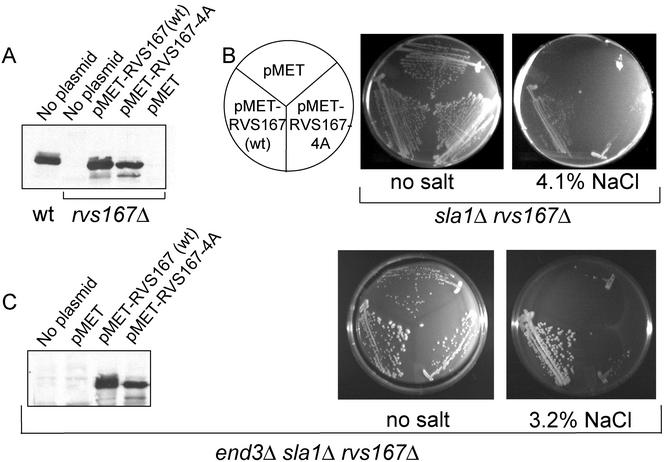
We next sought to discover the biological significance of Rvs167p phosphorylation by searching for conditions in which phosphorylation of Rvs167p was essential in vivo. We constructed plasmids encoding versions of Rvs167p that had serine or threonine to alanine substitutions in the residues that we had shown to be phosphorylated. pMET-RVS167-3A encodes Rvs167p with S299A, S321A, and S379A substitutions, and pMET-RVS167-4A encodes Rvs167p with these three changes plus a T323A substitution. T323 is a minor phosphorylation site (Figure 2B), and we have seen no differences in complementation between pMET-RVS167-3A and pMET-RVS167-4A (our unpublished results). Western blots revealed that, as expected, Rvs167-4A was underphosphorylated relative to wild-type Rvs167p in log-phase cells, as seen by the absence of slower-migrating phosphoforms (Figure 6A).
Figure 6.
Biological role of phosphorylation of Rvs167p. (A) Western blot showing levels of expression and phosphoforms of Rvs167p as expressed from its endogenous promoter compared with rvs167Δ cells containing p416MET-RVS167 or p416MET-RVS167-4A. All cells were grown in synthetic medium containing 0.17 mM methionine. (B) An sla1Δ rvs167Δ strain is complemented by wild-type RVS167 but not by RVS167-4A or by vector. Transformants containing vector, p413MET-RVS167-4A, or p413MET-RVS167 (wild-type) were streaked on SD-His or SD-His + 4.1% NaCl as indicated and grown at 34°C for 4 d. (C) An end3Δ sla1Δ rvs167Δ strain is complemented by wild-type RVS167 but not by RVS167-4A or by vector. Western blot showing levels of expression and phosphoforms of RVS167 from p416MET-RVS167 and p416MET-RVS167-4A in an end3Δ sla1Δ rvs167Δ strain (left). Transformants containing vector, p416MET-RVS167-4A, or p416MET-RVS167 (wild-type) were streaked on SD-Ura or SD-Ura + 3.2% NaCl as indicated and grown at 30°C for 4 d (right).
To assess genetically the consequences of a failure to phosphorylate Rvs167p, we needed to confront the genetic redundancy of the actin cytoskeleton. RVS167 is not an essential gene, although it is required for viability under certain growth conditions (Bauer et al., 1993; Colwill et al., 1999) and in strains deleted for a number of other actin cytoskeleton genes (Lila and Drubin, 1997; Singer-Krüger and Ferro-Novick, 1997; Breton and Aigle, 1998; Breton et al., 2001). Furthermore, expression of the RVS167 BAR domain alone is able to largely complement rvs167Δ defects, even though the Rvs167p SH3 domain is known to bind biologically important ligands and presumably plays a significant role in wild-type cells (Lila and Drubin, 1997; Bon et al., 2000; Tong et al., 2002). Because the Rvs167p phosphorylation sites that we have identified all lie in the GPA region, outside the BAR domain, we reasoned that a requirement for phosphorylation would probably only be apparent in genetic backgrounds that are compromised for actin cytoskeletal function and thus would be sensitized to defects in Rvs167p function. Consistent with this idea, we found that in rvs167Δ strains, expression of wild-type RVS167 or RVS167-4A complemented defects in growth on carbon and nitrogen starvation media and on medium containing salt, as well as defects in endocytosis and sporulation, with equal efficiency (our unpublished results). In addition, overexpression of RVS167-4A resulted in inhibition of growth to the same extent as did overexpression of wild-type RVS167 (our unpublished results).
To overcome the problem of functional redundancy, we tested whether the RVS167 phosphorylation site mutant was able to complement the synthetic lethality of various strains, both under standard conditions and in the presence of NaCl at 34°C, conditions that have been shown to exacerbate the phenotype of some rvs167 double mutants (Lila and Drubin, 1997) No difference in complementation by wild-type RVS167 and RVS167-4A was seen in rvs167Δ sac6Δ, rvs167Δ srv2Δ, or rvs167Δ sla2Δ strains (our unpublished results). In contrast, the growth defect of the rvs167Δ sla1Δ strain was complemented more efficiently by wild-type RVS167 than by the RVS167 phosphorylation site mutant on medium containing 4.1% NaCl at 34°C (Figure 6B). Thus, in at least one strain background, under certain growth conditions, phosphorylation of Rvs167p is important for viability. To screen for other genetic backgrounds in which phosphorylation of Rvs167p is essential, we used a modified version of the SGA technique (Tong et al., 2001; see MATERIALS AND METHODS). This screen yielded two genetic backgrounds that were not efficiently complemented for growth on 4% NaCl at 34°C by RVS167-3A but were complemented by wild-type RVS167: sla1Δ rvs167Δ (as already found by the candidate approach; see above) and end3Δ rvs167Δ.
The finding that both SLA1 and END3 had synthetic growth defects in combination with the RVS167 phosphorylation site mutant was of particular interest because End3p and Sla1p have recently been shown to form a complex with a third protein, Pan1p (Tang et al., 2000). PAN1 is an essential gene (Tang and Cai, 1996), whereas a sla1 end3 double mutant strain has a reduced growth rate (Tang et al., 2000). Tang et al. (2000) have suggested that the Pan1p-Sla1p-End3p complex may be essential but that the contributions of Sla1p and End3p are partially redundant. We constructed an end3Δ sla1Δ rvs167Δ strain (which had a more severe growth defect than the end3Δ sla1Δ strain) and found that this strain was complemented for growth on salt-containing medium by wild-type RVS167 but not by RVS167-4A (Figure 6C). Western blot analysis showed that Rvs167p was present in the strain expressing RVS167-4A (Figure 6C). These data show that under certain physiological circumstances, phosphorylation of Rvs167p is important for cell viability.
Role of Phosphorylation In Vitro
We next explored the biochemical consequences of Rvs167p phosphorylation in vitro. Our experiment was based on several genetic observations. Pan1p has recently been shown to be an activator of the Arp2/3 complex, which stimulates actin nucleation, in an in vitro system (Duncan et al., 2001). Because Pan1p, End3p, and Sla1p form a trimeric complex (Tang et al., 2000), the Arp2/3-activation activity may be due to this complex. Another activator of the Arp2/3 complex in yeast is Las17p (Madania et al., 1999; Winter et al., 1999), which binds to the SH3 domain of Rvs167p (Bon et al., 2000; Tong et al., 2002). When combined with a las17-6 allele (which encodes a protein with a truncation of the Arp2/3 activation domain), a truncation of the Pan1p carboxy terminus (which is thought to be involved in Arp2/3 activation) leads to temperature sensitivity, suggesting that the Arp2/3 activation function carried out by Las17p may be redundant with that of Pan1p (Duncan et al., 2001). These findings, coupled with our observation that a version of Rvs167p that cannot be phosphorylated is lethal under some growth conditions in the absence of Sla1p and End3p, suggest that phosphorylation of Rvs167p may be important in some function that involves Las17p.
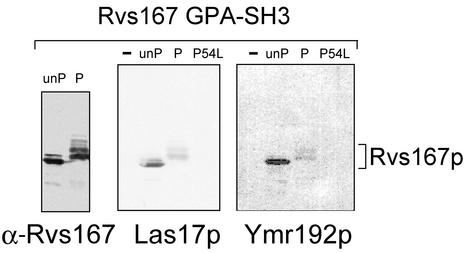
To determine whether phosphorylation of Rvs167p had any effect on its interactions with Las17p, we tested Rvs167 GPA-SH3 that had been quantitatively phosphorylated or mock phosphorylated in vitro (see MATERIALS AND METHODS) for its ability to bind to Las17p, using a Far Western assay. Phosphorylated and unphosphorylated Rvs167p were run on a gel and blotted to nitrocellulose, and the blot was hybridized with in vitro–translated Las17p that had been labeled with [35S]methionine (Figure 7, middle panel). Controls established that there were equal amounts of phosphorylated and unphosphorylated GPA-SH3 on the filter (Figure 7, left panel) and that the in vitro translation reaction hybridized with the Western blot contained no phosphatase activity, which could have removed phosphates from the phosphorylated Rvs167p (see MATERIALS AND METHODS). Phosphorimager analysis revealed that 6.0 times more Las17p bound to mock-phosphorylated Rvs167p GPA-SH3 than to phosphorylated GPA-SH3. Thus, phosphorylation of Rvs167p inhibited its interaction with Las17p.
Figure 7.
Far Western analysis of the effects of Rvs167p phosphorylation. Rvs167 GPA-SH3-His was quantitatively phosphorylated by Pcl2p-Pho85p in vitro (lanes marked P) or mock-phosphorylated (lanes marked unP). After acrylamide gel electrophoresis and transfer to nitrocellulose filters, one filter was hybridized with anti-Rvs167p antiserum (left panel) and the others were hybridized with in vitro-translated 35S-labeled Las17p or Ymr192p. The blots hybridized with Las17p and Ymr192p show an additional control: GPA-SH3-His with a substitution of leucine at position 54 of the SH3 domain, a change that is known to disrupt ligand binding in other SH3 domains.
We next asked if the effect of phosphorylation of the GPA region of Rvs167p on binding to the SH3 domain was specific for Las17p. Many proteins have been reported to interact with Rvs167p in two-hybrid (Bon et al., 2000; Drees et al., 2001; Tong et al., 2002) and phage display (Tong et al., 2002) assays, but few of these interactions have been confirmed in vitro. Among these interactions, the SH3 domain of Rvs167p has been reported to bind to an uncharacterized gene product, Ymr192p. Using affinity chromatography, we have shown that Ymr192p from yeast extracts binds to the SH3 domain of Rvs167p (Friesen, Colwill, and Andrews, manuscript in preparation). We tested Rvs167p GPA-SH3 that had been phosphorylated or mock-phosphorylated in vitro for its ability to bind to Ymr192p using a Far Western assay. As with Las17p, binding of Ymr192p to the SH3 domain of Rvs167p was inhibited by phosphorylation (Figure 7, right panel). Quantification by phosphorimager revealed a 5.8-fold difference. Thus, phosphorylation of the GPA region by Pcl2p-Pho85p inhibits binding of at least two proteins to the SH3 region of Rvs167p.
DISCUSSION
Phosphorylation Regulates Many Actin Cytoskeleton Proteins
Recent experiments have identified a number of actin cytoskeleton and endocytosis proteins in Saccharomyces that are regulated by phosphorylation. Clathrin light chain, Clc1p, is a phosphoprotein and is hyperphosphorylated in the presence of mating pheromone in a Fus3p-dependent manner (Chu et al., 1999). The actin-regulating kinase, Prk1p, phosphorylates Pan1p and Sla1p, leading to disruption of the End3p/Pan1p/Sla1p complex (Zeng and Cai, 1999; Zeng et al., 2001). Phosphorylation of the yeast epsin homologue, Ent1p, by Ark1p or Prk1p negatively regulates its interaction with Pan1p (Watson et al., 2001) In this work, we have shown that Pcl-Pho85p phosphorylates the actin cytoskeleton protein, Rvs167p.
Interestingly, the mammalian homologue of Rvs167p, amphiphysin I, is also a phosphorylated protein with its phosphorylation sites clustered in the center of the protein (Floyd et al., 2001). Amphiphysin is phosphorylated in vitro by p35-Cdk5, the mammalian functional homologue of Pcl-Pho85p (Huang et al., 1999a). Analogous to the situation with Rvs167p, for which we have shown that phosphorylation inhibits binding of two proteins to the SH3 domain, phosphorylation of amphiphysin inhibits its interaction with AP-2 and clathrin (Slepnev et al., 1998). This remarkable conservation of both a kinase and its substrate demonstrates a conserved pathway involving the regulation of an endocytosis protein.
Which Pho85 Cyclin Phosphorylates Rvs167p?
Several substrates for Pho85p kinase in conjunction with different cyclins have been identified: Pho80p-Pho85p phosphorylates Pho4p with the consensus SPXL/I (O'Neill et al., 1996); Pcl10p-Pho85p phosphorylates Gsy2p at S/TPXDL (Huang et al., 1996), Pcl5p-Pho85p phosphorylates Gcn4p at TPVL (Meimoun et al., 2000; Shemer et al., 2002), and Pho85p complexed with an unidentified cyclin phosphorylates Sic1p at TPPR (Nishizawa et al., 1998). We have shown that in a strain deleted for the five genes encoding the Pcl1,2 subfamily of cyclins, phosphorylation of Rvs167p at S379 and S299 is substantially reduced (Figure 3). One Pcl-Pho85p-dependent site on Rvs167p, S379, has the sequence SPPL, consistent with the requirement for a hydrophobic residue at position +3 seen with Pho4p and Gcn4p, whereas the other site, S299, has the sequence SPVS.
Because of functional redundancy among the Pcls (Measday et al., 1994, 1997; Lee et al., 1998), we cannot say which Pcl or Pcls are responsible for phosphorylation of Rvs167p. Preliminary Western blot analysis of Rvs167p from samples taken at various times in the cell cycle has shown that Rvs167p is specifically phosphorylated in the G1 phase of the cell cycle (J. Moffat, personal communication). This suggests that Rvs167p is likely to be phosphorylated in vivo by one or more of the Pcls that is expressed specifically in G1: Pcl1p, Pcl2p, or Pcl9p (Measday et al., 1997). Pho85p, in association with its G1 cyclins, has previously been demonstrated to have a role in cell integrity and polarity (Lee et al., 1998; Lenburg and O'Shea, 2001; Huang et al., 2002). Rvs167p is the first substrate that has been identified for Pho85p in association with a G1 cyclin.
Phosphorylation of Rvs167p by the MAP Kinase Fus3p
Our data indicate that one or more kinases, in addition to Pcl-Pho85p, are required for complete phosphorylation of Rvs167p during vegetative growth. Because Pcl2p-Pho85p can phosphorylate Rvs167p in vitro at all of the three sites that are known to be phosphorylated in vivo, we cannot say whether in a wild-type strain phosphorylation at these sites is redundant or whether the S321 site is phosphorylated only by another kinase in vivo. We have found that the MAP kinase Fus3p is required for full phosphorylation of Rvs167p in cells treated with α factor.
Fus3p and Kss1p are MAP kinases that associate tightly with the MAPK kinase (MEK) Ste7p. A putative binding site for Fus3p/Kss1p has been mapped to the N-terminal 22 amino acids of Ste7p (Bardwell et al., 1996, 2001; Bardwell and Thorner, 1996). A region with a high degree of sequence similarity to the N terminus of Ste7p is found within the BAR domain of Rvs167p, extending from amino acid 148–156 (Aaron Neiman, personal communication). This suggests the possibility that Fus3p could bind to and phosphorylate Rvs167p. We have found that Fus3p, but not the related kinase Kss1p, phosphorylates Rvs167p in vitro. The putative Fus3p/Kss1p docking site on Rvs167p is not required for this phosphorylation (Figure 5A); however, this may be due to our in vitro phosphorylation conditions. Our experiments do not reveal whether Fus3p is the kinase that is redundant with Pcl-Pho85 for phosphorylation of Rvs167p during vegetative growth. It is possible that Fus3p phosphorylates Rvs167p in the presence of α factor and that another (as yet unidentified) kinase phosphorylates Rvs167p during log phase in a manner redundant with Pcl-Pho85p.
In phosphopeptide mapping experiments with in vivo-labeled Rvs167p, we do not detect any Fus3p-dependent phosphopeptides of Rvs167p in log-phase cells (our unpublished results). Since Fus3p activity is stimulated upon treatment with mating pheromone (Peter et al., 1993), it is possible that Fus3p phosphorylates Rvs167p specifically upon activation by mating pheromone; however, we have been unable to test this because under the conditions we require for in vivo labeling of Rvs167p (low phosphate medium containing galactose compounded by the fact that RVS167 overexpression is toxic), cells do not mount a response to pheromone (our unpublished results).
Although Rvs167p is hyperphosphorylated in cells treated with α factor (Figure 4), no clear role for Rvs167p in mating has been identified (Brizzio et al., 1998). One possible role for phosphorylation of Rvs167p during mating is revealed by ultrastructural studies that showed that rvs167 mutants are slowed in the process of digesting the septum, suggesting a role for Rvs167p in cell fusion during mating (Breton et al., 2001). Thus, one role for phosphorylation of Rvs167p by Pcl-Pho85p and/or by Fus3p may be in regulating the actin cytoskeleton during mating. One member of the G1 class of Pho85p cyclins, PCL2, is induced upon treatment with mating pheromone. It is clear, however, that Rvs167p is also phosphorylated during vegetative growth.
Phosphorylation Inhibits Interaction of Rvs167p with Las17p and Ymr192p
Phosphorylation has been shown to disrupt protein-protein interactions between the SH3 domain of amphiphysin I and dynamin and between the central domain of amphiphysin and AP-2 (Slepnev et al., 1998). We have shown, using Far Western analysis, that phosphorylation of Rvs167p inhibits its interaction in vitro with two proteins that bind to the SH3 domain of Rvs167p: Las17p and Ymr192p. In contrast, we have mapped the phosphorylation sites of Rvs167p to amino acids S299, S321, and S379 in the GPA region, amino-terminal to the SH3 domain (Figure 2). The mechanism by which phosphorylation at a distant site affects binding to the SH3 domain of Rvs167p remains unclear. One possibility is that phosphorylation affects Rvs167p binding activity by inducing a conformational change due to the introduction of negative charges in the uncharged GPA region. Deletion of the GPA region has no detectable phenotype, suggesting that the role of the GPA may be simply to link the N- and C-terminal domains (Sivadon et al., 1997). A conformational change in the GPA could alter the way the Rvs167p domains are linked and could affect their interactions with other proteins. This type of phosphoregulation is believed to exist in the case of the mammalian cytoskeleton protein moesin (Huang et al., 1999b). In another example, regulation of the Src kinase Hck by a phosphorylation-induced conformational change has been demonstrated in structural studies (Sicheri et al., 1997).
A second possible explanation for our finding that phosphorylation in the GPA region inhibits binding to the SH3 domain of Rvs167p is that Las17p and Ymr192p may make interactions with both the GPA and the SH3 domains of Rvs167p. Rvs167p could have two independent binding faces for these proteins with the SH3 interaction providing the bulk of the binding energy but the interaction with the GPA, which would be inhibited by phosphorylation, still making a substantial contribution. Results from two-hybrid assays support this latter possibility (Colwill et al., 1999; Madania et al., 1999; Bon et al., 2000).
A Model to Suggest a Biological Role for Phosphorylation of Rvs167p
Our biochemical and genetic studies provide evidence for conserved Cdk and MAP kinase regulation of protein complex formation by the amphiphysin homologue Rvs167p. To explain our data, we suggest a model for phosphorylation of Rvs167p. We propose that the major role for phosphorylation of Rvs167p is to prevent binding or to release its interaction with Las17p. This model is based on the following observations: 1) We have found that under certain growth conditions, the gene encoding a version of Rvs167p that cannot be phosphorylated leads to synthetic growth defects in combination with deletions of SLA1 and/or END3. 2) Sla1p and End3p form a complex with the essential protein Pan1p (Tang et al., 2000). 3) PAN1 has a synthetic growth defect with LAS17 with respect to their roles as activators of the Arp2/3 complex (Duncan et al., 2001). We suggest that Las17p must be free of Rvs167p in order to carry out its function as an activator of the Arp2/3 complex. This function is redundant with some important function carried out by Sla1p and/or End3p (as indicated by our synthetic lethal data). We suggest that in the absence of Las17p-mediated activation of the Arp2/3 complex, the Pan1p/Sla1p/End3p complex becomes essential for activating Arp2/3. Thus in a cell containing a version of Rvs167p that cannot be phosphorylated, Rvs167p inappropriately binds to Las17p, which therefore cannot activate Arp2/3, making Sla1p and End3p (in a complex with Pan1p) essential. In support of this model, we found that phosphorylation of Rvs167p inhibits its interaction with Las17p (Figure 7).
Rvs167p is believed to be an adaptor protein like its mammalian homologue amphiphysin. Our model must represent only one example of how Rvs167p functions in the cell, a role that is regulated by the activity of the Cdk Pcl-Pho85p. We have observed that RVS167-4A is not able to complement an rvs167Δ sla1Δ end3Δ strain for growth on salt-containing medium, but is able to partially complement for growth on salt-free medium (Figure 6C). This is not surprising, because biochemical and genetic data indicate that Rvs167p has roles in multiple complexes in the cell, some of which are predicted to be independent of Pcl-Pho85p phosphorylation. We note that the model we have proposed does not take into account our finding that phosphorylation of Rvs167p also inhibits binding to Ymr192p, whose function is unknown. We can hypothesize that in its putative role as an adaptor protein, a certain population of Rvs167p in the cell is localized in a complex that allows it to sequester Las17p via its SH3 domain, whereas another population of Rvs167p is localized in a complex that allows it to bind Ymr192p in an analogous phosphorylation-dependent manner. Future studies will be directed toward identifying which Rvs167p complexes are formed in vivo in response to a variety of regulatory stimuli to control cell polarity and other functions of the actin cytoskeleton.
Acknowledgments
We are grateful to Aaron Neiman for communicating his observation that Rvs167p contained a consensus Fus3/Kss1 binding site. We thank Chris Nelson and Ivan Sadowski for technical advice and Julie Guzzo for making GST-Rvs167p and Charlie Boone and Christine Humphries for reading the manuscript. This work was supported by an operating grant to B.A. from the National Cancer Institute of Canada with funds from the Canadian Cancer Society. K.M. was supported by a studentship from the Canadian Institute of Health Research (CIHR) and by funds from a Premier's Research Excellence Award (PREA) to B.A. Our SGA laboratories are supported by a Collaborative Genomics Special Project grant from the CIHR and by funds from Genome Canada through the Ontario Genomics Institute.
References
- Amberg, D.C., Basart, E., and Botstein, D. (1995). Defining protein interactions with yeast actin in vivo. Nat. Struct. Biol. 2, 28-35. [DOI] [PubMed] [Google Scholar]
- Antal, J. Pal, G., Asboth, B., Buzas, Z., Patthy, A., and Graf, L. (2001). Specificity assay of serine proteinases by reverse-phase high-performance liquid chromatography analysis of competing oligopeptide substrate library. Anal. Biochem. 288, 156-167. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.M., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1994). Current Protocols in Molecular Biology. New York: John Wiley & Sons.
- Balguerie, A., Sivadon, P., Bonneu, M., and Aigel, M. (1999). Rvs167p, the budding yeast homolog of amphiphysin, colocalizes with actin patches. J. Cell Sci. 112, 2529-2537. [DOI] [PubMed] [Google Scholar]
- Bardwell, A.J., Flatauer, L.J., Matsukuma, K., Thorner, J., and Bardwell, L. (2001). A conserved docking site in MEKs mediates highaffinity binding to MAP kinases and cooperates with a scaffold protein to enhance signal transmission. J. Biol. Chem. 276, 10374-10386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell, L., Cook, J.G., Chang, E.C., Cairns, B.R., and Thorner, J. (1996). Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol. Cell. Biol. 16, 3637-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell, L., and Thorner, J. (1996). A conserved motif at the amino termini of MEKs might mediate high-affinity interaction with the cognate MAPKs. Trends Biochem. Sci. 21, 373-374. [PubMed] [Google Scholar]
- Bauer, F., Urdaci, M., Aigle, M., and Crouzet, M. (1993). Alteration of a yeast SH3 protein leads to conditional viability and defects in cytoskeletal and budding patterns. Mol. Cell. Biol. 13, 5070-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke, J.D., Lacroute, F., and Fink, G.R. (1984). A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197, 345-346. [DOI] [PubMed] [Google Scholar]
- Bon, E., Recordon-Navarro, P., Durrens, P., Iwase, M., Toh-e, A., and Aigle, M. (2000). A network of proteins around Rvs167p and Rvs161p, two proteins related to the yeast actin cytoskeleton. Yeast 16, 1229-1241. [DOI] [PubMed] [Google Scholar]
- Boyle, W., van der Geer, P., and Hunter, T. (1991). Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 201, 110-152. [DOI] [PubMed] [Google Scholar]
- Brachmann, C.B., Davies, A., Cost, G.J., Caputo, E., Li, J., Hieter, P., and Boeke, J.D. (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115-132. [DOI] [PubMed] [Google Scholar]
- Breitkreutz, A., and Tyers, M. (2002). MAPK signaling specificity: it takes two to tango. Trends Cell Biol. 12, 254-257. [DOI] [PubMed] [Google Scholar]
- Breitkreutz, A., Boucher, L., and Tyers, M. (2001). MAPK specificity in the yeast pheromone response independent of transcriptional activation. Curr. Biol. 11, 1266-1271. [DOI] [PubMed] [Google Scholar]
- Breton, A., and Aigle, M. (1998). Genetic and functional relationship between Rvsp, myosin and actin in Saccharomyces cerevisiae. Curr. Genet. 34, 280-286. [DOI] [PubMed] [Google Scholar]
- Breton, A.M., Schaeffer, J., and Aigle, M. (2001). The yeast Rvs161 and Rvs167 proteins are involved in secretory vesicles targeting the plasma membrane and in cell integrity. Yeast 18, 1053-1068. [DOI] [PubMed] [Google Scholar]
- Brizzio, V., Gammie, A.E., and Rose, M.D. (1998). Rvs161p interacts with Fus2p to promote cell fusion in Saccharomyces cerevisiae. J. Cell Biol. 141, 567-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, A.S., and O'Shea, E.K. (2002). Pho85 and signaling environmental conditions. Trends Biochem. Sci. 27, 87-93. [DOI] [PubMed] [Google Scholar]
- Chu, D.S., Pishvaee, B., and Payne, G.S. (1999). A modulatory role for clathrin light chain phosphorylation in Golgi membrane protein localization during vegetative growth and during the mating response of Saccharomyces cerevisiae. Mol. Biol. Cell 10, 713-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colwill, K., Field, D., Moore, L., Friesen, J., and Andrews, B. (1999). In vivo analysis of the domains of yeast Rvs167p suggests Rvs167p function is mediated through multiple protein interactions. Genetics 152: 881-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees, B.L. et al. (2001). A protein interaction map for cell polarity development. J. Cell Biol. 154, 549-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, M.C., Cope, M.J., Goode, B.L., Wendland, B., and Drubin, D.G. (2001). Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat. Cell Biol. 3, 687-690. [DOI] [PubMed] [Google Scholar]
- Figeys, D., McBroom, L.D., and Moran, M.F. (2001). Mass spectrometry for the study of protein-protein interactions. Methods 24, 230-239. [DOI] [PubMed] [Google Scholar]
- Floyd, S.R., Porro, E.B., Slepnev, V.I., Ochoa, G.C., Tsai, L.H., and De Camilli, P. (2001). Amphiphysin 1 binds the cyclin-dependent kinase (cdk) 5 regulatory subunit p35 and is phosphorylated by cdk5 and cdc2. J. Biol. Chem. 276, 8104-8110. [DOI] [PubMed] [Google Scholar]
- Goldstein, A.L., and McCusker, J.H. (1999). Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast 15, 1541-1553. [DOI] [PubMed] [Google Scholar]
- Guichet, A. et al. (1997). The nuclear receptor homologue Ftz-F1 and the homeodomain protein Ftz are mutually dependent cofactors. Nature 385, 548-552. [DOI] [PubMed] [Google Scholar]
- Gustin, M.C., Albertyn, J., Alexander, M., and Davenport, K. (1998). MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62, 1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, S.N., Hunt, H.D., Horton, R.M., Pullen, J.K., and Pease, L.R. (1989). Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77, 51-59. [DOI] [PubMed] [Google Scholar]
- Huang, D., Farkas, I., and Roach, P.J. (1996). Pho85p, a cyclindependent protein kinase, and the Snf1p protein kinase act antagonistically to control glycogen accumulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 4357-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D., Moffat, J., and Andrews, B. (2002). Dissection of a complex phenotype by functional genomics reveals roles for the yeast cyclin-dependent protein kinase Pho85 in stress adaptation and cell integrity. Mol. Cell. Biol. 22, 5076-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, D., Patrick, G., Moffat, J., Tsai, L.-H., and Andrews, B. (1999a). Mammalian Cdk5 is a functional homologue of the budding yeast Pho85 cyclin-dependent protein kinase. Proc. Natl. Acad. Sci. USA 96, 14445-14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L., Wong, T.Y., Lin, R.C., and Furthmayr, H. (1999b). Replacement of threonine 558, a critical site of phosphorylation of moesin in vivo, with aspartate activates F-actin binding of moesin. Regulation by conformational change. J. Biol. Chem. 274, 12803-12810. [DOI] [PubMed] [Google Scholar]
- Lee, J., Colwill, K., Aneliunas, V., Tennyson, C., Moore, L., Ho, Y., and Andrews, B. (1998). Interaction of yeast Rvs167 and Pho85 cyclin-dependent kinase complexes may link the cell cycle to the actin cytoskeleton. Curr. Biol. 8, 1310-1321. [DOI] [PubMed] [Google Scholar]
- Lenburg, M.E., and O'Shea, E.K. (2001). Genetic evidence for a morphogenetic function of the Saccharomyces cerevisiae Pho85 cyclin-dependent kinase. Genetics 157, 39-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lila, T., and Drubin, D. (1997). Evidence for physical and functional interactions among two Saccharomyces cerevisiae SH3 domain proteins, an adenylyl cyclase-associated protein and the actin cytoskeleton. Mol. Biol. Cell 8, 367-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie, A., DeMarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953-961. [DOI] [PubMed] [Google Scholar]
- Madania, A., Dumoulin, P., Grava, S., Kitamoto, H., Scharer-Brodbeck, C., Soulard, A., Moreau, V., and Winsor, B. (1999). The Saccharomyces cerevisiae homologue of human Wiskott-Aldrich syndrome protein Las17p interacts with the Arp2/3 complex. Mol. Biol. Cell 10, 3521-3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Measday, V., Moore, L., Ogas, J., Tyers, M., and Andrews, B. (1994). The Pcl2 (ORFD)-Pho85 cyclin-dependent kinase complex: a cell cycle regulator in yeast. Science 266, 1391-1395. [DOI] [PubMed] [Google Scholar]
- Measday, V., Moore, L., Retnakaran, R., Lee, J., Donoviel, M., Neiman, A., and Andrews, B. (1997). A family of cyclin-like proteins that interact with the cyclin-dependent kinase, Pho85. Mol. Cell. Biol. 17, 1212-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meimoun, A., Holtzman, T., Weissman, Z., McBride, H.J., Stillman, D.J., Fink, G.R., and Kornitzer, D. (2000). Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCF-CDC4 ubiquitin-ligase complex. Mol. Biol. Cell 11, 915-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat, J., Huang, D., and Andrews, B.J. (2000). Functions of Pho85 cyclin-dependent kinases in budding yeast. Prog. Cell Cycle Res. 4, 97-106. [DOI] [PubMed] [Google Scholar]
- Navarro, P., Durrens, P., and Aigle, M. (1997). Protein-protein interaction between the RVS161 and RVS167 gene products of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1343, 187-192. [DOI] [PubMed] [Google Scholar]
- Nishizawa, M., Kawasumi, M., Fujino, M., and Toh-e, A. (1998). Phosphorylation of Sic1, a cyclin-dependent kinase (Cdk) inhibitor, by Cdk including Pho85 kinase is required for its prompt degradation. Mol. Biol. Cell 9, 2393-2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill, E.M., Kaffman, A., Jolly, E.R., and O'Shea, E.K. (1996). Regulation of Pho4 nuclear localization by the Pho80-Pho85 cyclin-CDK complex. Science 271, 209-212. [DOI] [PubMed] [Google Scholar]
- Pawson, T., and Scott, J.D. (1997). Signaling through scaffold, anchoring, and adaptor proteins. Science 278, 2075-2080. [DOI] [PubMed] [Google Scholar]
- Peter, M., Gartner, A., Horecka, J., Ammerer, G., and Herskowitz, I. (1993). FAR1 links the signal transduction pathway to the cell cycle machinery in yeast. Cell 73: 747-760. [DOI] [PubMed] [Google Scholar]
- Ronicke, V., Graulich, W., Mumberg, D., Muller, R., and Funk, M. (1997). Use of conditional promoters for expression of heterologous proteins in Saccharomyces cerevisiae. Methods Enzymol. 283, 313-322. [DOI] [PubMed] [Google Scholar]
- Shemer, R., Meimoun, A., Holtzman, T., and Kornitzer, D. (2002). Regulation of the transcription factor Gcn4 by Pho85-Cyclin Pcl5. Mol. Cell. Biol. 22, 5395-5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F. (1991). Getting started with yeast. Methods Enzymol. 194, 3-21. [DOI] [PubMed] [Google Scholar]
- Sicheri, F., Moarefi, I., and Kuriyan, J. (1997). Crystal structure of the Src family tyrosine kinase Hck. Nature 385, 602-609. [DOI] [PubMed] [Google Scholar]
- Singer-Krüger, B., and Ferro-Novick, S. (1997). Use of a synthetic lethal screen to identify yeast mutants impaired in endocytosis, vacuolar protein sorting and the organization of the cytoskeleton. Eur. J. Cell Biol. 74, 365-375. [PubMed] [Google Scholar]
- Sivadon, P., Crouzet, M., and Aigle, M. (1997). Functional assessment of the yeast Rvs161 and Rvs167 protein domains. FEBS Lett 417, 21-27. [DOI] [PubMed] [Google Scholar]
- Slepnev, V.I., Ochoa, G.C., Butler, M.H., Grabs, D., and De Camilli, P.D. (1998). Role of phosphorylation in regulation of the assembly of endocytic coat complexes. Science 281, 821-824. [DOI] [PubMed] [Google Scholar]
- Tang, H.Y., and Cai, M. (1996). The EH-domain-containing protein Pan1 is required for normal organization of the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Cell. Biol. 16, 4897-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, H.Y., Xu, J., and Cai, M. (2000). Pan1p, End3p, and S1a1p, three yeast proteins required for normal cortical actin cytoskeleton organization, associate with each other and play essential roles in cell wall morphogenesis. Mol. Cell. Biol. 20, 12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, A.H. et al. (2002). A combined experimental and computational strategy to define protein interaction networks for peptide recognition modules. Science 295, 321-324. [DOI] [PubMed] [Google Scholar]
- Tong, A.H. et al. (2001). Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294, 2364-2368. [DOI] [PubMed] [Google Scholar]
- van der Geer, P., and Hunter, T. (1994). Phosphopeptide mapping and phosphoamino acid analysis by electrophoresis and chromatography on thin-layer cellulose plates. Electrophoresis 15, 544-554. [DOI] [PubMed] [Google Scholar]
- Warner, J.R. (1991). Labeling of RNA and phosphoproteins in Saccharomyces cerevisiae. Methods Enzymol. 194, 423-434. [DOI] [PubMed] [Google Scholar]
- Watson, H.A., Cope, M.J., Groen, A.C., Drubin, D.G., and Wendland, B. (2001). In vivo role for actin-regulating kinases in endocytosis and yeast epsin phosphorylation. Mol. Biol. Cell 12: 3668-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter, D.C., Choe, E.Y., and Li, R. (1999). Genetic dissection of the budding yeast Arp2/3 complex: a comparison of the in vivo and structural roles of individual subunits. Proc. Natl. Acad. Sci. USA 96, 7288-7293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E.A. et al. (1999). Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901-906. [DOI] [PubMed] [Google Scholar]
- Zeng, G., and Cai, M. (1999). Regulation of the actin cytoskeleton organization in yeast by a novel serine/threonine kinase Prk1p. J. Cell Biol. 144, 71-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, G., Yu, X., and Cai, M. (2001). Regulation of yeast actin cytoskeleton-regulatory complex Pan1p/Sla1p/End3p by serine/threonine kinase Prk1p. Mol. Biol. Cell 12, 3759-3772.Singer-Kruger and Ferro-Novick 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]