Abstract
Two prominent members of the ATP-binding cassette superfamily of transmembrane proteins, multidrug resistance 1 (MDR1) P-glycoprotein and multidrug resistance protein 1 (MRP1), can mediate the cellular extrusion of xenobiotics and (anticancer) drugs from normal and tumor cells. The MRP subfamily consists of at least six members, and here we report the functional characterization of human MRP5. We found resistance against the thiopurine anticancer drugs, 6-mercaptopurine (6-MP) and thioguanine, and the anti-HIV drug 9-(2-phosphonylmethoxyethyl)adenine (PMEA) in MRP5-transfected cells. This resistance is due to an increased extrusion of PMEA and 6-thioinosine monophosphate from the cells that overproduce MRP5. In polarized Madin–Darby canine kidney II (MDCKII) cells transfected with an MRP5 cDNA construct, MRP5 is routed to the basolateral membrane and these cells transport S-(2,4-dinitrophenyl)glutathione and glutathione preferentially toward the basal compartment. Inhibitors of organic anion transport inhibit transport mediated by MRP5. We speculate that MRP5 might play a role in some cases of unexplained resistance to thiopurines in acute lymphoblastic leukemia and/or to antiretroviral nucleoside analogs in HIV-infected patients.
Cancer cells that overproduce drug-transporting proteins may become resistant to a wide spectrum of drugs with different structures or cellular targets, a phenomenon called multidrug resistance (MDR) (1). ATP-dependent transmembrane drug transporters such as the MDR1 P-glycoprotein (P-gp, ABCB1) (1) and the multidrug-resistance protein (MRP1, ABCC1) (2) can render cells multidrug resistant in vitro, and it is likely that these proteins contribute to the (intrinsic or acquired) resistance against anticancer drugs used in patients.
MRP1 is a glutathione (GSH) conjugate (GS-X) pump or multispecific organic anion transporter (MOAT) (3). MRP1 can mediate the transport of negatively charged conjugated hydrophilic compounds with a large hydrophobic moiety such as glutathione S-, glucuronide, and sulfate conjugates of drugs (3–6). It can extrude neutral and basic organic compounds if the cell contains normal levels of GSH (7–10), probably by cotransport of the drug with GSH (7, 8). Whereas the function of P-glycoprotein in normal tissues appears to be limited to defense against drugs and other xenotoxins, MRP1 not only is involved in reducing the passage of drugs across some specialized epithelia (11, 12) but also is the major transporter for endogenous leukotriene C4 (LTC4), an important mediator of the inflammatory response (13, 14).
Several other MRP family members (MRP2–6, ABCC2–6) may play a role in MDR (15–17): MRP2 has been shown to confer low-level resistance to the anticancer drugs cisplatin, etoposide, vincristine, and methotrexate (MTX) (18–20), MRP3 to etoposide, vincristine, and MTX (15, 21, 22), and MRP4 to acyclic nucleoside phosphonates [e.g., 9-(2-phosphonylmethoxyethyl)guanine (PMEG)] and the anti-HIV drug 9-(2-phosphonylmethoxyethyl)adenine (PMEA) (23). No resistance against (anticancer or antiviral) drugs has been reported for MRP5 or MRP6.
We generated cell lines overexpressing human MRP5 and studied the (anticancer/antiviral) drug resistance spectrum, the transport characteristics, and the intracellular localization of MRP5 in polarized epithelial cells (24). We find that MRP5 is an organic anion transporter with the remarkable ability to confer resistance to base and nucleotide analogs.
Materials and Methods
Materials.
6-Mercaptopurine (6-MP) 14C-labeled at position 8 and bis(pivaloyloxymethyl)-PMEA (bis-POM-PMEA) 3H-labeled at position 8 were obtained from Moravek Biochemicals (Brea, CA). 1-Chloro-2,4-dinitro[14C]benzene ([14C]CDNB) was from Amersham. PMEA was kindly provided by M. Bijsterbosch (Leiden/Amsterdam Center for drug research, Leiden, The Netherlands), and PMEA and bis-POM-PMEA were from N. Bischofberger (Gilead Sciences, Foster City, CA).
Cell Lines.
293 Human Embryonic Kidney (HEK) cells (25) and Madin–Darby canine kidney II (MDCKII) cells (26) were grown in DMEM (GIBCO/BRL) containing 10% FCS and penicillin/streptomycin.
Cloning and Sequencing of MRP5 cDNA.
Two PCR primers [base pairs 18–37 and 345–326 from expressed sequence tag (EST) H69466 containing the first ATP-binding domain of a MRP family member] were used to generate a 327-bp cDNA probe from a human fetal brain λ DR2 cDNA library (CLONTECH). This probe was used to screen a 5′-stretch human fetal brain λgt11 cDNA library (CLONTECH). We obtained several long cDNA clones containing the first and second ATP-binding domains as well as the complete 3′ untranslated region. However, about 1 kb of 5′ coding and noncoding sequences was not retrieved from this library. Using total RNA from the human ovarian carcinoma cell line 2008A and a 5′-RACE protocol (GIBCO/BRL), we isolated the missing 5′ end. MRP5 cDNA was sequenced with the Applied Biosystems ABI377 automatic sequencer. The GenBank accession number is U83661 (16).
Overproduction of MRP5 in 293 HEK and MDCKII Cells.
The MRP5 cDNA was assembled in pGEM5Zf to generate pGEM5Zf-MRP5. We modified the retroviral expression vector pBABE-puro (27), by insertion of a blunted 500-bp HindIII–HindIII fragment containing the cytomegalovirus (CMV) promoter from pCMV-neo-cjun (28) into the blunted BamHI site of pBABE-puro, resulting in pBABE-CMV-puro. A blunted EcoRI–HindIII fragment containing the complete MRP5 cDNA was subcloned behind the CMV promoter in the blunted BamHI site of the retroviral vector pBABE-CMV-puro to generate pBABE-CMV-MRP5-puro. By using a calcium phosphate precipitation cell transfection kit (GIBCO/BRL), the retroviral vector was transfected into the packaging cell line Phoenix (29), and supernatants containing retrovirus particles were used to transduce 293 and MDCKII cells. MRP5-overproducing cells were identified on immunoblots using monoclonal antibodies (mAbs) directed against human MRP5 (M5I-1 or M5II-54).
mAbs.
For the generation of mAbs against MRP5, we subcloned the MRP5 cDNA coding for amino acids 722–910, or for amino acids 82–168, into the pmal-c vector (New England Biolabs) for production of a fusion protein of Escherichia coli maltose-binding protein (MBP) with MRP5. The affinity-purified protein was injected into rats, and mAbs were isolated as described (30, 31). mAb M5I-1 against amino acids 82–168, and M5II-54 against amino acids 722–910 specifically recognize the full-length human MRP5 protein in MRP5-overproducing 293 and MDCKII cells. Both mAbs are specific for MRP5 on Western blots, and they do not crossreact with MRP1, -2, or -3 (data not shown).
Protein Analysis and Immunocytochemistry.
For immunolocalization of MRP5, MDCKII cells were grown for 3 days on microporous polycarbonate filters (3-μm pore size, 24-mm diameter, Transwell 3414: Costar, Cambridge, MA). MRP5 was detected with mAbs M5I-1 and M5II-54 as primary antibodies and FITC-labeled rabbit anti-rat IgG (1:50, Nordic Immunology, Tilburg, The Netherlands) as the secondary antibody. Protein analyses on Western blots using mAbs M5I-1 and M5II-54 as primary antibodies and horseradish peroxidase-labeled rabbit anti-rat IgG (1:1000, Dako) as the secondary antibody and chemiluminescent detection were performed as described (30).
Cytotoxicity Assays.
293 and 293/MRP5 cells (1,800 cells in 100 μl of conditioned medium per 96 wells) were plated in triplicate and incubated for 24 h at 37°C under 5% CO2/95% air. Dilution series of drugs in 100 μl of conditioned medium were added to the cells and incubated for 5 days at 37°C. Medium was removed and cells were frozen at −80°C. Cells were thawed and the total number of cells was determined fluorimetrically by using the CyQuant Cell Proliferation Assay Kit (Molecular Probes) and the CytoFluor 4000 fluorescence plate reader (PerSeptive Biosystems, Framingham, MA). The relative resistance was calculated as the ratio of 50% inhibition of growth (IC50) of the resistant cell line to the IC50 of the parental cell line.
S-(2,4-dinitrophenyl)glutathione (DNP-GS) Transport and Inhibition Assays.
MDCKII or MDCKII/MRP5 cells were grown for 3–4 days on microporous polycarbonate membrane filters at a plating density of 3 × 106 cells per well in a six-well plate. Excretion of [14C]DNP-GS from cells was determined by incubating cells with 2 μM [14C]CDNB in Hanks' balanced salt solution (HBSS) as described previously (32). Briefly, cells were washed in HBSS and incubated at room temperature with 2 μM [14C]CDNB in HBSS applied to both the apical and basal compartments. Samples of medium from each compartment were taken at several time points. The water-soluble [14C]DNP-GS was separated from the hydrophobic [14C]CDNB by extraction of the samples with ethyl acetate, and radioactivities of samples were quantitated in a liquid scintillation counter.
Accumulation and Efflux Assays.
For accumulation studies (n = 6), 293 and 293/MRP5 cells were plated in triplicate at a density of 5 × 105 cells per well of a poly(l-lysine)-treated 12-well plate overnight. Cells were incubated with 0.5 ml of 0.4, 2.0, or 10 μM [8-14C]-6-MP (54 Ci/mol; 1 Ci = 37 GBq) for 30 min at 37°C in prewarmed DMEM/10% FCS medium, equilibrated under 5% CO2, and containing penicillin/streptomycin. After drug accumulation, medium was removed, and cells were put on ice, washed three times with 2 ml of ice-cold PBS, treated in 0.2 ml of trypsin solution, and added to 4 ml of scintillation mix to measure radioactivity.
For efflux studies, 293 and 293/MRP5 cells were plated in triplicate at a density of 2 × 106 cells per well of a poly(l-lysine)-treated 12-well plate overnight. Drug loading under ATP-depleting conditions was performed for 2 h in glucose-free, pyruvate-free DMEM (GIBCO/BRL) containing 5% dialyzed FCS, 10 mM deoxyglucose, 10 mM sodium azide, and 10 μM [8-14C]-6-MP (54 Ci/mol) or 1 μM bis-POM-[3H]PMEA (3 Ci/mmol). After loading, the wells were washed quickly with PBS, and incubated with prewarmed complete medium at 37°C. Medium samples were taken at timed intervals for analysis.
HPLC Analyses.
6-Thioinosine monophosphate (tIMP) for peak identification was synthesized for 4 h at 37°C in a 1.5-ml reaction by mixing 500 μl of 15 μM 6-MP or [8-14C]-6-MP, 500 μl of 8 mM phosphoribosyl pyrophosphate (Sigma), 480 μl of Tris/MgCl2 (0.5 M Tris⋅HCl/50 mM MgCl2, pH 7.4) and 20 μl of hypoxanthine-guanine phosphoribosyltransferase (250 units per 500 μl in Tris⋅HCl buffer; Sigma). Medium samples containing 6-MP radioactivity were separated by HPLC (Sphereclone SAX, 150 × 3.2 mm, 5-μm particle size; Phenomenex, Torrance, CA) by using a solvent system of 0.01 M KH2PO4 adjusted to pH 3.1 at a flow rate of 0.7 ml/min. Radioactivity was quantitated by scintillation counting.
Medium samples containing PMEA radioactivity were separated by HPLC for analysis of PMEA, PMEA-monophosphate (PMEAp), and PMEA-diphosphate (PMEApp) as described (33, 34). For determination of intracellular PMEA levels, cells were lysed in ice-cold 60% methanol before HPLC analysis.
Results
Cloning and Sequencing of Human MRP5 cDNA.
Starting with a partial cDNA designated MRP5 (16), we obtained a full-length sequence encoding a protein of 1,437 amino acids, for which we obtained GenBank accession no. U83661 (16). Our coding sequence is similar to the MOAT-C sequence (ref. 35; GenBank AF104942), except for a nonconservative substitution of a serine in MRP5 for a glycine in MOAT-C at amino acid position 400. Our deduced protein sequence is identical to the pABC11 sequence (ref. 36; AF146074). The identity of human MRP5 with the mouse Mrp5 (ref. 37; GenBank AB019003) is 94%, and with the other MRP subfamily members (MRP1, -2, -3, -4, and -6) it is 31–36% (38). The features of the MRP5 sequence (16) have been described (35, 39). Our hydrophobicity analysis of MRP5 largely agrees with the published data (35, 36).
Generation of MRP5-Overproducing Cell Lines.
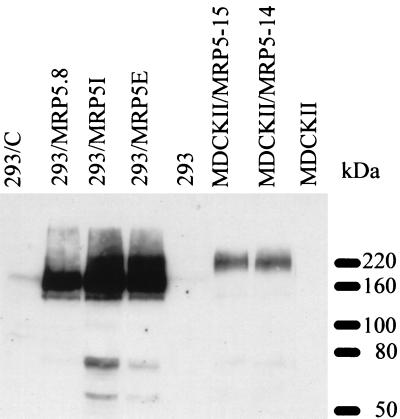
We generated MRP5 retrovirus-overproducing cells by insertion of the MRP5 cDNA behind the CMV promoter/enhancer of a retroviral expression vector and subsequent transfection into the packaging cell line Phoenix (29). When immunoblotting was used, no MRP5 was detected in MDCKII or 293 cells and the retrovirus was therefore used to transduce these cells. MRP5-overproducing puromycin-resistant clones were identified with our mAbs M5II.54 and M5I.1. Both mAbs recognized a protein of the expected size, 160–180 kDa, on Western blots in MDCKII/MRP5 or 293/MRP5 cells (Fig. 1). Much higher levels of MRP5 were obtained in the 293/MRP5 cells than in the MDCKII/MRP5 cells. The apparent molecular mass of MRP5 in 293/MRP5 cells is lower than in MDCKII cells, suggesting that more extensive glycosylation occurs in MDCKII cells.
Figure 1.
Western blot analysis of MRP5 protein in 293 and MDCKII cells transfected with MRP5 cDNA. Cell lysate proteins were fractionated on a 7.5% polyacrylamide gel containing 0.5% SDS and transferred to a nitrocellulose membrane by electroblotting. MRP5 was detected with mAb M5I.1.
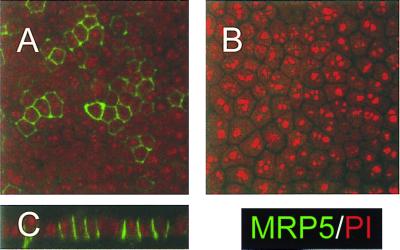
Immunolocalization of MRP5 in MDCKII/MRP5 Cells.
The subcellular localization of MRP5 in cell clones MDCKII/MRP5–14, -15, and -4 was determined by immunocytochemistry using mAb M5I.1 and confocal laser scanning microscopy. In polarized monolayers on microporous membrane filters MRP5 is mainly localized in the plasma membranes. Little intracellular staining was found in MDCKII/MRP5–14 and -15 cells, but more in MDCKII/MRP5-4 cells (Fig. 2 A and B, and data not shown). The vertical x/z section of the monolayer shows that MRP5 is localized in the (baso)lateral plasma membranes of these epithelial cells (Fig. 2C).
Figure 2.
Immunolocalization of MRP5 in MDCKII cell monolayers by confocal laser scanning microscopy. MRP5 is detected by indirect immunofluorescence (green signal) with mAb M5I.1. Nucleic acids were counterstained by propidium iodide (red signal). (×800.)(A) MDCKII/MRP5–14 cells. (B) Parental MDCKII cells. (C) Vertical y/z section of the monolayer shown in A.
In contrast, in the nonpolarized cell clones 293/MRP5I and 5E, staining for MRP5 with mAb M5I.1 was predominantly intracellular with some staining of the plasma membrane (data not shown).
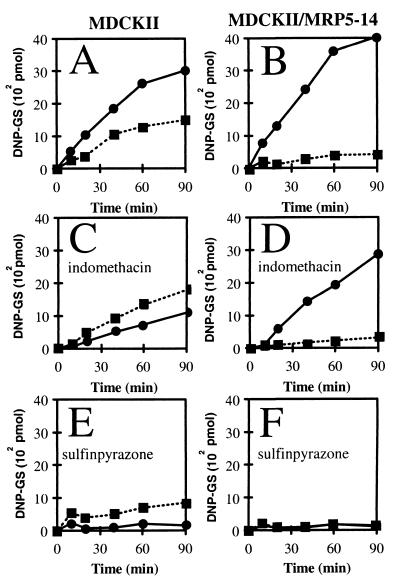
DNP-GS Excretion from MRP5-Overproducing MDCKII Cells.
A classical substrate for GS-X pumps is DNP-GS (19, 21, 32, 40). This charged compound does not enter intact cells, but it can be generated intracellularly by incubating cells with CDNB. The hydrophobic CDNB passively diffuses into the cells and is converted intracellularly into the hydrophilic DNP-GS by the action of glutathione S-transferases. We tested the MDCKII/MRP5 clones for DNP-GS transport and found increased transport of DNP-GS to the basal compartment (Fig. 3), in accordance with the basolateral localization of MRP5 in these cells. A complication is that parental cells (Fig. 3A) export considerable amounts of DNP-GS in the basolateral (presumably by canine MRP1) and apical (presumably by canine MRP2) directions. Increased basolateral DNP-GS export by MRP5 suppresses the endogenous apical DNP-GS export (Fig. 3B), probably by competition for substrate, as MDCKII/MRP5–14 cells accumulated less DNP-GS (60% ± 5%; n = 3 independent experiments in duplicate, P < 0.005) than the parental cells. Indomethacin, which inhibits MRP1 more effectively than MRP2, inhibits basolateral, but not apical, transport in parental cells, but it has little effect on MRP5-mediated export (Fig. 3 C and D). Sulfinpyrazone (Fig. 3 E and F) inhibits all transporters. MDCKII/MRP5–15 cells gave results similar to those with the MDCKII/MRP5–14 cells.
Figure 3.
Export of [14C]DNP-GS from MDCKII and MDCKII/MRP5 cells grown in monolayers. MDCKII (A, C, and E) and MDCKII/MRP5–14 (B, D, and F) cells were incubated at room temperature with 2 μM [14C]CDNB in both the apical and basal medium compartments, and export of [14C]DNP-GS was determined. Similar results were obtained with the other MDCKII/MRP5 clones. This transport was inhibited by sulfinpyrazone (IC50 < 0.5 mM) and benzbromarone (IC50< 5 μM), and less effectively by probenecid (IC50 > 5 mM), indomethacin (IC50 > 100 μM), sulfobromophthalein (IC50 > 100 μM), nitrobenzylthioinosine (NBTI; IC50 > 10 μM) or dipyridamole (IC50 > 10 μM). Variation between representative duplicate experiments was mostly within the size of the symbols. ● and ■ represent transport to basal and apical medium compartments, respectively. (A and B) Without inhibitor. (C and D) Indomethacin present at 100 μM. (E and F) Sulfinpyrazone present at 2.5 mM.
MDCKII cells transfected with MRP1 or MRP2 gene constructs secrete GSH and cannot maintain intracellular GSH levels in simple salt media (40). We also found this for the MRP5-transfected cells. The MDCKII/MRP5–14, -15, and -4 clones secreted more GSH to the basal compartment (on average 2.2-fold after 2, 3, and 4 h; P < 0.05) and contained less intracellular GSH (on average 61% ± 2% after 4 h; P < 0.05; results not shown).
Cytotoxicity Assays Using 293 Cells.
To determine whether MRP5 can confer drug resistance by acting as a plasma membrane drug efflux pump, we used several 293/MRP5 clones in growth inhibition assays with continuous exposure to drugs for 5 days. We found resistance in at least two independent clones against 6-MP, its prodrug azathioprine (data not shown), thioguanine, 5-hydroxypyridine-2-carboxaldehyde thiosemicarbazone (5-HP), and PMEA but not against other purine, pyrimidine, or nucleoside analogs or other compounds tested (Table 1).
Table 1.
Growth inhibition of 293 and 293/MRP5 cells by cytotoxic agents
| Drug | 293
|
293/C
|
293/MRP5E
|
293/MRP5I
|
||||
|---|---|---|---|---|---|---|---|---|
| IC50, μM | RF | IC50, μM | RF | IC50, μM | RF | IC50, μM | RF | |
| 6-MP | 1.7 ± 0.2 | 1 | 2.3 ± 0.3 | 1.3 | 4.2 ± 0.4 | 2.5* | 5.3 ± 0.2 | 3.1* |
| Thioguanine | 1.2 ± 0.1 | 1 | 1.1 ± 0.1 | 0.8 | 2.7 ± 0.3 | 2.3* | 2.4 ± 0.1 | 2.1* |
| PMEA | 79 ± 13 | 1 | 94 ± 15 | 1.2 | 126 ± 12 | 1.6† | 201 ± 33 | 2.5* |
| 5-HP | 4.5 ± 0.3 | 1 | 4.6 ± 0.2 | 1.0 | 6.3 ± 0.3 | 1.4† | 9.1 ± 0.3 | 2.0* |
| Etoposide | 0.50 ± 0.02 | 1 | ND | ND | 0.57 ± 0.05 | 1.1 | 0.73 ± 0.05 | 1.5* |
| Teniposide | 36 ± 1 | 1 | ND | ND | 41 ± 20 | 1.1 | 59 ± 1 | 1.6* |
The IC50 values shown are the means ± SE, for at least three experiments, except for two experiments with teniposide, each experiment performed in triplicate. RF, resistance factor; ND, not done. *, P < 0.01;
, P < 0.05 compared with 293 cells. No resistance was detected for doxorubicin, daunorubicin, epirubicin, vincristine, vinblastine, CPT-11, SN-38, topotecan, MTX, trimetrexate, pyrimethamine, cisplatin, mitoxantrone, Taxol, tamoxifen, acyclovir, 8-azaadenosine, azathioprine, 8-bromoadenosine, cordycepin, 5-fluorouracil, carbenicillin, hygromycin, neomycin, benzbromarone, dipyridamole, indomethacin, probenecid, sulfinpyrazone, arsenite, cadmium, potassium antimonyl tartrate, acetaminophen, caffeine, Colcemid, ethacrynic acid, quinidine, sodium deoxycholate, or uric acid.
Because MRP1, -2, and -3 confer high resistance to MTX in short-term drug exposure experiments, and low resistance in long-term exposures (21, 41), we repeated our experiments with 4-h exposures to MTX at high concentrations. Under these conditions we did not detect resistance of MRP5-overproducing cells to MTX either.
McAleer et al. (36) recently described a single transfected 293 cell clone overproducing a fusion of MRP5 (pABC11) and green fluorescent protein (GFP). This 293/MRP5-GFP clone showed low-level resistance to CdCl2 and potassium antimonyl tartrate and increased export of the organic anion fluorochrome 5-chloromethylfluorescein diacetate. We also observed increased fluorochrome export from MDCKII/MRP5 cells (unpublished data). However, our three independent 293/MRP5 clones did not show any resistance to CdCl2 or potassium antimonyl tartrate. This result could be simply a matter of the concentration of MRP5 in the plasma membrane, since most of the MRP5-GFP was in the plasma membrane (36), whereas in our transfectants most of the MRP5 was intracellular. More puzzling is that McAleer et al. (36) found only a “minimal effect” of sulfinpyrazone on transport by MRP5-GFP, whereas we find this drug to be an excellent inhibitor of MRP5 (Fig. 3F; Fig. 4D).
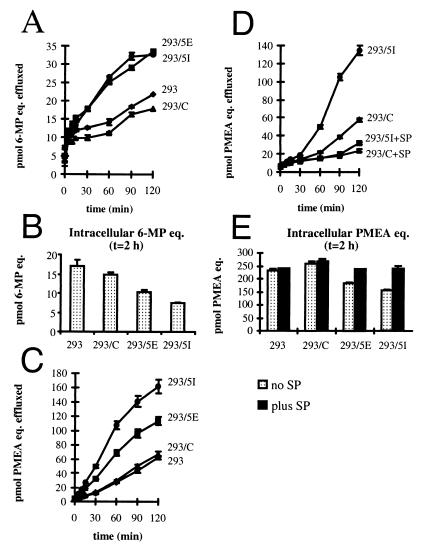
Figure 4.
Drug efflux from 293 and 293/MRP5 cells. (A) Efflux of 6-MP radioactivity from cells loaded with 10 μM [14C]-6-MP. Cells were loaded for 2 h in the presence of azide and deoxyglucose. (B) Retention of intracellular 6-MP radioactivity after 2-h efflux. (C) Efflux of PMEA radioactivity from cells loaded with 1 μM bis-POM-[8-3H]PMEA. Loading conditions as in B. (D) Inhibition of PMEA radioactivity efflux by 2.5 mM sulfinpyrazone (SP). (E) Retention of intracellular PMEA in the presence or absence of sulfinpyrazone after 2-h efflux. Radioactivity was determined by scintillation counting. Data are expressed as pmol of radiolabeled PMEA equivalents (eq.) effluxed as a function of time.
Reduced Drug Accumulation and Increased Efflux.
To determine whether the resistance to thiopurines in 293/MRP5 cells is caused by altered accumulation of the drugs, we studied the accumulation of [8-14C]-6-MP in transfected cells not expressing MRP5 (293/C) and transfected cells expressing MRP5 (293/MRP5I) after exposure to various concentrations of [8-14C]-6-MP. The accumulation of [8-14C]-6-MP in 293/MRP5I cells was consistently lower (76% ± 4%, 78% ± 6%, 76% ± 6%, at 0.4 μM, 2 μM, and 10 μM 6-MP for 30-min exposure, respectively; n = 6, P < 0.01) than in 293/C cells (100%).
To test whether this decreased accumulation is due to increased efflux of 6-MP or cellular conversion products of 6-MP, we preloaded cells with [14C]-6-MP under ATP-depleting conditions to inhibit export and incorporation of 6-MP into nucleic acids. No effect of MRP5 was found on 6-MP loading. The MRP5-containing cells (293/MRP5E and 293/MRP5I) effluxed radioactivity more rapidly than did 293 cells or the transfected cells not expressing MRP5 (293/C) (Fig. 4A), whereas the MRP5 cells retained less radioactivity (Fig. 4B).
Similar results were obtained with PMEA, an acyclic nucleoside phosphonate containing a nonhydrolyzable phosphonate bond and resembling a nucleoside 5′-monophosphate. Because PMEA enters cells rather slowly (42), we preloaded the cells with the hydrophobic precursor bis-POM-[8-3H]PMEA, which is rapidly converted into PMEA intracellularly (43). Fig. 4C shows that the rate of 3H efflux is highest for the 293/MRP5I clone, intermediate for the 293/MRP5E clone, and not significantly different from 293 cells for the 293/C clone, which has no detectable MRP5 expression. Efflux was nearly completely inhibited by sulfinpyrazone (IC50 < 0.1 mM; Fig. 4D and data not shown), as was the low efflux in cells without MRP5. Fig. 4E shows that the MRP5 cells retained less radioactivity than the control cells in the absence of sulfinpyrazone but not in the presence of the inhibitor.
To test which radioactive components the cells preloaded with bis-POM-PMEA effluxed or retained, the relevant PMEA metabolites were analyzed by HPLC (33, 34). More than 98% of radioactivity effluxed from 293/MRP5 cells and control cells was associated with [3H]PMEA (data not shown). Table 2 shows that the bis-POM-PMEA was efficiently converted into PMEA in 293 cells, and that the cells effluxed PMEA (a phosphonate) itself rather than phosphorylated forms of PMEA. Interestingly, levels of PMEApp, the active metabolite that is responsible for the biological activities of the drug (34, 44), were lower in 293/MRP5 cells. Our results indicate that MRP5 transports nucleoside monophosphate analogs rather than base or nucleoside analogs and suggested that the resistance of the 293/MRP5 cells to 6-MP might be due to efflux of tIMP, the nucleoside monophosphate product of intracellular 6-MP metabolism. To test this possibility, we set up an HPLC analysis separating tIMP from 6-MP and analyzed medium samples (t = 1 h) from cells loaded with [14C]-6-MP as in Fig. 4B. About half of the radioactivity (45 ± 2%) in the supernatants of 293/MRP5E and 293/MRP5I cells comigrated with 6-MP, 36% ± 3% with tIMP, and 19% ± 1% with an unidentified peak X of higher retention on the column, possibly a disulfide dimer of tIMP or methyl-tIMP. Only 1% ± 1% tIMP and 2% ± 2% X were found in the supernatants of the 293 and 293/C cells, which do not significantly express MRP5.
Table 2.
Intracellular amounts of PMEA, PMEAp, and PMEApp (in pmol per 106 cells) in cells incubated with bis-POM-[3H]PMEA
| Cell line | PMEA, pmol
|
PMEAp, pmol
|
PMEApp, pmol
|
Total, pmol
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 h | 1 h | 2 h | 0 h | 1 h | 2 h | 0 h | 1 h | 2 h | 0 h | 1 h | 2 h | |
| 293 | 138 | 104 | 75 | 0.7 | 3.2 | 6.2 | 0.4 | 15.2 | 27.6 | 139 | 122 | 109 |
| 293/C | 138 | 111 | 81 | 0.5 | 3.5 | 6.2 | 0.4 | 12.5 | 22.3 | 139 | 127 | 109 |
| 293/MRP5E | 136 | 81 | 39 | 0.5 | 2.4 | 4.2 | 0.3 | 11.8 | 16.1 | 137 | 95 | 60 |
| 293/MRP5I | 128 | 61 | 19 | 0.5 | 3.0 | 3.4 | 0.3 | 11.4 | 12.6 | 128 | 76 | 35 |
Discussion
Our results show that MRP5 is a multispecific organic anion pump able to transport nucleotide analogs. MRP5 is a GS-X pump, because it transports DNP-GS and is inhibited by typical inhibitors of organic anion transport, such as sulfinpyrazone. Interestingly, MRP5 also transports organic anions in which the anionic moiety is a phosphate/phosphonate group, resulting in the ability to confer resistance against the anticancer thiopurine drugs 6-MP and thioguanine, and the anti-HIV drug PMEA. The MRP5-mediated PMEA efflux from resistant cells can be inhibited by low concentrations of sulfinpyrazone linking the GS-X pump function of MRP5 to its ability to transport nucleotide analogs.
MRP1, -2, -3, and -5 are all GS-X pumps, but they differ in tissue distribution (16, 38) and in substrate preference. MRP1 has the highest affinity for GSH conjugates, such as leukotriene C4 or S-glutathionyl aflatoxin B1, but can also transport 17β-glucuronosyl estradiol with micromolar affinity. In addition, MRP1 transports organic sulfate and monoglutamate (e.g., MTX) conjugates with lower affinity (45, 46). The substrate specificity of MRP2 is similar to that of MRP1, but its preference for GSH conjugates is less and it has the interesting ability to confer resistance to cisplatin, probably by transporting a cisplatin-(GS)2 complex (18). MRP3 has a low affinity for GSH conjugates (47), and it is the only MRP known to date that does not induce increased GSH export in transfected cells (21). MRP3 does not have a high affinity for glucuronide conjugates or MTX (47), but it transports bile salt sulfate-conjugates with micromolar affinity (48). Whether MRP1, -2, or -3 can transport nucleotide analogs as MRP4 and MRP5 do remains to be tested. We have not observed 6-MP or PMEA resistance in 2008 ovarian carcinoma cells transfected with MRP1, MRP2, or MRP3 constructs (unpublished results), but a more systematic analysis will be required to test whether these transporters bind nucleotide analogs at all. More work is also required to sort out which type of conjugate (GSH, glucuronide, sulfate, or phosphate) is preferentially used by MRP5. The fact that we have not observed any resistance to MDR drugs in MRP5-expressing cells does not mean that these drugs are not transported by MRP5. Because most of the MRP5 present in our transfected 293 cells is intracellular rather than in the cell membrane, most of the MRP5 may not contribute to resistance. We may therefore have seen only the tip of the resistance iceberg thus far.
Within the MRP family, MRP5 is most closely related to MRP4, both proteins lack the first five membrane-spanning regions (35), and MRP4 also appears able to transport nucleotides. The MRP4 gene was recently found to be overexpressed and amplified (23) in a PMEA-resistant cell line (49), crossresistant to 9-(2-phosphonylmethoxyethyl)guanine (PMEG) and azidothymidine (AZT). The overexpression of MRP4 correlated well with an increased ATP-dependent efflux of PMEA and AZT monophosphate from the drug-resistant cell line (23). Whether MRP4 is also a GS-X pump and able to transport compounds other than nucleotide analogs remains to be determined.
Nothing is known yet about the physiological function of MRP5. Although MRP5 RNA is found in all tissues analyzed thus far (16, 36), it is still not known in which subpopulation of cells the protein is located, as our currently available mAbs do not detect MRP5 in tissues. We have generated knockout mice with disrupted Mrp5 alleles (24), and these mice are healthy and fertile. This result does not necessarily mean that MRP5 has no physiological function and is required only for defense against nucleotide analogs. Mrp1(−/−) mice are also doing fine in the protected environment of an animal facility (14, 50), even though Mrp1 is the major transporter for endogenous leukotriene C4, a mediator of the inflammatory response (13, 14). An obvious question is whether MRP5 is able to transport any natural purine nucleotides, and this remains to be tested. It is interesting that MRP5 RNA is present in the brain (16) and even various segments of the brain (36). The only other MRP family member known to reside in the brain is MRP1, which is restricted to the choroid plexus (12, 51). The possibility that MRP5 is part of the arrays of pumps used to protect the brain against drugs should therefore be explored.
Whether MRP5 contributes to clinical resistance to 6-MP or PMEA remains to be seen. Resistance to thiopurines is most often caused by alterations in hypoxanthine-guanine phosphoribosyltransferase or other enzymes in the purine salvage pathway, but diminished intracellular drug accumulation has been described as well (52, 53). Clinical resistance to PMEA often results from mutation of the reverse transcriptase in the HIV genome (reviewed in ref. 54), but it is conceivable that drug efflux pumps might be involved in creating cellular sanctuaries (reviewed in ref. 55) poorly accessible to nucleoside/nucleotide analogs (23, 33, 49, 56).
Acknowledgments
We thank Drs. Tohru Saeki, Peter Wielinga, and Noam Zelcer (Amsterdam) for critical advice on the manuscript, Dr. Lieve Naesens (Leuven) for helpful discussions, and Mrs. Ria Van Berwaer (Leuven) for her important technical assistance. We thank Lauran Oomen (Department of Biophysics, The Netherlands Cancer Institute) for assistance with the confocal laser scanning microscopy. We thank the Drug Synthesis and Chemistry Branch, Developmental Therapeutics Program, Division of Cancer Treatment (National Cancer Institute, Bethesda, MD) for providing 5-HP. This work was supported in part by grants from the Dutch Cancer Society.
Abbreviations
- GSH
glutathione
- GS-X
GSH conjugate
- DNP-GS
S-(2,4-dinitrophenyl)glutathione
- CDNB
1-chloro-2,4-dinitrobenzene
- CMV
cytomegalovirus
- HEK
human embryonic kidney cells
- 5-HP
5-hydroxypyridine-2-carboxaldehyde thiosemicarbazone
- MDCKII cells
Madin–Darby canine kidney II cells
- MDR
multidrug resistance
- 6-MP
6-mercaptopurine
- MTX
methotrexate
- MRP1
MDR protein 1
- PMEA
9-(2-phosphonylmethoxyethyl)adenine
- bis-POM-PMEA
bis(pivaloyloxymethyl)-PMEA
- PMEAp
PMEA monophosphate
- PMEApp
PMEA diphosphate
- tIMP
6-thioinosine monophosphate
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. U83661).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.120159197.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.120159197
References
- 1.Ambudkar S V, Dey S, Hrycyna C A, Ramachandra M, Pastan I, Gottesman M M. Annu Rev Pharmacol Toxicol. 1999;39:361–398. doi: 10.1146/annurev.pharmtox.39.1.361. [DOI] [PubMed] [Google Scholar]
- 2.Cole S P, Deeley R G. BioEssays. 1998;20:931–940. doi: 10.1002/(SICI)1521-1878(199811)20:11<931::AID-BIES8>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 3.Ishikawa T, Li Z S, Lu Y P, Rea P A. Biosci Rep. 1997;17:189–207. doi: 10.1023/a:1027385513483. [DOI] [PubMed] [Google Scholar]
- 4.Leier I, Jedlitschky G, Buchholz U, Cole S P, Deeley R G, Keppler D. J Biol Chem. 1994;269:27807–27810. [PubMed] [Google Scholar]
- 5.Jedlitschky G, Leier I, Buchholz U, Barnouin K, Kurz G, Keppler D. Cancer Res. 1996;56:988–994. [PubMed] [Google Scholar]
- 6.Muller M, Meijer C, Zaman G J, Borst P, Scheper R J, Mulder N H, de Vries E G, Jansen P L. Proc Natl Acad Sci USA. 1994;91:13033–13037. doi: 10.1073/pnas.91.26.13033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loe D W, Deeley R G, Cole S P. Cancer Res. 1998;58:5130–5136. [PubMed] [Google Scholar]
- 8.Loe D W, Almquist K C, Deeley R G, Cole S P. J Biol Chem. 1996;271:9675–9682. doi: 10.1074/jbc.271.16.9675. [DOI] [PubMed] [Google Scholar]
- 9.Zaman G J, Lankelma J, van Tellingen O, Beijnen J, Dekker H, Paulusma C, Oude Elferink R P, Baas F, Borst P. Proc Natl Acad Sci USA. 1995;92:7690–7694. doi: 10.1073/pnas.92.17.7690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rappa G, Lorico A, Flavell R A, Sartorelli A C. Cancer Res. 1997;57:5232–5237. [PubMed] [Google Scholar]
- 11.Wijnholds J, Scheffer G L, van der Valk M, van der Valk P, Beijnen J H, Scheper R J, Borst P. J Exp Med. 1998;188:797–808. doi: 10.1084/jem.188.5.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wijnholds J, de Lange E C, Scheffer G L, van den Berg D J, Mol C A, van der Valk M, Schinkel A H, Scheper R J, Breimer D D, Borst P. J Clin Invest. 2000;105:279–285. doi: 10.1172/JCI8267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jedlitschky G, Leier I, Buchholz U, Center M, Keppler D. Cancer Res. 1994;54:4833–4836. [PubMed] [Google Scholar]
- 14.Wijnholds J, Evers R, van Leusden M R, Mol C A, Zaman G J, Mayer U, Beijnen J H, van der Valk M, Krimpenfort P, Borst P. Nat Med. 1997;3:1275–1279. doi: 10.1038/nm1197-1275. [DOI] [PubMed] [Google Scholar]
- 15.Young L C, Campling B G, Voskoglou-Nomikos T, Cole S P, Deeley R G, Gerlach J H. Clin Cancer Res. 1999;5:673–680. [PubMed] [Google Scholar]
- 16.Kool M, de Haas M, Scheffer G L, Scheper R J, van Eijk M J, Juijn J A, Baas F, Borst P. Cancer Res. 1997;57:3537–3547. [PubMed] [Google Scholar]
- 17.Allikmets R, Gerrard B, Hutchinson A, Dean M. Hum Mol Genet. 1996;5:1649–1655. doi: 10.1093/hmg/5.10.1649. [DOI] [PubMed] [Google Scholar]
- 18.Cui Y, Konig J, Buchholz J K, Spring H, Leier I, Keppler D. Mol Pharmacol. 1999;55:929–937. [PubMed] [Google Scholar]
- 19.Evers R, Kool M, van Deemter L, Janssen H, Calafat J, Oomen L C, Paulusma C C, Oude Elferink R P, Baas F, Schinkel A H, Borst P. J Clin Invest. 1998;101:1310–1319. doi: 10.1172/JCI119886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koike K, Kawabe T, Tanaka T, Toh S, Uchiumi T, Wada M, Akiyama S, Ono M, Kuwano M. Cancer Res. 1997;57:5475–5479. [PubMed] [Google Scholar]
- 21.Kool M, van der Linden M, de Haas M, Scheffer G L, de Vree J M, Smith A J, Jansen G, Peters G J, Ponne N, Scheper R J, et al. Proc Natl Acad Sci USA. 1999;96:6914–6919. doi: 10.1073/pnas.96.12.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng H, Bain L J, Belinsky M G, Kruh G D. Cancer Res. 1999;59:5964–5967. [PubMed] [Google Scholar]
- 23.Schuetz J D, Connelly M C, Sun D, Paibir S G, Flynn P M, Srinivas R V, Kumar A, Fridland A. Nat Med. 1999;5:1048–1051. doi: 10.1038/12487. [DOI] [PubMed] [Google Scholar]
- 24.Wijnholds J, Mol C A, Scheffer G L, Scheper R J, Borst P. Proc Am Assoc Cancer Res. 1999;40:315. [Google Scholar]
- 25.Graham F L, Smiley J, Russell W C, Nairn R. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 26.Louvard D. Proc Natl Acad Sci USA. 1980;77:4132–4136. doi: 10.1073/pnas.77.7.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgenstern J P, Land H. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bengal E, Ransone L, Scharfmann R, Dwarki V J, Tapscott S J, Weintraub H, Verma I M. Cell. 1992;68:507–519. doi: 10.1016/0092-8674(92)90187-h. [DOI] [PubMed] [Google Scholar]
- 29.Grignani F, Kinsella T, Mencarelli A, Valtieri M, Riganelli D, Lanfrancone L, Peschle C, Nolan G P, Pelicci P G. Cancer Res. 1998;58:14–19. [PubMed] [Google Scholar]
- 30.Flens M J, Zaman G J, van der Valk P, Izquierdo M A, Schroeijers A B, Scheffer G L, van der Groep P, de Haas M, Meijer C J, Scheper R J. Am J Pathol. 1996;148:1237–1247. [PMC free article] [PubMed] [Google Scholar]
- 31.Scheffer, G. L., Kool, M., Heijn, M., de Haas, M., Pijnenborg, A. C., Wijnholds, J., van Helvoort, A., de Jong, M. C., Hooijberg, J. H., Mol, C. A., et al. (2000) Cancer Res., in press. [PubMed]
- 32.Evers R, Zaman G J, van Deemter L, Jansen H, Calafat J, Oomen L C, Oude Elferink R P, Borst P, Schinkel A H. J Clin Invest. 1996;97:1211–1218. doi: 10.1172/JCI118535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hatse S, De Clercq E, Balzarini J. Mol Pharmacol. 1998;54:907–917. doi: 10.1124/mol.54.5.907. [DOI] [PubMed] [Google Scholar]
- 34.Balzarini J, Hao Z, Herdewijn P, Johns D G, De Clercq E. Proc Natl Acad Sci USA. 1991;88:1499–1503. doi: 10.1073/pnas.88.4.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Belinsky M G, Bain L J, Balsara B B, Testa J R, Kruh G D. J Natl Cancer Inst. 1998;90:1735–1741. doi: 10.1093/jnci/90.22.1735. [DOI] [PubMed] [Google Scholar]
- 36.McAleer M A, Breen M A, White N L, Matthews N. J Biol Chem. 1999;274:23541–23548. doi: 10.1074/jbc.274.33.23541. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki T, Sasaki H, Kuh H J, Agui M, Tatsumi Y, Tanabe S, Terada M, Saijo N, Nishio K. Gene 2000. 2000;242:167–173. doi: 10.1016/s0378-1119(99)00529-6. [DOI] [PubMed] [Google Scholar]
- 38.Borst P, Evers R, Kool M, Wijnholds J. Biochim Biophys Acta. 1999;1461:347–357. doi: 10.1016/s0005-2736(99)00167-4. [DOI] [PubMed] [Google Scholar]
- 39.Cole S P. J Natl Cancer Inst. 1999;91:888–889. doi: 10.1093/jnci/91.10.888. [DOI] [PubMed] [Google Scholar]
- 40.Paulusma C C, van Geer M A, Evers R, Heijn M, Ottenhoff R, Borst P, Oude Elferink R P. Biochem J. 1999;338:393–401. [PMC free article] [PubMed] [Google Scholar]
- 41.Hooijberg J H, Broxterman H J, Kool M, Assaraf Y G, Peters G J, Noordhuis P, Scheper R J, Borst P, Pinedo H M, Jansen G. Cancer Res. 1999;59:2532–2535. [PubMed] [Google Scholar]
- 42.Palu G, Stefanelli S, Rassu M, Parolin C, Balzarini J, De Clercq E. Antiviral Res. 1991;16:115–119. doi: 10.1016/0166-3542(91)90063-w. [DOI] [PubMed] [Google Scholar]
- 43.Srinivas R V, Robbins B L, Connelly M C, Gong Y F, Bischofberger N, Fridland A. Antimicrob Agents Chemother. 1993;37:2247–2250. doi: 10.1128/aac.37.10.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pisarev V M, Lee S H, Connelly M C, Fridland A. Mol Pharmacol. 1997;52:63–68. doi: 10.1124/mol.52.1.63. [DOI] [PubMed] [Google Scholar]
- 45.Hipfner D R, Deeley R G, Cole S P. Biochim Biophys Acta. 1999;1461:359–376. doi: 10.1016/s0005-2736(99)00168-6. [DOI] [PubMed] [Google Scholar]
- 46.Konig J, Nies A T, Cui Y, Leier I, Keppler D. Biochim Biophys Acta. 1999;1461:377–394. doi: 10.1016/s0005-2736(99)00169-8. [DOI] [PubMed] [Google Scholar]
- 47.Hirohashi T, Suzuki H, Sugiyama Y. J Biol Chem. 1999;274:15181–15185. doi: 10.1074/jbc.274.21.15181. [DOI] [PubMed] [Google Scholar]
- 48.Hirohashi T, Suzuki H, Takikawa H, Sugiyama Y. J Biol Chem. 2000;275:2905–2910. doi: 10.1074/jbc.275.4.2905. [DOI] [PubMed] [Google Scholar]
- 49.Robbins B L, Connelly M C, Marshall D R, Srinivas R V, Fridland A. Mol Pharmacol. 1995;47:391–397. [PubMed] [Google Scholar]
- 50.Lorico A, Rappa G, Finch R A, Yang D, Flavell R A, Sartorelli A C. Cancer Res. 1997;57:5238–5242. [PubMed] [Google Scholar]
- 51.Rao V V, Dahlheimer J L, Bardgett M E, Snyder A Z, Finch R A, Sartorelli A C, Piwnica-Worms D. Proc Natl Acad Sci USA. 1999;96:3900–3905. doi: 10.1073/pnas.96.7.3900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Baskin F, Rosenberg R N. J Pharmacol Exp Ther. 1975;193:293–300. [PubMed] [Google Scholar]
- 53.Bemi V, Turchi G, Margotti E, Giorgelli F, Pesi R, Sgarrella F, Tozzi M G, Camici M. Int J Cancer. 1999;82:556–561. doi: 10.1002/(sici)1097-0215(19990812)82:4<556::aid-ijc14>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 54.Balzarini J. Biochem Pharmacol. 1999;58:1–27. doi: 10.1016/s0006-2952(99)00029-5. [DOI] [PubMed] [Google Scholar]
- 55.Finzi D, Silliciano R F. Cell. 1998;93:665–671. doi: 10.1016/s0092-8674(00)81427-0. [DOI] [PubMed] [Google Scholar]
- 56.Shen D W, Akiyama S, Schoenlein P, Pastan I, Gottesman M M. Br J Cancer. 1995;71:676–683. doi: 10.1038/bjc.1995.134. [DOI] [PMC free article] [PubMed] [Google Scholar]