Abstract
CYP51 sterol demethylases are the only cytochrome P450 enzymes with a conserved function across the animal, fungal, and plant kingdoms (in the synthesis of essential sterols). These highly conserved enzymes, which are important targets for cholesterol-lowering drugs, antifungal agents, and herbicides, are regarded as the most ancient member cytochrome P450 family. Here we present a report of a CYP51 enzyme that has acquired a different function. We show that the plant enzyme AsCYP51H10 is dispensable for synthesis of essential sterols and has been recruited for the production of antimicrobial compounds (avenacins) that confer disease resistance in oats. The AsCyp51H10 gene is synonymous with Sad2, a gene that we previously had defined by mutation as being required for avenacin synthesis. In earlier work, we showed that Sad1, the gene encoding the first committed enzyme in the avenacin pathway (β-amyrin synthase), had arisen by duplication and divergence of a cycloartenol synthase-like gene. Together these data indicate an intimate evolutionary connection between the sterol and avenacin pathways. Sad1 and Sad2 lie within 70 kb of each other and are expressed specifically in the epidermal cells of the root tip, the site of accumulation of avenacins. These findings raise intriguing questions about the recruitment, coevolution, and regulation of the components of this specialized defense-related metabolic pathway.
Keywords: Avena, disease resistance, oat, metabolic diversity, gene duplication
Plants synthesize a diverse range of natural products. Many of these compounds are specialized metabolites that are produced only by certain taxonomic groups (1). Plant-derived natural products have important ecological functions, often serving as attractants or deterrents in interactions with other organisms (1, 2). The ability to synthesize particular natural products therefore is likely to be a consequence of niche colonization and adaptive evolution (2, 3). Currently, we know very little about how metabolic pathways arise. A better understanding of the origin and nature of the genes and enzymes that comprise natural product pathways will enable us to probe the mechanisms underpinning the generation of metabolic diversity.
Avenacins are antimicrobial triterpene glycosides (saponins) that accumulate in the roots of oats (Avena spp.) (4, 5). The ability to synthesize avenacins is restricted to members of the genus Avena (4) and has arisen relatively recently, since the divergence of oats from other cereals and grasses (6). The major avenacin, A-1, contains the fluorophore N-methyl anthranilic acid and so confers a bright blue fluorescence on the roots of oat seedlings under ultraviolet illumination (Fig. 1). In previous work we have exploited this fluorescence as a screen to isolate saponin-deficient (sad) mutants of diploid oat (Avena strigosa) after chemical (sodium azide) mutagenesis (5). sad mutants are compromised in disease resistance to a range of fungal pathogens, demonstrating that avenacins confer broad-spectrum protection against microbial attack (5). These experiments have provided direct evidence for a role for preformed antimicrobial compounds in plant defense.
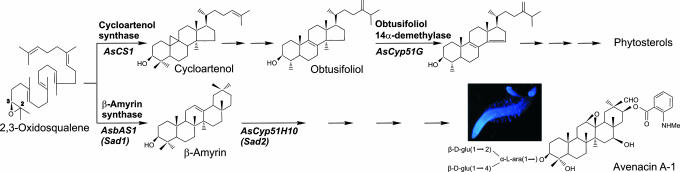
Fig. 1.
Synthesis of sterols and defense-related triterpenes in oats. The sterol and triterpene pathways branch after 2,3-oxidosqualene. Key genes and enzymes are indicated. Inset shows fluorescence of roots of an oat seedling, viewed on an ultraviolet transilluminator.
Avenacins are synthesized from the isoprenoid pathway and share a common biogenetic origin with sterols, the two pathways diverging after 2,3-oxidosqualene (Fig. 1) (4, 7–9). In primary sterol biosynthesis, 2,3-oxidosqualene is cyclized to cycloartenol by cycloartenol synthase. Cycloartenol then is converted to other sterols via a series of intermediates that includes obtusifoliol. The first committed step in the avenacin pathway is the cyclization of 2,3-oxidosqualene to the triterpene precursor β-amyrin, catalyzed by the oxidosqualene cyclase enzyme β-amyrin synthase (7–9). β-Amyrin is not antimicrobial but is converted to the biologically active avenacins by a series of uncharacterized modifications that are predicted to involve oxidation, glycosylation, and acylation (9). From genetic analysis of our mutant collection, we originally defined eight loci for avenacin synthesis (Sad1–8). We have cloned Sad1, the gene encoding β-amyrin synthase (Fig. 1) (8). Our data indicate that Sad1 is likely to have been recruited from sterol metabolism by duplication and divergence of a plant cycloartenol-synthase like gene and that this is a relatively recent evolutionary event (6, 8). Remarkably, six of the seven other Sad loci that we have defined by mutation (Sad2, Sad3, Sad5, Sad6, Sad7, and Sad8) cosegregate with Sad1, indicating that the genes for avenacin biosynthesis are clustered (5, 6). Although many examples of clustered genes for natural product pathways have been reported in microbes, gene clusters of this kind are not a common phenomenon in plants (2, 6). The reason for clustering of avenacin biosynthetic genes is not yet known.
In this article, we report the cloning and characterization of a second gene in the avenacin pathway, Sad2 (AsCyp51H10), which encodes a cytochrome P450 enzyme belonging to the CYP51 sterol demethylase family. The CYP51 sterol demethylases are regarded as the most ancient cytochrome P450 family. They are highly conserved across the animal, fungal, and plant kingdoms and are known only to have a single strictly conserved function, in the synthesis of essential sterols (10–13). AsCYP51H10 belongs to a unique subfamily of divergent plant CYP51 enzymes (CYP51H) that until now has been defined only by rice sequences of unknown function (11). This subfamily is not represented in Arabidopsis or other dicots. Our data indicate that AsCYP51H10 has undergone neofunctionalization and is required for the synthesis of defense-related antimicrobial triterpene glycosides (avenacins) but is dispensable for primary sterol biosynthesis. Thus, this is a report of a CYP51 enzyme that has acquired a unique function. Our demonstration that both Sad1 (6, 8) and Sad2 (AsCyp51H10) (this work) have been recruited from plant primary sterol metabolism indicates an intimate evolutionary connection between the sterol and avenacin pathways. However, the expression patterns of Sad1 and Sad2 have been refined. While their sterol biosynthesis counterparts (the cycloartenol synthase and obtusifoliol 14α-demethylase genes, respectively) are expressed constitutively throughout the plant, expression of Sad1 and Sad2 (which are 70 kb apart) is tightly regulated and is restricted to the epidermal cells of the root tip, the site of accumulation of avenacins.
Results and Discussion
Cloning of AsCyp51H10.
The avenacin gene cluster maps to the distal part of linkage group AswC of diploid oat in a region of the genome that is not conserved in other cereals (6). We have shown that the uncharacterized restriction fragment length polymorphism (RFLP) probe isu441, which is derived from a hexaploid oat cDNA library, maps within the avenacin gene cluster (6). We sequenced this 480-nt cDNA and found homology with cytochrome P450 monoxygenases, the closest match being with wheat obtusifoliol 14α-demethylase (CYP51) (52% amino acid sequence identity) (14). Because cytochrome P450s are implicated in avenacin biosynthesis (9), this gene became a candidate pathway gene. We isolated and sequenced the full-length cDNA and gene corresponding to isu441 from the diploid oat accession A. strigosa S75 (the wild-type parent of the sad mutants). The gene was designated AsCyp51H10.
The AsCyp51H10 cDNA was used as a probe to screen a bacterial artificial chromosome (BAC) library that we constructed for A. strigosa S75. Six BAC clones spanning the Sad1 region were identified. Sequence analysis established that AsCyp51H10 is 66,828 bp from Sad1 and that the gap between these two genes contains repetitive sequences but no other obvious ORFs (Fig. 2A). A seventh BAC clone mapped to a different linkage group (AswG) and contained a homologue of AsCyp51H10 (designated AsCyp51H11). AsCyp51H10 and AsCyp51H11 share 74% nucleotide sequence identity. The presumed oat obtusifoliol 14α-demethylase gene was not detected in our BAC screen. However, we were able to identify sequences corresponding to this in an expressed sequence tag (EST) database of >16,000 sequences that we previously had generated from oat roots (8). We then cloned and sequenced the full-length cDNA and the corresponding gene (designated AsCyp51G1). AsCyp51H10 and AsCyp51H11 have 53% and 54% nucleotide sequence identity with AsCYP51G, respectively. AsCyp51G1 maps to a third linkage group, AswB.
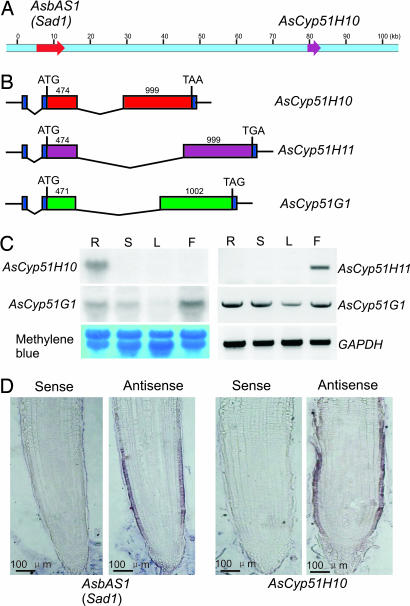
Fig. 2.
Isolation and characterization of AsCyp51H10. (A) BAC clone #B460D15 contains Sad1 and AsCyp51H10. (B) Gene structures of AsCyp51H10, AsCyp51H10, and AsCyp51G1. (C) Northern blot analysis of AsCyp51H10 and AsCyp51G1 transcripts (Left) and RT-PCR analysis of AsCyp51H11 and AsCyp51G1 transcripts (Right) in oat roots (R), shoots (S), leaves (L), and flowers (F). The oat glyceraldehyde 3-phosphate dehydrogenase gene (GAPDH) was used as a control for RT-PCR. (D) In situ mRNA analysis of Sad1 and AsCyp51H10 transcripts in the root tips of A. strigosa.
Expression of AsCyp51H10 Is Restricted to the Root Epidermis.
The gene structures of AsCyp51H10, AsCyp51H11, and AsCyp51G1 are very similar (Fig. 2B). All three genes are predicted to encode products 490 aa in length. The exon sizes of AsCyp51H10 and AsCyp51H11 are identical but differ from those of the obtusifoliol 14α-demethylase gene AsCyp51G, suggesting a closer evolutionarily relationship between the former two genes (Fig. 2B). The obtusifoliol 14α-demethylase gene AsCyp51G1 is expressed in all plant organs examined, consistent with a role in primary sterol metabolism (Fig. 2C). In contrast, AsCyp51H10 is expressed specifically in the roots, whereas AsCyp51H11 transcripts were detected only in the flowers (Fig. 2C). Synthesis of avenacin A-1 is under tight regulation and is restricted to the epidermal cells of the root tip (8). Previously we have shown by mRNA in situ hybridization that expression of Sad1 is restricted to this cell type (8). AsCyp51H10 showed a very similar pattern of expression (Fig. 2D).
AsCYP51H10 Is Synonymous with Sad2.
The above data are suggestive of a role for AsCYP51H10 in avenacin biosynthesis. We therefore sequenced the AsCyp51H10 gene in our original collection of ten sad mutants (5) to establish whether this gene was likely to correspond to any of the loci that we had defined by mutation. As expected, we found no differences in the sequence of AsCyp51H10 in the two characterized sad1 mutants within the collection. The sequence of AsCyp51H10 in six other mutants (single mutants for each of the loci, Sad3–Sad8) also was unaffected. However, nonsynonymous point mutations were found in the coding region of AsCyp51H10 in the two independent sad2 mutants within the collection (#791 and #1027) (Table 1). Preliminary experiments suggested that the sad2 mutants #791 and #1027 accumulate β-amyrin, whereas mutants affected at other Sad loci do not (15). This finding was confirmed by quantitative GC/MS analysis (Fig. 3A and Table 1). These data are consistent with a block in a cytochrome P450-mediated step early in the pathway and suggest that AsCyp51H10 is synonymous with Sad2.
Table 1.
Characterization of sad2 mutants
| Mutant | Mutation event | Predicted amino acid change | Region of protein | Mean β-amyrin content (μg/g of fresh roots) |
|---|---|---|---|---|
| Wild type | ||||
| S75 | — | — | — | 1.4 ± 0.1 |
| Original sad2 | ||||
| 791 | C2360 → T | Pro463 → Ser | Near SRS6 | 40.2 ± 1.9 |
| 1027 | C371 → T | Ala124 → Val | SRS1 | 50.4 ± 1.2 |
| New sad2 | ||||
| 283 | G2277 → A | Gly435 → Asp | Heme binding | 47.4 ± 8.5 |
| 500 | G475 → A | Splicing error | — | 41.3 ± 0.6 |
| 638 | G1922 → A | Glu317 → Lys | Conserved amino acid in αJ helix | 48.2 ± 2.9 |
| 698 | G1670 → A | Ala233 → Thr | SRS3 | ND |
| 1325 | C1866 → T | Ser298 → Phe | SRS4 | 37.1 ± 1.1 |
| 1412 | C338 → T | Thr113 → Ile | SRS1 | 41.5 ± 1.3 |
SRSs, predicted substrate recognition sites (16). ND, not determined.
Fig. 3.
Identification and characterization of sad2 mutants. (A) GC analysis of root extracts from S75 (WT) and sad1 and sad2 mutants. The vertical bar on the bottom left indicates relative mass abundance. sad2 mutants accumulate β-amyrin, whereas sad1 mutants (such as mutant #109) accumulate 2,3-oxidosqualene. The identity of the accumulated intermediates was determined by MS. (B) Northern blot analysis of AsCyp51H10 transcript levels in sad2 mutants and the WT S75.
We then screened an extended collection of 92 reduced root fluorescence mutants, with the objective of isolating more sad2 mutants, by using TLC analysis as a preliminary screen. This screen identified six more candidate sad2 mutants (#283, #500, #638, #698, #1325, and #1412). Allelism tests confirmed that these were indeed unique mutant alleles of Sad2. Five of these mutants had nonsynonymous point mutations in the AsCyp51H10 gene, whereas the sixth mutant (#500) had a point mutation in an exon–intron boundary (Table 1). Significantly, AsCyp51H10 transcript levels were substantially reduced in mutant #500 (Fig. 3B). Quantitative GC/MS confirmed that, like #791 and #1027, these newly isolated sad2 mutants had elevated levels of β-amyrin (Table 1). These data provide compelling evidence that AsCyp51H10 corresponds to Sad2.
AsCYP51H10 Is a Divergent Member of the CYP51 Family.
Comparisons of the amino acid sequences of sterol 14α-demethylase (CYP51) sequences from diverse organisms indicate 34 conserved amino acid residues across bacteria, protozoa, fungi, animals, and plants. Six of these residues are not conserved in the oat AsCYP51H10 protein (Fig. 5, which is published as supporting information on the PNAS web site). The predicted amino acid changes in the seven sad2 mutants with normal levels of AsCYP51H10 transcript were all within conserved substrate recognition sites or in other regions that are likely to be critical for structure and/or activity (Table 1). An alignment of selected regions of 36 representative CYP51 amino acid sequences across substrate recognition sites 1, 4, 5, and 6 (16) is shown in Fig. 4A. Modeling of the 3D structures of AsCYP51H10 and the oat sterol 14α-demethylase AsCYP51G1 by using the Mycobacterium tuberculosis MtCYP51B1 crystal structure (16) as a template predicted the shapes and sizes of the active site cavities of MtCYP51B1 and AsCYP51G1 to be very similar, whereas that of AsCYP51H10 is quite different (Fig. 4B). Residues that are predicted to significantly affect the size and shape of the active site cavity are shown in Fig. 4B. The ensemble-averaged active site volume in the model of AsCYP51H10 is 568 ± 96 Å3, whereas that of AsCYP51G1 is 346 ± 108 Å3, very similar to the active site volume determined from the crystal structure of MtCYP51B1 (343 ± 62 Å3). These observations are consistent with acquisition of a different function by AsCYP51H10.
Fig. 4.
Sequence and structure of AsCYP51H10. (A) Alignment of selected regions of 36 representative CYP51 sequences from diverse organisms. The predicted substrate recognition sites (SRS) (16) are framed. Completely conserved amino acids are shown on a black background and those that are conserved in all except AsCYP51H10 are shown on a gray background. Mutations in sad2 mutants in these regions are shown (mutant number preceded by “#”; changes marked with black dots). Residues that line the active site cavity are indicated by triangles. The filled triangles denote the subset of these that are likely to be key determinants in modulating the size and shape of the cavity in AsCYP51H10. (B) Modeling of the active site cavity of AsCYP51H10 (Bottom) and the oat sterol 14α-demethylase AsCYP51G1 (Middle) based on the Mycobacterium tuberculosis MtCYP51B1 crystal structure (Top). (C) Phylogenetic analysis of CYP51 amino acid sequences. The numbers indicate the percentage of bootstrap replicates (out of 1,000) in which the given branching was observed. Accession nos. for the sequences used in alignments, modeling, and phylogenetic analysis are given in Table 2.
Fig. 4C shows conserved subfamilies of CYP51 sterol 14α-demethylases from animals (CYP51A; pink), bacteria (CYP51B; light blue), protozoa (CYP51E; turquoise), fungi (CYP51F; orange), and plants (CYP51G; dark green). AsCyp51G1, the predicted obtusifoliol 14α-demethylase from oat, falls within the CYP51G (dark green) subfamily. Yeast expression experiments have confirmed that this gene does indeed encode functional obtusifoliol 14α-demethylase (data not shown). However, we were unable to express AsCYP51H10 and AsCYP51H11 in active form with the standard yeast expression system used for conserved plant CYP51G enzymes (17). The CYP51H subfamily appears to be restricted to oats and rice and is not represented in Arabidopsis.
The position of Chlamydomonas reinhardtii CYP51G (CrCYP51G1) in the phylogenetic tree (Fig. 4C) implies that the CYP51H family was derived from an ancient CYP51G-like sequence during the evolution of green plants. Tajima's relative rate test (18) using C. reinhardtii CYP51G as an outgroup indicates that the AsCYP51H10 and AsCYP51H11 branches are significantly longer than that of AsCYP51G1 (χ2 = 39.68 and 41.67, respectively; P < 0.0001). The branches of the rice sequences within the CYP51H subgroup also are significantly longer than that of rice CYP51G1 (data not shown). There is greater mean diversity in the rice and oat CYP51H subfamily than in the monocot CYP51G subfamily (0.653 ± 0.028 versus 0.088 ± 0.009, respectively). Collectively, these results indicate that the CYP51H subfamily is evolving at a much higher rate than the conserved CYP51G subfamily, consistent with acquisition of a different function(s), as suggested by Nelson et al. (11). Our data confirm that AsCYP51H10 has indeed acquired a different function in the synthesis of secondary metabolites required for plant defense. This finding has broad significance for understanding the mechanisms of action and potential evolutionary plasticity of the CYP51 family as a whole.
Coevolution of Sad1 and Sad2.
Previously we reported that Sad1 has arisen by duplication and divergence of a cycloartenol synthase-like gene (6, 8). The data presented here indicate that a second gene in the avenacin pathway, Sad2 (AsCyp51H10), has been recruited from an ancient CYP51G-like sequence. These results indicate an intimate evolutionary connection between sterol and triterpene biosynthesis. The first step in the pathway for the synthesis of a different group of defense-related compounds produced by maize (benzoxazinoids) also has been shown to be recruited from primary metabolism, in this case from tryptophan biosynthesis (19, 20), and there is an increasing body of evidence to indicate that gene duplication, neofunctionalization, and positive selection drive metabolic diversification in plants (e.g., refs. 21 and 22). Unlike their sterol pathway counterparts AsCS1 (cycloartenol synthase) and AsCyp51G1 (obtusifoliol 14α-demethylase), which are expressed throughout the plant, expression of Sad1 and Sad2 is highly tissue-specific and is restricted to the epidermal cells of the root tips. AsCS1 and AsCyp51G1 are not genetically linked to each other or to the Sad gene cluster. Sad1 and Sad2 are physically linked and cosegregate with other genetically defined loci in the pathway that are required for clearly distinct biochemical functions (6). The biochemical function of AsCYP51H10 is as yet unknown. Conversion of β-amyrin to avenacin A-1 will require oxidation at five different sites (Fig. 1), and all of these conversions potentially could involve cytochrome P450 enzymes. AsCYP51H10 therefore may be required for hydroxylation of β-amyrin (or a modified derivative of β-amyrin) at one or more positions. Elucidation of the precise biochemical function of AsCYP51H10, coupled with further investigation of the nature and origin of the avenacin gene cluster, will shed light on mechanisms underpinning the evolution of metabolic diversity in plants and on the selective pressures that drive this process.
Materials and Methods
Plant Material.
Wild-type and mutant A. strigosa lines are as described previously (5).
AsCyp51H10, AsCyp51H11, and AsCyp51G1 cDNA and Gene Isolation.
Full-length cDNAs were defined by 5′ and 3′ RACE with a GeneRacer kit (Invitrogen, Carlsbad, CA), amplified by PCR and cloned into the pCR4-TOPO plasmid (Invitrogen). Genes were characterized by direct sequencing of PCR products generated from genomic DNA and/or by sequencing of BAC clones (see below).
BAC Library Construction and Screening.
A BAC library of A. strigosa accession no. S75 was constructed by using established methods (23). Approximately 150,000 colonies with an average insert size of ≈110 Kb (ca. 4.2× genome coverage) were stored in 384-well microtiter plates and gridded onto high-density filters. Filters were screened with32P-labeled cDNA probes. Hybridization and washing were conducted at stringencies of either 60°C or 65°C following standard methods (24). BAC fingerprinting was conducted by digestion of BAC DNA with HindIII and BamHI, and manual comparison of the restriction fragments was performed after agarose gel electrophoresis. Subcloning of BAC inserts and sequencing was carried out by using standard methods (24).
Transcript Analysis.
For Northern blot analysis, total RNA was extracted by using TRI-REAGENT (Sigma, St. Louis, MO). Hybridizations with biotin-labeled (Biotin-16-dUTP; Roche, Indianapolis, IN) antisense RNA probes for AsCyp51H10 were carried out at high stringency (68°C) with signal detection by using BrightStar BioDetect (Ambion, Austin, TX). For RT-PCR, first-strand cDNA synthesis was carried out by using SuperScript II Reverse Transcriptase (Invitrogen) according to the manufacturer's instructions, and cDNA was amplified by standard PCR with 30 cycles. For mRNA in situ analysis, biotin-labeled sense and antisense RNA probes from AsbAS1 and AsCyp51H10 were used. Tissue preparation and hybridization was carried out as described in ref. 25.
Metabolite Analysis.
Roots from individual 6-day-old seedlings were harvested, freeze-dried, ground in liquid nitrogen, and extracted with methanol. Extracts were centrifuged, and the supernatant was removed and dried down before extraction with 100 μl of CHCl3/MeOH (7:3 vol/vol). Extracts and a β-amyrin standard were spotted onto silica gel 60 TLC plates (Merck & Co., Whitehouse Station, NJ), and the TLC was developed with hexane:acetone (80:20 vol/vol). β-Amyrin and other compounds were detected with iodine vapor. Qualitative and quantitative GC/MS was conducted by using an Agilent (Santa Clara, CA) 5973 Electron Ionization mass selective detector coupled to an Agilent 6890 gas chromatograph. Trimethylsilyl (TMS) derivatives of the extracted samples were separated on a J&W DB-5MS capillary column (30-m long, 0.25-mm i.d., 0.25-μm film thickness; Agilent). The GC oven temperature was maintained at 250°C for 1 min after injection, then programmed to 325°C at a rate of 5°/min and held for 10 min at the final temperature. Helium carrier gas was used at a flow of 1.0 ml/min, and 2-μl samples were injected in hexane in split mode (10:1) at an injector temperature of 250°C. The mass spectrometer ion source was maintained at 250°C. β-Amyrin was supplied by Apin Chemicals Ltd. (Oxon, U.K.), and 2,3-oxidosqualene and 5β-cholestan-3β-ol standards were supplied by Sigma-Aldrich.
Sequence Comparisons and Homology Modeling.
Protein sequences (Table 2, which is published as supporting information on the PNAS web site) were aligned by using CLUSTAL X, Version 1.8 (http://bips.u-strasbg.fr), manually adjusted according to Lepesheva et al. (26), and displayed by using ESPript (version 2.1) (27). MEGA3.1 software (28) was used for phylogenetic analysis, assessment of sequence diversity, and Tajima's relative rate test (18). Gaps in the alignment were excluded from our analysis (complete-deletion option). The neighbor-joining method was used to construct the phylogenetic tree.
For homology modeling, sequence alignments of AsCYP51H10 and AsCYP51G1 with MtCYP51B1 utilized the structural information available in the Protein Data Bank entry 1EA1 and were generated by using Fugue (29). Modeler (version 8.2) (30) was used to generate homology models of the two enzymes based on the MtCYP51B1 crystal structure. The models were subjected to stereochemical validation by using appropriate routines in Modeler (30). To more fully explore alternative active site residue conformations in our models other than those delivered directly by Modeler, we used the non-Newtonian ensemble generator CONCOORD (31). The pocket definitions included residues 72–85, 95–103, 253–263, 319–324, and 433–435 in MtCYP51B1. Similar residues were included in simulations of AsCYP51H10 and AsCYP51G1. This approach allows prediction of the range of likely configurations adopted by the residues of the substrate-binding pocket. An energy-based method QsiteFinder (www.bioinformatics.leeds.ac.uk) then was used to characterize the active site cavities in the ensembles of modeled structures (32).
Supplementary Material
Acknowledgments
We thank Mike Lee (Iowa State University, Ames, IA) for restriction fragment length polymorphism (RFLP) probes, Stephane Deschamps and Karlene Butler (DuPont Agricultural Products, Wilmington, DE) for BAC sequence analysis, David Nelson (University of Tennessee, Memphis, TN) for guidance on cytochrome P450 nomenclature, and Guo Jiemao (John Innes Centre, Norwich, U.K.) for technical assistance with in situ analysis. We also thank Enno Krebbers (DuPont/Pioneer Crop Genetics) for constructive comments throughout the project and the Biotechnology and Biological Sciences Research Council U.K. and the Gatsby Charitable Foundation for funding.
Footnotes
References
- 1.Dixon RA. Nature. 2001;411:843–847. doi: 10.1038/35081178. [DOI] [PubMed] [Google Scholar]
- 2.Field B, Jordán F, Osbourn A. New Phytologist. 2006;172:193–207. doi: 10.1111/j.1469-8137.2006.01863.x. [DOI] [PubMed] [Google Scholar]
- 3.Pichersky E, Gang DR. Trends Plants Sci. 2000;5:439–445. doi: 10.1016/s1360-1385(00)01741-6. [DOI] [PubMed] [Google Scholar]
- 4.Hostettmann V, Marston A. Saponins. Cambridge, UK: Cambridge Univ Press; 1995. [Google Scholar]
- 5.Papadopoulou K, Melton RE, Leggett M, Daniels MJ, Osbourn AE. Proc Natl Acad Sci USA. 1999;96:12923–12928. doi: 10.1073/pnas.96.22.12923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qi X, Bakht S, Leggett M, Maxwell C, Melton R, Osbourn A. Proc Natl Acad Sci USA. 2004;101:8233–8238. doi: 10.1073/pnas.0401301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chappell J. Curr Opin Plant Biol. 2002;5:151–157. doi: 10.1016/s1369-5266(02)00241-8. [DOI] [PubMed] [Google Scholar]
- 8.Haralampidis K, Bryan G, Qi X, Papadopoulou K, Bakht S, Melton R, Osbourn A. Proc Natl Acad Sci USA. 2001;98:13431–13436. doi: 10.1073/pnas.231324698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haralampidis K, Trojanowska M, Osbourn AE. Adv Biochem Eng Biotechnol. 2002;75:31–49. doi: 10.1007/3-540-44604-4_2. [DOI] [PubMed] [Google Scholar]
- 10.Aoyama Y, Noshiro M, Gotoh O, Imaoka S, Funae Y, Kurosawa N, Horiuchi T, Yoshida Y. J Biochem. 1996;119:926–933. doi: 10.1093/oxfordjournals.jbchem.a021331. [DOI] [PubMed] [Google Scholar]
- 11.Nelson DR, Schuler MA, Paquette SM, Werck-Reichhart D, Bak S. Plant Physiol. 2004;135:756–772. doi: 10.1104/pp.104.039826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aoyama Y. Front Biosci. 2005;10:1546–1557. doi: 10.2741/1639. [DOI] [PubMed] [Google Scholar]
- 13.Waterman R, Lepesheva GI. Biochem Biophys Res Commun. 2005;338:418–422. doi: 10.1016/j.bbrc.2005.08.118. [DOI] [PubMed] [Google Scholar]
- 14.Cabello-Hurtado F, Zimmerlin A, Rahier A, Taton M, DeRose R, Nedelkina S, Batard Y, Durst F, Pallett KE, Werck-Reichhart D. Biochem Biophys Res Commun. 1997;230:381–385. doi: 10.1006/bbrc.1996.5873. [DOI] [PubMed] [Google Scholar]
- 15.Trojanowska MR, Osbourn AE, Daniels MJ, Threlfall DR. Phytochemistry. 2001;56:121–129. doi: 10.1016/s0031-9422(00)00399-x. [DOI] [PubMed] [Google Scholar]
- 16.Podust LM, Poulos TL, Waterman MR. Proc Natl Acad Sci USA. 2001;98:3068–3073. doi: 10.1073/pnas.061562898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cabello-Hurtado F, Taton M, Forthoffer N, Kahn R, Bak S, Rahier A, Werck-Reichhart D. Eur J Biochem. 1999;262:435–446. doi: 10.1046/j.1432-1327.1999.00376.x. [DOI] [PubMed] [Google Scholar]
- 18.Tajima F. Genetics. 1993;135:599–607. doi: 10.1093/genetics/135.2.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gierl A, Frey M. Planta. 2001;213:493–498. doi: 10.1007/s004250100594. [DOI] [PubMed] [Google Scholar]
- 20.Osbourn AE, Qi X, Townsend B, Qin B. New Phytologist. 2003;159:101–108. doi: 10.1046/j.1469-8137.2003.00759.x. [DOI] [PubMed] [Google Scholar]
- 21.Hartmann T, Kutchan TM, Strack D. Phytochemistry. 2005:1198–1199. doi: 10.1016/j.phytochem.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 22.Benderoth M, Textor S, Windsor AJ, Mitchell-Olds T, Gershenzon J, Kroymann J. Proc Natl Acad Sci USA. 2006;103:9118–9123. doi: 10.1073/pnas.0601738103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allouis S, Moore G, Bellec A, Sharp R, Faivre Rampant P, Mortimer K, Pateyron S, Foote TN, Griffiths S, Caboche M, Chalhoub B. Cereal Res Commun. 2003;31:331–338. [Google Scholar]
- 24.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd Ed. New York: Cold Spring Harbor Lab Press; 2001. [Google Scholar]
- 25.Mao G, Buschmann H, Doonan JH, Lloyd CW. J Cell Sci. 2006;119:753–758. doi: 10.1242/jcs.02813. [DOI] [PubMed] [Google Scholar]
- 26.Lepesheva GI, Virus C, Waterman MR. Biochemistry. 2003;42:9091–9101. doi: 10.1021/bi034663f. [DOI] [PubMed] [Google Scholar]
- 27.Gouet P, Courcelle E, Stuart DI, Metoz F. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 28.Kumar S, Tamura K, Nei M. Brief Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- 29.Shi J, Blundell TL, Mizuguchi K. J Mol Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- 30.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 31.de Groot BL. Proteins. 1997;29:240–251. doi: 10.1002/(sici)1097-0134(199710)29:2<240::aid-prot11>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 32.Laurie AT, Jackson RM. Bioinformatics. 2005;21:1908–1916. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.