Abstract
Objective
To develop and test an auditing methodology for detecting errors in medical terminologies satisfying systematic inheritance. This methodology is based on various abstraction taxonomies that provide high-level views of a terminology and highlight potentially erroneous concepts.
Design
Our auditing methodology is based on dividing concepts of a terminology into smaller, more manageable units. First, we divide the terminology’s concepts into areas according to their relationships/roles. Then each multi-rooted area is further divided into partial-areas (p-areas) that are singly-rooted. Each p-area contains a set of structurally and semantically uniform concepts. Two kinds of abstraction networks, called the area taxonomy and p-area taxonomy, are derived. These taxonomies form the basis for the auditing approach. Taxonomies tend to highlight potentially erroneous concepts in areas and p-areas. Human reviewers can focus their auditing efforts on the limited number of problematic concepts following two hypotheses on the probable concentration of errors.
Results
A sample of the area taxonomy and p-area taxonomy for the Biological Process (BP) hierarchy of the National Cancer Institute Thesaurus (NCIT) was derived from the application of our methodology to its concepts. These views led to the detection of a number of different kinds of errors that are reported, and to confirmation of the hypotheses on error concentration in this hierarchy.
Conclusion
Our auditing methodology based on area and p-area taxonomies is an efficient tool for detecting errors in terminologies satisfying systematic inheritance of roles, and thus facilitates their maintenance. This methodology concentrates a domain expert’s manual review on portions of the concepts with a high likelihood of errors.
Introduction
Terminologies are increasingly used in biomedical and health care applications. With the awareness of their value, more institutions, in government, academia, and industry, are allocating resources needed for the design of various terminologies. The belief that these investments are more than made up for, in operational cost-savings and improved quality of care, is spreading.
Much of the time and effort in terminology development is spent on increasing coverage of various biomedical areas with the addition of new concepts and relationships. Further resources are invested in classification tools, user-friendly interfaces, servers, etc. But very little has been directed toward auditing of terminology content. While we have recently seen a surge in papers on auditing techniques for terminologies (see Background), there does not seem to be a wide-spread commitment in the terminology industry to making auditing an important part of the design, development, and maintenance phases. We believe that recognizing the importance of auditing as an integral part of the terminology design life cycle is critical for the terminology industry (see Background).
Auditing large terminologies is a major challenge facing the medical informatics community. Due to the size and complexity of terminologies, it is unavoidable that errors will occur. To demonstrate this point, we set out to search for errors in one of the new generation of terminologies, the National Cancer Institute Thesaurus (NCIT). For details, see Background. For this purpose a proper auditing methodology is required.
In this paper, we present an auditing methodology for terminologies satisfying systematic inheritance of roles (relationships). Our auditing methodology comprises two major phases: (1) the automated preparatory phase; and (2) the manual guided-discovery phase. Phase (1) consists of 4 steps. First, the terminology is divided into groups of concepts with the same roles. This division provides structurally uniform collections of concepts. From this division, the second step constructs a compact abstraction network, called an area taxonomy. Third, the division is refined into groups of concepts called p-areas that are both structurally uniform and singly rooted. Finally, an enhanced abstraction network, called the p-area taxonomy, is derived.
It is very difficult to comprehend terminologies because they are typically huge in size (number of concepts) and have high complexity (proportional to the number of relationships). 1 Auditing, which requires comprehension, is even more difficult since it is like finding needles in a haystack, as the target of the search is unknown and usually no semantic or structural guidance exists. The two taxonomies derived in Phase (1) provide compact, comprehensible views of the terminology, and tend to highlight its relevant features, while hiding unimportant details.
In Phase (2)—the actual auditing phase—elements of the p-area taxonomy are used to guide the auditor to suspicious parts of the terminology. For example, our previous experience with a related methodology 2 in the context of the MED 3 has shown that areas of small size tend to denote irregularities in the terminology and therefore may reveal errors. The p-area taxonomy readily exposes such situations to the auditor. An application of our methodology to a sample hierarchy of the 2004 NCIT is presented. An analysis of the errors found is also presented.
Background
Importance of Auditing Terminologies
The common perception in the terminology “industry,” reflected in anecdotal evidence, is that customers want to increase coverage, and this is what they are willing to pay for. Note that when we refer to the terminology “industry,” we include departments in corporations, government agencies, hospitals, and academic institutions that design, maintain, and use terminologies. If a customer discovers an error and complains about it, it will be fixed. But undertaking an extensive auditing effort is typically not what the customer wants.
Such a situation is common in emerging industries, but not in mature ones. For example, as the software industry matured, different models for software life cycle processes have been developed. However, in recent years it has become clear that no such model is complete without activities dedicated to assuring the correctness of the software. Typically, software life cycle models list auditing as part of quality assurance, one of the support activities. 4 It is normally assumed that a software quality assurance audit is performed by a team that is independent of the development team. Life cycle models have also been expanded to knowledge-based systems, such as in an application of auditing in the development of knowledge-based expert systems for business and finance. 5 We observe that auditing has been typically absent from the life cycle of many ontologies, terminologies, and controlled vocabularies, and that this omission needs to be rectified. Auditing is essential since terminologies underlie decision-support systems, clinical patient records, health care administrative systems, etc., and errors in a terminology will propagate to errors in these systems, which in turn may result in endangering the life or quality of life of a patient.
In our view, terminologies are now being created by a maturing industry. To support this claim, we note the recent emergence of a generation of medical terminologies satisfying the desiderata of Cimino. 6 These terminologies have sound theoretical models such as description logics 7,8 and frames. 9 Examples include the Systematized Nomenclature of Medicine Clinical Terms (SNOMED CT), 10 the Foundational Model of Anatomy (FMA), 11 the National Cancer Institute Thesaurus (NCIT), 12 Lab LOINC, 13 and the Medical Entities Dictionary (MED). 3 They all have accompanying software tools—either commercial or of commercial quality—that provide users with convenient interfaces: e.g., Protégé 14 used for FMA, and Apelon’s Terminology Development Environment (TDE) 15 used for SNOMED and NCIT. These terminologies tend to be of substantial size and complexity, and they keep growing; e.g., the recent version (January ’06) of SNOMED contains over 366,000 concepts, while the January ’03 version contained only about 344,000 concepts. Similarly, NCIT has grown in two years from about 25,000 concepts to 42,404 concepts.
Our research group, jointly with J. J. Cimino and G. Hripcsak of Columbia University, distributed a questionnaire about Unified Medical Language System (UMLS) 16,17,18 users’ applications and priorities to the UMLS users mailing list. We received 70 responses. Three questions dealt with auditing. Two asked to what extent the user is bothered by a list of twelve kinds of errors, with the choice of answers: “not at all,” “a little,” “moderately,” and “a lot,” coded by the values 0, 1, 2, and 3, respectively. It became clear from the results that there is a demand for high-quality auditing. For example, the average user is “bothered moderately” by incorrect hierarchical relationships (1.97), incorrect associative relationships (2.11), incorrect semantic-type assignments (2.15), missing hierarchical relationships (1.86), and missing semantic-type assignments (1.76). For the other kinds of errors, the average user was bothered to an extent that is between “a little” and “moderately” (1.46).
Furthermore, the responding UMLS users clearly saw auditing as a high priority since, on average, they would allocate 35% of a putative NLM budget to auditing, the highest of all given options by a large margin. The three trailing categories, “designing a derived terminology,” “improving interfaces,” and “extending coverage,” were assigned only 24%, 20%, and 16% of the budget, respectively.
In summary, the results of our study showed that users of the UMLS care about eliminating errors and would like to see a substantial portion of the available budget allocated to auditing activities for quality assurance. These results confirm our claims that UMLS users are demanding serious auditing efforts so they can rely on the represented knowledge with a reasonable level of confidence. It is a research issue whether users of other medical terminologies share similar opinions to those expressed by the UMLS users in our study.
Terminology Auditing Methodologies
Researchers have developed different methodologies to help with auditing terminologies. For the UMLS alone, we find a variety of approaches. For example, semantic methods 19 have been used to detect classification errors. Techniques have been developed for finding cycles, 20 reverse hierarchical relationships, 21 concept redundancy and ambiguity, 22 and redundant categorization. 23 The UMLS Semantic Network (SN) 24 was revised through the reclassification of the semantic types. 25 Object-oriented models have been employed to support navigation, maintenance, and auditing. 26,27 A method to find undetected synonymy in the UMLS has been developed. 28 A technique, 29 based on the notion of an SN metaschema, 30 has been applied to the UMLS.
Furthermore, terminological, ontological and linguistic techniques were utilized to audit the NCIT and SNOMED. 31–34 A technique based on an object-oriented database representation 2 has been developed for auditing the MED. 3 Other techniques have been used to find errors caused by design problems in the Gene Ontology. 35–38 Error detection 39 for the Diagnoses for Intensive Care Evaluation (DICE) System 40 is based on migration to description logic.
NCI Thesaurus
The National Cancer Institute Thesaurus (NCIT) was designed in response to a need for a consistent, shared vocabulary for the various projects and initiatives at the NCI, as well as in the broader cancer research community. The NCIT covers clinical and basic research as well as administrative terminology.
The NCIT’s design is based on description logic. It has a tool for automatic classification couched in this model. The NCIT has defined and inferred versions. The defined version is the one containing the assertions made about each concept by the editors. The inferred version additionally includes assertions and tree placements inherited during DL classification. In this paper, we used the inferred version of the NCIT to do our analysis.
The NCIT’s data model consists of four basic elements: concepts, kinds, roles, and properties. 41 The foundational unit of information is the concept. There are 42,404 concepts organized into 21 disjoint hierarchies, covering different subject areas such as Biological Process, Genes, and Gene Products. Each hierarchy consists of IS-A relationships between child and parent concepts forming a directed acyclic graph (DAG). The largest hierarchy, Diseases, Disorders, and Findings, contains 9,613 concepts. Roles describe semantic (non IS-A) relationships between concepts and are inherited by a child concept from a parent concept along the IS-A hierarchy. For example, the concept Malignant Breast Neoplasm ∗ has the role located in connecting it to Breast. Since Breast Ductal Carcinoma IS-A Malignant Breast Neoplasm, it inherits the role located in with the target Breast.
Each concept is associated with exactly one of 21 disjoint sets called kinds, representing major subdivisions of the NCIT, e.g., the Biological Process Kind and the Gene Kind. Properties are used to describe a concept. Examples include: definition, preferred name, synonyms, and semantic type.
Methods
Dividing a Terminology into Areas
Our terminology-dividing methodology is based on the notions of area, structure, 30 and root, defined as follows:
Definition 1 (Area): An area is a group of all concepts that have exactly the same roles.
Definition 2 (Structure): The structure of a concept (and an area) is the set of its roles.
Hence, an area of a terminology is structurally uniform.
Definition 3 (Root of an Area): A concept X in an area A is called a root of A if no parents of X are in A.
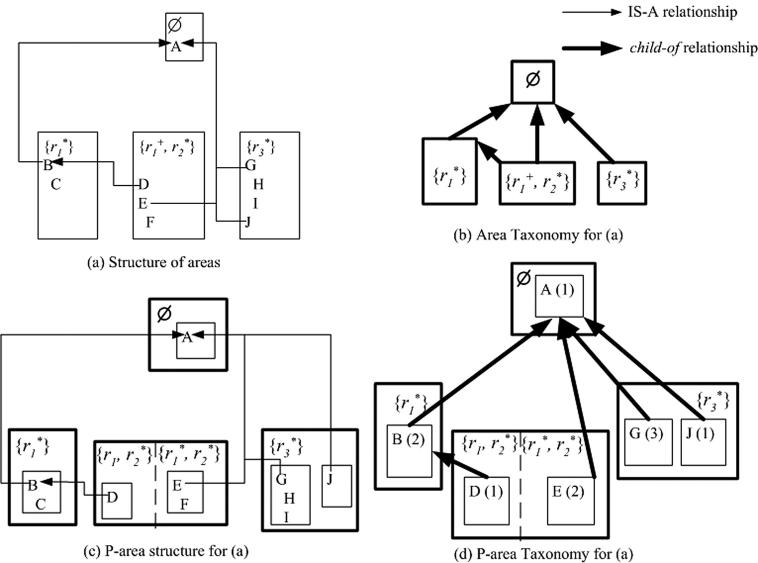
A terminology is divided so that each concept belongs to one and only one area according to its structure. We assign a name to an area that consists of the list of its roles inside braces. The area with no roles is named Ø (empty set). ▶(a) shows the division of a sample abstract terminology into four areas. Areas are shown as boxes. An IS-A link from a concept to another concept in the same area is indicated by indentation. B and C are grouped into the area with the name {r 1} since both of them have only the role r 1. C is indented relative to B, because C IS-A B. Similarly, D, E, and F are in the area with the name {r 1, r 2}, and G, H, I, and J are in the area {r 3}. An IS-A link from a concept to another concept in a different area is indicated by a thin arrow. Thus there is a thin arrow from D to B. Concept A is in the area Ø because it has no roles at all. The symbols “*” and “+” will be explained later. In ▶(a), D and E are the roots of {r 1, r 2}. The concept A is the root of Ø and the concept B is the root of {r 1}. G and J are the roots of {r 3}.
Figure 1.
Examples of an area taxonomy and p-area taxonomy.
We repeat that all the concepts within an area’s box share the same structure. This is due to the inheritance of roles along the IS-A hierarchy that enables the structural division into areas. If inheritance can be interrupted, e.g., by a blocking mechanism as in the UMLS Semantic Network, 24 then the division into groups of concepts with identical roles is more problematic. 30
The division into areas lends itself to a useful kind of abstraction diagram that we call the area taxonomy (AT). The AT serves as a compact, high-level representation of the terminology and shows the distribution of its roles. Specifically, the AT is a DAG of area nodes (▶(b)). An area (a node) is displayed as a bold box and is named by the list of roles of all its concepts. Nodes can be connected to other nodes via child-of relationships, which serve as abstractions of the underlying IS-As in the terminology. Specifically, one node X is child-of another node Y if a root of X has an IS-A relationship to some concept in Y. A child-of link is shown as a bold arrow. Note that the concept in Y need not be a root. The area {r 1} is a child-of Ø because B IS-A A, where B is the root of {r 1} and A is in Ø.
Definition 4 (Introducing concept): A concept at which one or more new roles are introduced into the terminology is called an introducing concept.
In every area, the root is, by definition, an introducing concept because it introduces one or more new roles. The only reason why this concept is a root—and is not in its parent’s area—is that it introduces new roles. Other roles may be inherited.
Role-introduction points are highlighted by placing a “*” next to the name of any role introduced at a root of the particular area. The concept B is an introducing concept for the role r 1 while both G and J are introducing the role r 3. We note that for some areas there are several introduction patterns for the same structure. In ▶(a), the role r 1 in {r 1, r 2} is inherited from B by root D but introduced by root E. Role r 2 is introduced at both roots D and E. In such a case of varying introduction patterns for a role (e.g., r 1), we mark the role with “+” instead of “*” in the area name. For example, in the area name {r 1 +, r 2*} in ▶(a), r 1 is marked by a plus sign.
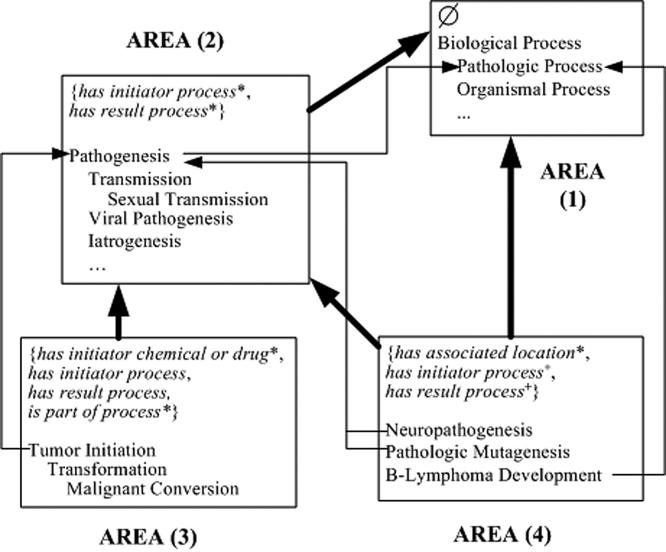
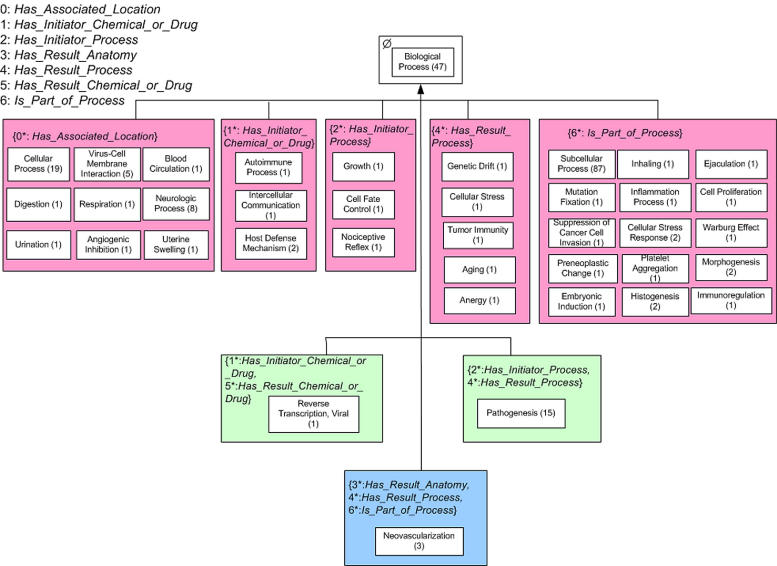
To illustrate the above definitions, we show the areas of a few concepts from the NCIT’s Biological Process hierarchy in ▶. Some concepts are not displayed, and their absence is marked by “…”. ▶ shows concepts in four areas. Due to the long area names we have also numbered the areas: AREA (1) Ø, AREA (2) {has initiator process*, has result process*}, AREA (3) {has initiator chemical or drug*, has initiator process, has result process, is part of process*}, and AREA (4) {has associated location*, has initiator process+, has result process +}.
Figure 2.
Area Taxonomy with a few areas and their concepts.
There are 15 concepts in AREA (2), all of which share the same structure comprising these two roles. Similarly, the three concepts Tumor Initiation, Transformation, and Malignant Conversion of AREA (3) share the same four roles. In fact, each area has a structurally uniform set of concepts. The child-of from AREA (3) to AREA (2) is due to the IS-A from the root Tumor Initiation to the root Pathogenesis, while the IS-A relationship from Pathogenesis to Pathological Process is responsible for the child-of from AREA (2) to AREA (1).
Dividing an Area into P-Areas
An area of a terminology is by definition structurally uniform. However, an area might not be semantically uniform in the sense of having a unique root concept that generalizes all its descendants in the area. A unique root can convey the semantics of the whole group. For example, the unique root Pathogenesis of AREA (2) conveys the general semantics of all concepts in its area. When a role can be introduced at multiple points in the IS-A hierarchy, as in the NCIT, then an area may have multiple roots.
As we shall see later, the NCIT area {has associated location*} has a group of 19 concepts rooted at Cellular Process and another group of eight rooted at Neurologic Process. Obviously, both these groups are semantically uniform, but the area is not.
Therefore, we further divide areas into concept groups, called partial areas (p-areas), that are structurally uniform and singly-rooted. A p-area is named after its unique root since the root generalizes all the p-area’s other concepts.
Definition 5 (P-area): A p-area in an area A is a group of concepts that contains only one root X and all descendants of X in A.
We can further divide the areas in ▶(a) into six p-areas according to the roots A, B, E, G, and J (▶(c)). The division of areas into p-areas leads to an expanded, two-level AT that we call the p-area taxonomy (PAT). The PAT is a DAG, with p-areas represented as nodes and connected to other p-areas via child-of relationships. One p-area X is a child-of another p-area Y if the root concept X has an IS-A relationship to some concept in Y. To capture the additional level of division, p-areas are grouped into the AT’s areas. In a PAT diagram, areas are displayed as boxes (▶(d)). A “small caps” font will be used for p-areas. The p-areas of an area are drawn as boxes inside their respective area boxes. The child-of links are displayed as bold arrows. The name of a p-area is identical to the name of its root. Therefore, a p-area box contains the name of its root, which conveys the essence (semantics) of the group. The number in parentheses represents the number of concepts in the respective p-area, including the root. The PAT offers a view that provides both relationship distribution information across the entire terminology and further hierarchical grouping information within areas.
As in the area taxonomy, we use a “*” to indicate the p-area where a role is introduced. The lack of a “*” means the role is inherited. Note that the “+” in the AT is disambiguated in the PAT. Each root of a p-area has a distinct introduction pattern. Areas with a “+” in their names are divided into several parts of a specific introduction pattern, separated from one another in the PAT diagram by a dashed line. Each of these parts includes the p-areas of the corresponding introduction pattern. ▶(c) shows that the area {r 1 +, r 2*} in ▶(a) is separated (by the dashed line) into two parts: {r 1, r 2*} for the p-area D and {r 1*, r 2*} for the p-area E with child-of relationships to the p-areas B and A, respectively.
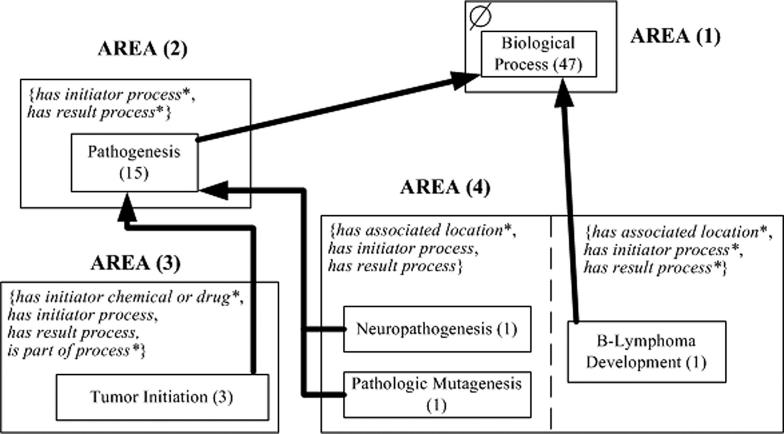
The PAT for ▶ appears in ▶. (Note that ▶ and ▶ are excerpts of the complete AT and PAT for the Biological Process hierarchy shown in ▶ and ▶, respectively.) For some areas, there is only one root implying that the concepts of such an area are not only structurally uniform but also semantically uniform due to the fact that all are specializations of the root. Such areas have only one p-area named after their respective roots. In other words, the area box contains only one p-area box. This is the case for AREA (1)—Biological Process, AREA (2)—Pathogenesis, and AREA (3)—Tumor Initiation. Some areas, however, have several roots. In AREA (4), each of the three concepts is a root and constitutes a p-area. Each of these three p-areas has a uniform semantics captured by its name. Moreover, we see two different role introduction patterns, graphically separated by a dashed line. For the left two p-areas, has associated location is introduced, while has initiator process and has result process are inherited. This is denoted as {has associated location*, has initiator process, has result process}. For B-Lymphoma Development on the right of the dashed line, all three roles are introduced, since Pathologic Process in Ø has no roles. This is denoted as {has associated location*, has initiator process*, has result process*}. Thus, the area is divided by the dashed line into two parts according to the introduction pattern. Each part contains the proper p-areas with separate child-of’s according to the parent of the root. In this way, the “+” notation in the area’s name is disambiguated. No plus sign appears in the PAT.
Figure 3.
Excerpt of PAT.
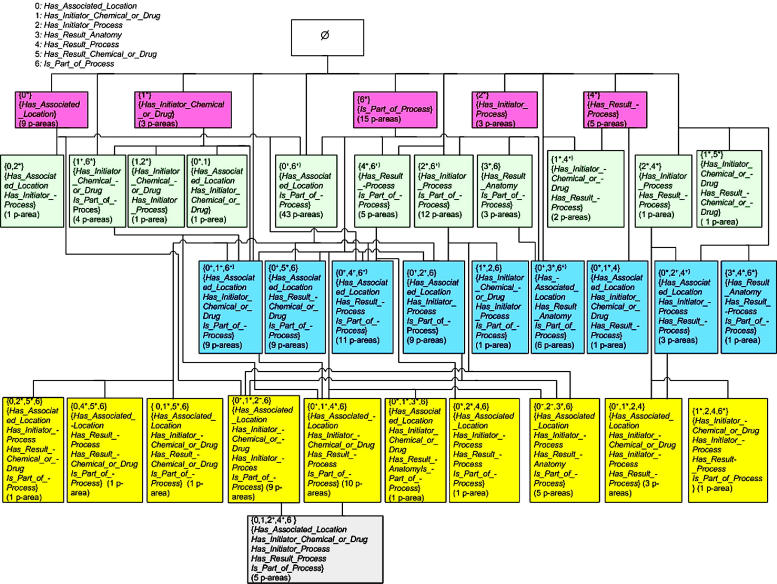
Figure 4.
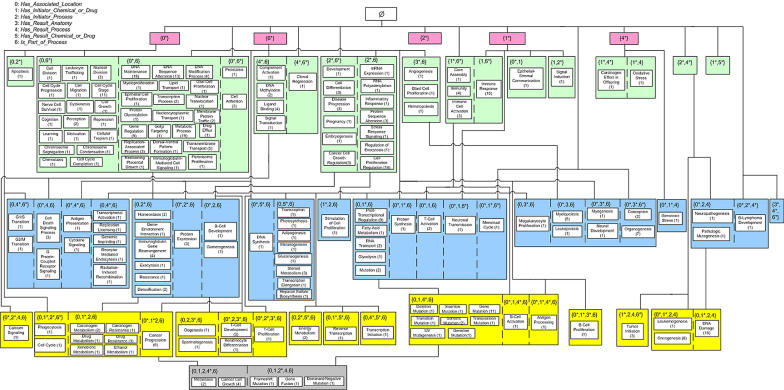
AT for the Biological Process hierarchy.
Figure 5.
PAT for the Biological Process hierarchy.
The process of dividing the terminology into areas and p-areas as well as the design of the AT and PAT was fully automated. The programs to carry out these tasks were written in PERL and contain 700 lines of code. They took two weeks to write, test, and debug. The AT and PAT diagrams were manually created using the Visio graphical tool. An advantage of the AT and PAT is in providing groupings of similar concepts into small collections. Furthermore, the taxonomies guide the auditor to concentrate on groups of concepts with higher likelihood of errors, as discussed below.
Auditing Methodology
Our “divide and conquer” auditing methodology first divides the terminology into areas and into p-areas. Then the conquer phase utilizes these p-areas to expose errors, otherwise buried undetected in the complex terminology. The AT and PAT are typically smaller than the original concept network. These compact views allow the terminologies’ auditors to see it in a new, different light. These views can help in the orientation to and navigation of the terminology in the auditing process. Looking at the concepts, grouped according to their structure and root, can help in exposing problems.
In the first part of the manual auditing phase, we utilize the notion of “concept similarity” to identify omissions and misplacements of concepts. Two concepts are structurally similar if they share the same set of roles and are thus in the same area. Two concepts in the same area are called semantically similar if they share the same root and are in the same p-area. If we find that two concepts, similar in their essence, are in different p-areas in the PAT (or, worse, in different areas), there may be some inconsistencies or errors for some of these concepts. For example, Inhaling and Respiration (see ▶) ▶ are similar in essence, but are in different areas. Also, if a concept similar in its essence to concepts of a p-area is missing from that p-area or from the terminology overall, this may indicate an unjustified absence. For example, the concept Exhaling, related to Inhaling, is missing from the NCIT.† It is easier and more effective for an auditor to detect irregularities when reviewing relatively small areas and p-areas of similar concepts, due to the limited capacity of human comprehension and memory.
Figure 6.
A portion of the PAT for the Biological Process hierarchy.
Figure 7.
Descendants of Organismal Process in (a) NCIT hierarchy indented format, and (b) Selected areas and p-areas.
Due to the limited resources available for auditing and the desire to optimize their impact, our methodology is intended to check a limited number of concepts whose probability of being erroneous is high. Our techniques are designed to use automated means to identify groups of concepts with high likelihoods of errors, where the manual review is to be concentrated.
The second part of the audit phase follows this approach and focuses on “small” p-areas, having very few concepts. Our previous experience 2,27,29 suggests that whenever we have small groups of “similar” concepts, there is a high likelihood that these groups represent irregularities that are manifestations of errors more severe than omissions and misplacements of concepts. The reason for this is as follows. If a p-area exists due to its legitimate structure and semantics, then there would probably be quite a few, or at least several concepts, in it. For example, the legitimate p-area Subcellular Process(87) (see ▶) contains the largest number of biological process concepts at the subcellular level. On the other hand, a p-area containing only one or two concepts may indicate an error where no concepts at all should be grouped in the particular manner. For example, in the p-areas Inhaling(1) and Ejaculation(1) (see ▶), the concepts are missing a role and therefore end up in erroneous p-areas by themselves. In our auditing methodology, we especially need to examine all concepts in the PAT’s small p-areas.
In the following, we will need to denote the number of p-areas within areas and concepts within p-areas.
Definition 6 (Size): The size of a p-area (area) is its number of concepts.
Definition 7 (Cardinality): The cardinality of an area is its number of p-areas.
Definition 8 (k̄-p-area): A k̄-p-area is a p-area of size k or less.
Note that we use overline k̄ to indicate integers and differentiate them from “p” (partial). Example: a 3̄-p-area is a p-area that has 1, 2, or 3 concepts.
Definition 9 (m̄-area): An m̄-area is an area of cardinality m or less.
Definition 10 (m̄-k̄ -area): An m̄-k̄-area is an m̄-area that consists of k̄-p-areas only.
In later sections, we will use 3̄-3̄-areas to test the following two hypotheses.
Hypothesis 1: The probability of erroneous concepts is higher for k̄-p-areas with small k than for k̄-p-areas with large k.
Hypothesis 2: The likelihood of errors in concepts of a k̄-p-area with small k is higher in an m̄-area with low m than in an m̄-area of high m.
In Hypothesis 2, we further differentiate between small p-areas in areas of high cardinality and low cardinality. In the first case, we have many concepts sharing the same structure and being hierarchically independent of one another, which is a likely configuration. An example of such an area is {has associated location+, is part of process +} (see ▶), which has 43 p-areas, 33 of which have only one or two concepts. Only one error was discovered in the 124 concepts of this area.
In the second case, we encounter one or very few hierarchically related concepts with a unique combination of roles. The rare occurrence of the structure of this p-area may indicate an error. Consider, for example, the single concept Transcription initiation in its p-area (see ▶). This p-area is the only one in {has associated location, has result process*, has result chemical or drug*, is part of process}. As a matter of fact, the role with the target Transcription should be is part of process instead of has result process (as is the case in the new release of the NCIT). After this change, this p-area will belong to {has associated location +, has result chemical or drug*, is part of process}, which already has nine p-areas (see ▶).
Following our two hypotheses, our auditing methodology concentrates the typically limited time available for an expert’s manual review on the p-areas with a relatively high likelihood of errors. To test these two hypotheses, we conducted an extensive audit of one of NCIT’s hierarchies, auditing all its p-areas, small and large.
Results
AT and PAT for an NCIT Hierarchy
We have chosen to demonstrate both the AT and PAT for the BP hierarchy of the NCIT’s 2004 release. ▶ shows the AT organized by levels according to the number of roles. There are 7 roles, numbered from 0 to 6, defined for the concepts of the BP hierarchy. The levels of the AT are labeled 0 to 5 according to the number of roles in each. The 589 concepts in the BP hierarchy are grouped into 37 areas (see ▶). For example, 38 concepts are grouped into {has associated location*}. For each area, we list the cardinality (i.e., its number of p-areas).
▶ shows the PAT for the BP hierarchy. Due to lack of space, ▶ does not show all p-areas. Omitted p-areas are in ▶ in a subhierarchy of the PAT used later for auditing. The numbers in parentheses are the total numbers of concepts in the respective p-areas. The previously mentioned area {has associated location*} (▶) is further divided into nine p-areas: cellular process(19), virus-cell membrane interaction(5), blood circulation(1), digestion(1), urination(1), respiration(1), neurologic process(8), uterine swelling(1), and angiogenic inhibition(1).
As for an example of the separation of an area according to different introduction patterns, the area {has associated location +, is part of process +} in ▶ is separated into three different parts: {has associated location, is part of process*} with 19 p-areas; {has associated location*, is part of process} with 22 p-areas; and {has associated location*, is part of process*} with two p-areas, whose roots introduce both roles since their parents are in Ø.
The child-of relationships in a PAT are defined from p-area to p-area (▶) as described in the Methods section. However, the number of such relationships in a large PAT makes their complete display impractical. The division of an area into different parts, according to the introduction pattern of its p-areas, enables a more compact display. Instead of many child-of relationships emanating from each p-area of a part of an area, we display only a single child-of from that part to the unique part of another area containing the target p-areas of all the original child-of’s. For example, see the child-of from the {0*, 3, 6} part with the two p-areas, myelopoiesis(5) and leukopoiesis(1), to the area {3*, 6}, with 3 p-areas, in ▶. This area, in turn, has a unique child-of to area {6*} since it has a unique introduction pattern. This graphical convention enables the display of a large, complex PAT without cluttering it with an excessive number of lines.
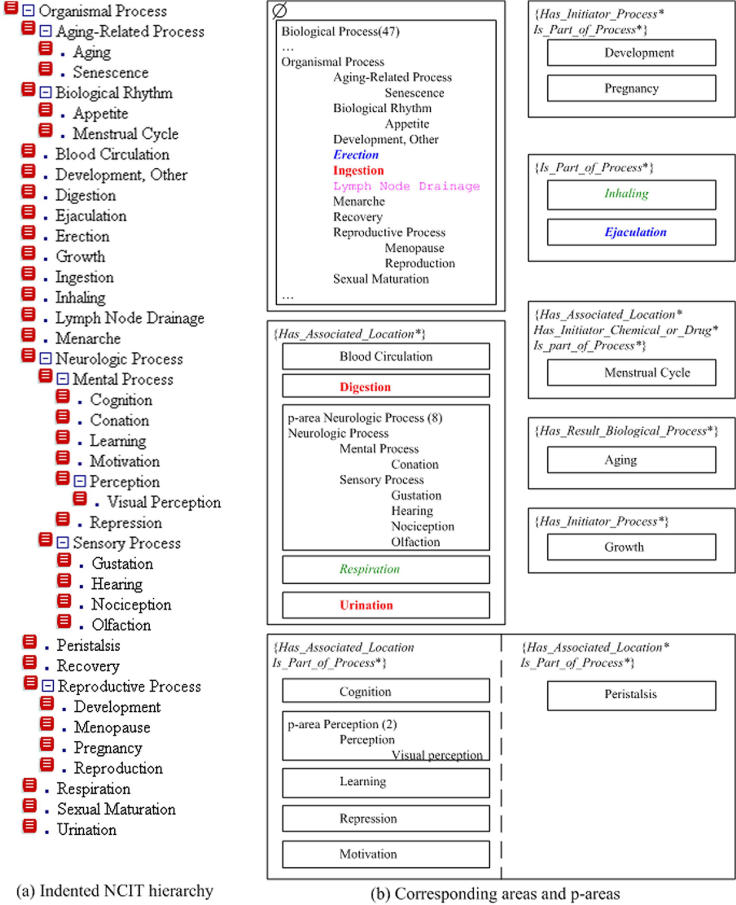
▶ shows Organismal Process and all its 40 descendants. ▶(a) displays them in an indented hierarchy format, provided by the NCIT interface, 12 and ▶(b) displays them as a collection of 19 p-areas grouped into nine areas. In the next subsection, we will utilize this figure to demonstrate different kinds of errors that we have found with the use of our auditing methodology. In the root area Ø of the PAT, the “…” denotes the fact that we only list concepts that are descendants of Organismal Process, not all the concepts. Note also that the other areas in ▶ may be incomplete since some of their concepts are not descendants of Organismal Process. The reader is invited to audit the indented list in ▶(a) to see how many of the errors can be found without our technique.
We use various fonts in ▶ to highlight concepts that are different from the rest of the concepts in the same p-area. We also use the same font to highlight groups of concepts similar in essence but in different areas, e.g., Inhaling and Respiration. These fonts will help to highlight errors described in the next subsection.
Errors Found in P-areas
We will demonstrate various kinds of modeling errors exposed by the small groups of similar concepts, represented by the AT and PAT. As mentioned above, it is easier to find missing concepts, missing roles, or erroneous concepts by comparing groups of structurally and semantically similar concepts. Furthermore, auditors can easily find inconsistencies among concepts if concepts, similar in their essence, are not in the same area or p-area. If for one concept a role exists while for a similar concept it does not, this may suggest that the latter is missing that role. Auditing was performed by H. Min and Y. Chen, both of whom studied medicine in China. Much of our demonstration is focused on ▶. However, in order to test our hypotheses, we have reviewed all p-areas, large and small, of all areas in the BP hierarchy. In our report, we emphasize explicitly the cases of 3̄-3̄-areas.
Missing Roles:
From the PAT (▶), we see that inhaling(1) in {is part of process*} contains only one concept Inhaling. The same is true for respiration(1) in {has associated location*} (see ▶(b) highlighted with italics). Respiration has the role has associated location to Lung. These two related concepts Inhaling and Respiration are in different areas. As noted, this may indicate some inconsistency or error. We see that inhaling is a part of the process of respiration. However, Inhaling is missing the role has associated location to Lung, which is the target of this role for Respiration. Inhaling will have two roles after we add this new role to it. Since its parent Organismal Process does not have any roles, both roles should have been introduced at Inhaling. The concept Inhaling should thus be moved from its original area to {is part of process*, has associated location*}.
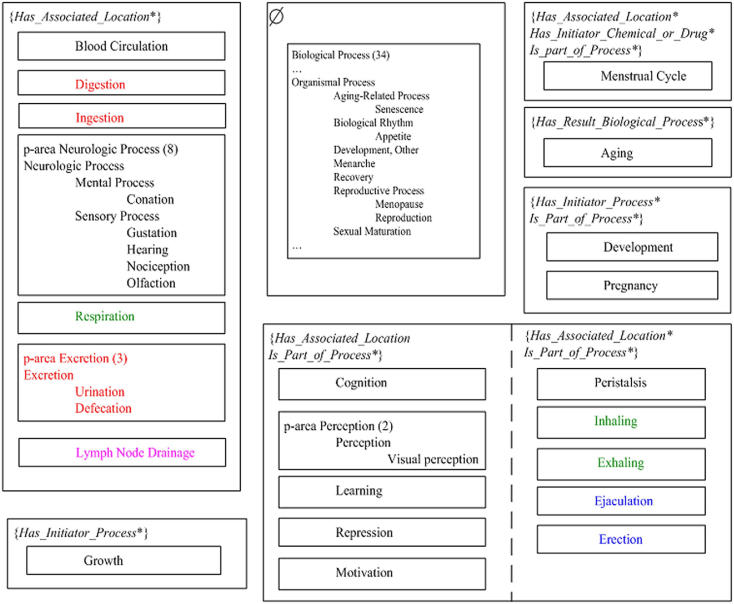
Another p-area in {is part of process*} contains only one concept Ejaculation which is part of Reproduction. But Ejaculation is also missing the role has associated location to the concept Male Reproduction System. After moving these two concepts to {is part of process*, has associated location*}, the area {is part of process*} in ▶(b) becomes empty and does not appear in the revised ▶, reflecting the changes. We note that this area will still exist in the AT and the PAT due to other p-areas.
Figure 8.
Areas and p-areas of the Organismal Process subhierarchy after corrections.
We found that seven concepts in four 3̄-p-areas, cellular stress(1) in {has result process*}, cellular stress response(2) in {is part of process*}, cancer cell growth regulation(3) in {has initiator process*, is part of process*}, and oxidative stress(1) in {has initiator chemical or drug*, has result process}, are missing the role has associated location with the value Cell. Cancer Cell Growth and Cellular Infiltration in Ø have the same mistake. It is interesting to note that the role has associated location is from a dependent process entity to an independent anatomical structure entity. We observe a parallel between the structural and ontological constraints. The issue of missing roles in SNOMED has been discussed previously. 34
Missing Synonyms:
The above concept Inhaling does not have any synonyms. However, inhaling is part of the concept Respiration in {has associated location*}. Thus Inspiration, obviously referring to the same part of respiration, should be a legitimate synonym of Inhaling.
This example was brought up since it is related to a previously discussed missing role error, and because it exposes an inconsistency in the choice of names for concepts. Altogether, we found 70 missing synonyms for the BP hierarchy of the NCIT. However, we are not counting those as errors, and do not include them in the error analysis tables in the Discussion section. As another example, G1 phase, G2 phase, and Interphase in the p-area cell cycle stage(11) are missing G1 period, G2 period, and Resting Phase as synonyms, respectively.
Missing Concepts:
Respiration consists of two parts, inspiration and expiration, which are synonyms for the concepts Inhaling and Exhaling, respectively. These last two concepts should be in one area since they are similar in essence. From the PAT, we see that exhaling(1) is missing from the area that inhaling(1) is located in, and in fact from the NCIT altogether. The concept Exhaling with the synonym Expiration should also be added as part of respiration to the same area as Inhaling.† As another example, the cell cycle includes interphase (which can be divided into four steps: G0 phase, G1 phase, S phase, and G2 phase) and cell division phase. After we examined all concepts in the p-area cell cycle stage(11), we found that G0 Phase is missing from the NCIT. As with synonyms, we do not include missing concepts in the error analysis tables of the Discussion section.
Concept Redundancy:
We mention the following redundancy error and missing synonyms, which are not from the BP hierarchy, due to their critical importance to NCI interests. In the Properties or Attributes hierarchy of the NCIT, there are two concepts, Benign and Non-Malignant, listed as children of Disease Morphology Modifier. They are synonyms, as both of them have an identical definition: “not cancerous.” So only one concept should appear. The other one should be a synonym. Furthermore, Not Cancerous and Noncancerous should appear as synonyms, too. As a matter of fact, there is in the NCIT a concept Mouse Noncancerous Conditions whose name contains such an extra synonym. Note that if a cancer researcher searches for all benign diagnoses, all those listed as Non-Malignant, Not Cancerous, and Noncancerous will be missed.
Missing Parent:
In the BP hierarchy, there are only four concepts with multiple parents in the following p-areas: leukocyte trafficking(1) in {has associated location, is part of process*}, tumor immunity(1) in {has result process*}, inflammation process(1) in {is part of process*}, and the root of cancer cell growth regulation(3) in {has initiator process*, is part of process*}. As the NCIT allows multiple parents, this low number raises concerns that there should probably be more concepts of this sort. This is especially true since the same process can have different aspects such as structural, functional, and clinical that can be reflected by the appropriate parents. For example, the parents of Inflammation Process are Multicellular Process (structural) and Pathologic Process (PP) (clinical). The parents of Cancer Cell Growth Regulation are Cell Proliferation Regulation (functional) and PP (clinical). The fact that three of these concepts have the same ancestor, PP, suggests that more descendants of PP may have more than one parent as well.
After we examined all children of PP, we found that according to the NCIT definition, Autoimmune Process is an immune response and should therefore also be a child of Immune Response. Another example occurs with Necrosis (in the p-area cellular process(19) of {has associated location*}). Necrosis is a pathological process caused by the progressive degradative action of enzymes and is generally associated with severe cellular trauma. Therefore, it is missing PP as another parent. For an alternative modeling approach, see the Discussion section.
Incorrect IS-A:
Senile Corneal Change, in the root area of the BP hierarchy, is a child of PP; but this is incorrect. Senile corneal change is part of the normal aging process. It is neither abnormal nor pathologic (a manifestation of disease). The correct placement of Senile Corneal Change is as a child of Aging-Related Process (and as a sibling of Aging) in the same area.†
The parent of Tumorigenesis in the p-area oncogenesis(6) is Oncogenesis, in the area {has associated location*, has initiator chemical or drug*, has initiator process, has result biological process} (▶). This represents an incorrect IS-A relationship because Tumorigenesis has Oncogenesis as a synonym.
Redundant Target:
The concept Phagocytosis of the p-area phagocytosis(1) is in the area {has associated location, has initiator chemical or drug*, has initiator process*, is part of process*}, with only two p-areas. It has two target values for the role has associated location, Cell and Phagocytic cell. The first target Cell should be removed from this role since the other target, Phagocytic cell, is more specific. This and some other errors were corrected in a later release of the NCIT independent of our work while some of our reported errors are still under consideration.††
Testing the Hypotheses
We formulated two hypotheses concerning the concentration of errors in specific kinds of p-areas. To test the hypotheses on a small portion of the NCIT, we audited all p-areas of the BP hierarchy. We concentrated our analysis on 3̄-3̄-areas, which often represent some kind of irregularity.
In ▶, we give a breakdown of the p-areas according to their size. In ▶, we show a breakdown of the areas by their cardinality. We further distinguish between areas with only 3̄-p-areas and other areas. In ▶, we concentrate just on the 3̄-p-areas. Thus, the last row in ▶, which shows the information regarding all such p-areas, reflects the sums of the first three rows of ▶. In ▶, we present the distribution of the 174 3̄-p-areas, between two kinds of areas, according to their cardinality. In the first row, we consider only 3̄-3̄-areas. There are 27 such p-areas in 18 3̄-3̄-areas (see first three rows in ▶, 13+1+4=18) consisting of a total of 33 concepts, ten of which (30%) are erroneous. In the second row, we consider all other areas. That is, cases where an area’s cardinality is larger than three (e.g., the area {has result biological process} contains five 3̄-p-areas; see ▶) or cases where an area contains k̄-p-areas with k larger than three (e.g., the area {has associated location +, has initiator chemical or drug*, has initiator process, has result process} has three p-areas, but one is a 6̄-p-area and another is an -p-area; see ▶). There are 147 such 3̄-p-areas with 189 concepts, 17 of which (9%) are erroneous.
Table 1.
Table 1. Analysis of errors by p-area size
| P-area size | # P-areas | Total # Concepts | Erroneous Concepts | Percentage of Errors |
|---|---|---|---|---|
| 1 | 141 | 141 | 18 | 13% |
| 2 | 18 | 36 | 3 | 8% |
| 3 | 15 | 45 | 6 | 13% |
| 4-6 | 10 | 47 | 1 | 2% |
| 7-15 | 10 | 112 | 0 | 0% |
| 16-20 | 4 | 74 | 1 | 1% |
| 21-50 | 1 | 47 | 14 | 30% |
| more than 50 | 1 | 87 | 1 | 1% |
| Total: | 200 | 589 | 44 | 7% |
Table 2.
Table 2. Distribution of areas by their cardinality and number of 3̄-p-areas
| Area cardinality m | # Areas | Areas with Small P-areas |
Other Areas |
||||||
|---|---|---|---|---|---|---|---|---|---|
| # m-3̄-areas | # Concepts | # Errors | % of Errors | # Other Areas | # Concepts | # Errors | % of Errors | ||
| 1 | 15 | 13 | 18 | 7 | 39% | 2 | 62 | 14 | 23% |
| 2 | 1 | 1 | 2 | 1 | 50% | 0 | 0 | 0 | 0% |
| 3 | 5 | 4 | 13 | 2 | 15% | 1 | 25 | 1 | 4% |
| 4 | 1 | 0 | 0 | 0 | 0% | 1 | 18 | 0 | 0% |
| 5 | 4 | 2 | 12 | 2 | 17% | 2 | 18 | 0 | 0% |
| 6-10 | 7 | 1 | 11 | 0 | 0% | 6 | 130 | 4 | 3% |
| 11-15 | 3 | 1 | 13 | 1 | 8% | 2 | 138 | 12 | 9% |
| 16-45 | 1 | 0 | 0 | 0 | 0% | 1 | 129 | 0 | 0% |
| Total: | 37 | 22 | 69 | 13 | 19% | 15 | 520 | 31 | 6% |
Table 3.
Table 3. Analysis of errors in 3̄-p-areas of different kinds of areas
| # P-areas | # Concepts | Erroneous Concepts | Percentage of Errors | |
|---|---|---|---|---|
| In 3̄-3̄-areas | 27 | 33 | 10 | 30% |
| In Other areas | 147 | 189 | 17 | 9% |
| Total: | 174 | 222 | 27 | 12% |
Discussion
Interpretation
Our division methodology relies on structural similarity of concepts, as it groups all concepts into areas and further divides areas into p-areas which display semantic similarity of their concepts. For example, the resulting division of the BP hierarchy of NCIT contains 37 areas and 200 p-areas. Based on this, we derived the AT and PAT of the BP hierarchy. The two compact, abstract diagrams help in comprehending and managing the terminology.
Auditing a whole terminology, or even substantial parts of it, is an overwhelming task due to its size and complexity. Also, auditing resources are typically limited. Thus, our auditing methodology is designed to focus the limited available resources for manual editing on relatively small parts of the terminology with high likelihood of errors. The purpose of such a focus is to maximize the impact of a limited auditing effort. This approach of our methodology was expressed by the two hypotheses of the Methods section, the limited testing of which in the small BP hierarchy is discussed below.
Our first hypothesis was that the probability of erroneous classifications and incorrect or incomplete modeling is higher for small p-areas than for large p-areas. As we see in ▶, the percentage of erroneous concepts for 3̄-p-areas (about 12%) is high. The percentage decreases for medium-sized p-areas (2%) and large p-areas (1%). (The one exception is discussed below.) The results in ▶, with the interpretation that small p-areas are those with up to three concepts, support our first hypothesis and show that for effective and economical auditing, we should concentrate our effort on smaller p-areas, where most of the errors are.
One exception is the top-level, singly rooted area Ø (47 concepts) with an error rate of 30%. This area contains concepts with no roles at all. However, we found that 13 out of the 47 concepts (three of which are highlighted in ▶(b), namely Erection, Ingestion and Lymph Node Drainage) are missing roles. After adding the missing roles, these concepts are moved to other areas, leaving this area with 34 concepts (▶) and one error. The error percentage of this area is thus reduced to 3%. We note that there is very little semantic similarity among concepts in this area because they are located at the top levels of the hierarchy. This observation is also true according to the information content matrices of Resnik, 42 since the information content of such concepts is low. This is a very special area, which contains many unrelated concepts, since there is no unifying structure to make them similar. That is, although technically all concepts of Ø share the same empty set of roles, the lack of common specific roles causes the lack of a unifying structure. We gather from this that our auditing methodology should be augmented and special attention paid to Ø.
In ▶, we explore the number of concepts and errors as a function of the cardinality of an area and the size of its p-areas. We see that the likelihood of errors is higher for areas with relatively small cardinality and small p-areas versus all other areas. (Note that the 14 errors in the first row are the exception we just discussed.) The combination of the two factors is considered in our following discussion of Hypothesis 2.
Our second hypothesis was motivated by the intention to further prioritize the auditing of concepts of small p-areas. Such priority is important when there are not enough resources to manually audit all the small p-areas. For example, in our case, the 3̄-p-areas add up to 222 concepts (last row of ▶) which is almost 38% of the concepts in the hierarchy. Hypothesis 2 means that we expect a higher likelihood of errors in m̄-k̄-areas, for small m and k values. As a consequence of the results in ▶, we interpret for the BP hierarchy that small p-areas are 3̄-p-areas. For limited testing of Hypothesis 2, we studied all 3̄-p-areas in the BP hierarchy. ▶ compares the percentages of errors for 3̄-3̄-areas versus areas with larger cardinality or with larger p-areas. As we see from ▶, by just checking 33 concepts of the 3̄-3̄-areas (about 15%) of the 222 concepts of 3̄-p-areas, we find about 37% of the 27 errors in those concepts. However, reviewing ▶, one can take a less strict interpretation of small cardinality of an area to be five. The results show a slight trade-off between the recall and the precision where erroneous concepts are considered relevant.
To demonstrate the impact of the correction of the errors, ▶ shows the division of the descendants of Organismal Process into areas and p-areas reflecting their structure after the corrections. Compared to ▶(b), the number of areas was reduced from eight to seven and the number of 2̄-2̄-areas in ▶ was reduced from 5 to 4. These changes reflect simplifications of the AT and PAT following the correction of errors. Another change is the reduction of size in the AT root area Ø from 47 to 34, due to the discovery of missing roles.
We also found that only four concepts have more than one parent in the BP hierarchy. This may be a reason for the relatively low number of errors we found in this hierarchy. Typically, many concepts with multiple parents are more complex due to the compound nature of the concepts and the multiple inheritance of roles from the different parents. Thus, one expects to find more errors in a hierarchy with more complex concepts. For comparison, we used our techniques to explore missing roles in the Experimental Organism Diagnosis hierarchy of the NCIT consisting of 1,097 concepts. It contains 237 concepts with two parents and five with three parents. Using our methodology, we have found 640 missing roles in 578 concepts, a much higher rate than in the BP hierarchy, where we found only 38 missing roles (the most common kind of error).
We note a philosophical difference between our team and the designers of the NCIT. As a design policy of the NCIT, all processes in the BP hierarchy that are not categorized as a PP are understood as normal biological processes. Hence, parents of concepts in the PP sub-hierarchy can only be concepts that are categorized as pathologic processes. In other words, any normal biological process is not an appropriate parent for the descendants of PP. According to this, instead of adding more multiple parents as we suggested, the NCIT team modified these four concepts to have only one parent. While we respect this approach, we suggest an alternative.
In order to solve this modeling problem, we suggest creating new concepts that are children of both PP and another normal process. These new concepts and their descendants can inherit roles from both the pathologic and normal processes. For example, we would create a new concept called Cellular Pathologic Process that is a child of both PP and Cellular Process. Then, we would add the concepts Cancer Cell Growth Regulation (with its two children) and B-Lymphoma Development as children of Cellular Pathologic Process. These concepts will inherit roles from both PP and Cellular Process as necessary. This modeling is according to the polyhierarchy characteristic of the desiderata. 6
Limitation
In auditing, one has to tailor the techniques to the properties of the terminologies. Different terminologies with different properties require different auditing techniques. For example, some auditing techniques for the UMLS use the assignment of semantic types of the Semantic Network (SN) to the concepts of the Metathesaurus. Since the SN is a unique feature of the UMLS, these techniques will be applicable only for the UMLS.
The auditing technique presented here requires systematic inheritance of a rich set of named roles along the IS-A hierarchy. Hence, it is applicable for popular terminologies enjoying such properties, e.g., NCIT, SNOMED CT, 10 which is the basis for the Veteran Administration’s (VA) internal Enterprise Reference Terminology and Kaiser’s Convergent Medical Terminology (CMT), FMA, 11 RxNorm, 43 MED, 3 and Vocabulary Server (VOSER) terminology, 44 which became the basis for the 3M Healthcare Data Dictionary. 45 Although, the number of such terminologies is not large, they tend to be recent terminologies which also satisfy the desiderata of Cimino. 6 Following this trend, we expect future terminologies to satisfy systematic inheritance of a rich set of named roles and thus be receptive to our structural auditing technique.
In our technique, because concepts are grouped by their structure, the attention of domain-expert auditors may be drawn preferentially to concepts with structure that especially stands out. Since structural similarity tends to go along with semantic similarity, reviewers may indeed find that these concepts are erroneous due to semantic or ontological reasons. (Errors due to ontological reasons have been found in the NCIT. 33 ) However, our technique will not uncover semantic or ontological errors for concepts whose structure is not particularly special.
Another limitation of the study was that the time to complete the actual reviews with the study’s new methodology was not measured. In addition, the study did not determine how long it would take to complete the review in the standard item-by-item manner, using just the IS-A hierarchy view. Thus, the study did not determine how much improvement, time wise, the new methodology accomplishes. Note that the purpose of our auditing techniques is not to speed up the auditing, but to improve the efficiency by detecting more errors.
We tested the hypotheses on error concentrations only for one small hierarchy of the NCIT. This preliminary testing confirms our hypotheses. However, a much more extensive study of more and larger hierarchies of the NCIT is needed to confirm or refute our hypotheses. Other hierarchies may show a different behavior. For example, we expect a terminology with a meaningful portion of its nodes with multiple parents to exhibit a higher ratio of errors than the Biological Process hierarchy with almost no nodes with multiple parents, since such nodes tend to be more complex due to their dual nature and multiple inheritance of roles. Would those errors still follow our hypotheses? We hope that the initial results in this paper will motivate further studies.
Conclusions
We have developed a methodology to divide a hierarchy of a medical terminology, satisfying systematic inheritance, into groups called areas and then further divide areas into p-areas. We obtained two abstraction taxonomies, the AT and PAT, from these divisions. These taxonomies can help audit the terminology since they help to highlight groups of concepts that tend to have higher proportions of errors. When we applied our auditing methodology to the small BP hierarchy of the NCIT, we encountered different kinds of errors highlighted by our structural analysis, e.g., missing roles, missing concepts, incorrect IS-As, etc. The results of our audit of the BP hierarchy show that 12% of the concepts in small p-areas have errors. Furthermore, the percentage of errors in areas with only a few small p-areas is high (30%). The limited results support both our hypotheses, which direct our auditing methodology to focus the auditing efforts on relatively small parts of the terminology that have a high chance of errors. More experiments with several larger NCIT hierarchies are needed to further assess the generality of our hypotheses or the need to modify them. At the same time, we have demonstrated, with the errors we exposed, the need to include auditing as an integral part of the terminology design life cycle, following similar actions taken in software engineering and knowledge-based systems. 4,5
Footnotes
The authors thank Frank Hartel, Director, NCI Enterprise Vocabulary Services, Nicole Thomas, an NCIT editor, and James J. Cimino of Columbia University, for their feedback regarding this paper.
A capitalized italic font is used for concepts. Role names will be italicized and start with a lowercase letter.
Corrected by Nicole Thomas, an NCIT editor, following our report.
Nicole Thomas, personal communication.
References
- 1.Gu H, Perl Y, Halper M, Geller J, Kuo F, Cimino JJ. Partitioning an object-oriented terminology schema Meth Inform Med 2001;40(3):204-212. [PubMed] [Google Scholar]
- 2.Gu H, Halper M, Geller J, Perl Y. Benefits of an object-oriented database representation for controlled medical terminologies JAMIA 1999;6(4):283-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cimino JJ, Clayton PD, Hripcsak G, Johnson SB. Knowledge-based approaches to the maintenance of a large controlled medical terminology JAMIA 1994;1(1):35-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Software Quality Assurance. Available at: http://satc.gsfc.nasa.gov/assure/assurepage.html. Accessed March 2006.
- 5.In: Watkins PR, Eliot LB, editors. Expert systems in business and finance: issues and applications. New York, NY: John Wiley & Sons, Inc; 1993.
- 6.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century Methods Inform Med 1998;37:394-403. [PMC free article] [PubMed] [Google Scholar]
- 7.Baader F. Restricted role-value-maps in a description logic with existential restrictions and terminological cyclesIn: Calvanese D, DeGiacomo G, Franconi E, editors. 2003 International Workshop on Description Logics DL2003. 2003. pp. 31-38Vol. 81 of CEUR Workshop Proceedings.
- 8.Nardi D, Brachman RJ. An introduction to description logicsIn: Baader F, Calvanese D, McGuinness DL, Nardi D, Patel-Schneider PF, editors. The Description Logic Handbook: Theory, Implementation, and Applications. Cambridge, UK: Cambridge University Press; 2003. pp. 1-40.
- 9.Minsky M. A framework for representing knowledge. MIT-AI Laboratory Memo. 1974. pp. 306.
- 10.SNOMED International. The systematized nomenclature of medicine. Available at: http://www.snomed.org. Accessed January 2006.
- 11.Rosse C, Mejino JL. A reference ontology for biomedical informatics: the foundational model of anatomy J Biomed Inform 2003;36(6):478-500. [DOI] [PubMed] [Google Scholar]
- 12.NCI. Available at: http://nciterms.nci.nih.gov/NCIBrowser/. Accessed May 2004.
- 13.McDonald CJ, Huff SM, Suico JG, et al. LOINC, a universal standard for identifying laboratory observations: a 5-year update Clin Chem 2003;49(4):624-633. [DOI] [PubMed] [Google Scholar]
- 14.Protégé. Available at: http://protege.stanford.edu. Accessed September 2005.
- 15.Apelon. Available at: http://www.apelon.com. Accessed October 2005.
- 16.Lindberg DA, Humphreys BL, McCray AT. The unified medical language system Methods Inform Med 1993;32(4):281-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Humphreys BL, Lindberg DA, Schoolman HM, Barnett GO. The unified medical language system: an informatics research collaboration JAMIA 1998;5(1):1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bodenreider O. The Unified medical language system (UMLS): integrating biomedical terminology Nucleic Acids Res 2004;1(32):D267-D270(Database issue.). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cimino JJ. Auditing the unified medical language system with semantic methods JAMIA 1998;5(1):41-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bodenreider O. Circular hierarchical relationships in the UMLS: etiology, diagnosis, treatment, complications and preventionIn: Bakken S, editor. Proc 2001 AMIA Annual Symposium. 2001. pp. 57-61Washington, DC. [PMC free article] [PubMed]
- 21.Cimino JJ, Min H, Perl Y. Consistency across the hierarchies of the UMLS semantic network and metathesaurus J Biomed Inform 2003;36(6):450-461. [DOI] [PubMed] [Google Scholar]
- 22.Cimino JJ. Battling scylla and charybdis: the search for redundancy and ambiguity in the 2001 UMLS metathesaurusIn: Bakken S, editor. Proc 2001 AMIA Annual Symposium. 2001. pp. 120-124Washington, DC. [PMC free article] [PubMed]
- 23.Peng Y, Halper M, Perl Y, Geller J. Auditing the UMLS for redundant classificationsIn: Kohane IS, editor. Proc. 2002 AMIA Annual Symposium. 2002. pp. 612-616San Antonio, TX. [PMC free article] [PubMed]
- 24.McCray AT. An upper-level ontology for the biomedical domain Comp Funct Genom 2003;4:80-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schulze-Kremer S, Smith B, Kumar A. Revising the UMLS semantic networkIn: Fieschi M, Coiera E, Li YC, editors. Proc Medinfo 2004. 2004. San Francisco, CA; 1700.
- 26.Bodenreider O. An object-oriented model for representing semantic locality in the UMLSIn: Rogers R, Haux R, Patel V, editors. Proc Medinfo2001. 2001. pp. 161-165London, UK. [PMC free article] [PubMed]
- 27.Gu H, Perl Y, Geller J, Halper M, Liu L, Cimino JJ. Representing the UMLS as an OODB: modeling issues and advantages JAMIA 2000;7(1):66-80Selected for reprint in R. Haux, C. Kulikowski, editors: Yearbook of Medical Informatics, International Medical Informatics Association, Rotterdam, 2001;271–285.10641964 [Google Scholar]
- 28.Hole WT, Srinivasin S. Discovering missed synonymy in a large concept-oriented metathesaurus JAMIA 2000;7:354-358. [PMC free article] [PubMed] [Google Scholar]
- 29.Gu H, Perl Y, Elhanan G, Min H, Zhang L, Peng Y. Auditing concept categorizations in the UMLS Artific Intell Med 2004;31(1):29-44. [DOI] [PubMed] [Google Scholar]
- 30.Perl Y, Chen C, Halper M, Geller J, Zhang L, Peng Y. The cohesive metaschema: A higher-level abstraction of the UMLS semantic network J Biomed Inform 2002;35(3):194-212. [DOI] [PubMed] [Google Scholar]
- 31.Ceusters W, Smith B, Kumar C, Dhaen C. Ontology-based error detection in SNOMED-CTIn: Fieschi M, Coiera E, Li YC, editors. Proc Medinfo 2004. 2004. pp. 482-486San Francisco, CA. [PubMed]
- 32.Ceusters W, Smith B, Kumar C, Dhaen C. Mistakes in medical ontologies: where do they come from and how can they be detected?In: Pisanelli DM, editor. Ontologies in Medicine: Proc. Workshop on Medical Ontologies. 2003. pp. 145-164Rome, Italy. [PubMed]
- 33.Ceusters W, Smith B, Goldberg L. A terminological and ontological analysis of the NCI thesaurus Methods Inform Med 2005;44:498-507. [PubMed] [Google Scholar]
- 34.Bodenreider O, Smith B, Kumar A, Burgun A. Investigating subsumption in DL-based terminologies: a case study in SNOMED CTIn: Hahn U, Schulz S, Cornet R, editors. Proceedings of the First International Workshop on Formal Biomedical Knowledge Representation (KR-MED 2004). 2004. pp. 12-20Whistler, Canada.
- 35.Kumar A, Smith B, Borgelt C. Dependence relationships between gene ontology terms based on TIGR gene product annotationsIn: Ananadiou S, Zweigenbaum P, editors. CompuTerm 2004, 3rd International Workshop on Computational Terminology. 2004. pp. 31-38Coling, Geneva.
- 36.Kumar A, Smith B. The unified medical language system and the gene ontology: some critical reflectionsIn: Günter A, Kruse R, Neumann B, editors. KI 2003, Advances in Artificial Intelligence (Lecture Notes in Artificial Intelligence 2821). Berlin: Springer; 2003. pp. 135-148.
- 37.Smith B, Williams J, Schulze-Kremer S. The ontology of the gene ontologyIn: Musen M, editor. Proc 2003 AMIA Annual Symposium. 2003. pp. 609-613Washington, DC. [PMC free article] [PubMed]
- 38.Smith B, Köhler J, Kumar A. On the application of formal principles to life science data: a case study in the gene ontology DILS 2004 (Data Integration in the Life Sciences), (Lecture Notes in Bioinformatics 2994). Berlin: Springer; 2004. pp. 79-94.
- 39.Cornet R, Abu-Hanna A. Description logic-based methods for auditing frame-based medical terminological systems Artif Intell Med 2005;34(3):201-217. [DOI] [PubMed] [Google Scholar]
- 40.DeKeizer NF, Abu-Hanna A, Cornet R, Zwersloot-Schonk JH, Stoutenbeek CP. Analysis and design of an ontology for intensive care diagnoses Methods Inform Med 1999;38(2):102-112. [PubMed] [Google Scholar]
- 41.caCORE Technical Guide. Available at: http://www.ncicb.nci.nih.gov. Accessed February 2005.
- 42.Resnik P. Using information content to evaluate semantic similarity in a taxonomy Proceedings of the 14th IJCAI. vol. 1. 1995. pp. 448-453Montreal, Canada.
- 43.RxNorm. A Guide for the Perplexed. Available at: http://www.nlm.nih.gov/research/umls/rxnorm-guide.pdf. Accessed June 2005.
- 44.Rocha RA, Huff SM, Haug PJ, Warner HR. Designing a controlled medical vocabulary server: The VOSER project Comp Biomed Res 1994;27(6):472-507. [DOI] [PubMed] [Google Scholar]
- 45.3M Worldwide. Available at: http://www.mmm.com/market/healthcare/his/product/hems/fctsheet/hdd.htm. Accessed July 2005.