Abstract
The high-mobility-group (HMG) SSRP1 protein is a member of a conserved chromatin-remodeling complex (FACT/DUF/CP) implicated in DNA replication, basal and regulated transcription, and DNA repair. To assist in the functional analysis of SSRP1, the Ssrp1 gene was targeted in murine embryonic stem cells, and the mutation was introduced into the germ line. Embryos homozygous for the targeted allele die soon after implantation, and preimplantation blastocysts are defective for cell outgrowth and/or survival in vitro. The Ssrp1 mutation was also crossed into a p53 null background without affecting growth and/or survival defects caused by loss of Ssrp1 function. Thus, Ssrp1 appears to encode nonredundant and p53-independent functions that are essential for cell viability.
In eukaryotic cells, DNA replication, transcription, and repair involve templates packaged into chromatin, a highly organized but structurally heterogeneous nucleic acid-protein complex. Chromatin typically suppresses enzymatic reactions involving DNA; therefore, mechanisms to remodel and maintain chromatin structures play essential roles in regulating DNA function. This is well illustrated by the functions of chromatin-remodeling complexes (e.g., SWI/SNF and ACF) and histone-modifying enzymes (e.g., histone acetyltransferases) in the regulation of gene transcription (1).
Another conserved and highly abundant (∼1,000,000 copies per nucleus) chromatin-remodeling complex contains orthologs of the yeast CDC68/SPT16 and POB3 proteins (6, 34, 35), designated FACT (human), DUF (Xenopus), and CP (Saccharomyces cerevisiae), depending on the source. Genetic and biochemical studies have implicated this complex in DNA replication (34, 46, 47), basal and regulated transcription (6, 14, 22, 35, 44), and DNA repair (24, 25, 48). For example, the mammalian FACT (for facilitates chromatin transcription) complex was initially characterized as an activity capable of suppressing the inhibitory effects of nucleosomes on transcriptional elongation in vitro (35). This activity involves the release of histones H2A and H2B from chromatin, apparently through stoichiometric interactions with nucleosomes rather than from a processive enzymatic activity. Both components of the yeast CP complex, SPT16/CDC68 and POB3, are essential for cell viability. However, alleles of SPT16/CDC68 have emerged from a variety of genetic screens and display widespread effects on transcription that are reminiscent of mutations in histones H2A and H2B (30, 31, 37). The Xenopus complex was purified as a duplex DNA-unwinding factor (DUF) that simulates DNA replication in oocyte extracts (34). Finally, FACT has also been reported to influence casein kinase 2-dependent phosphorylation and the activity of the p53 tumor suppressor (25). In this context, the complex has been postulated to activate p53 as part of a transcription- and/or replication-coupled mechanism for recognizing DNA damage.
SSRP1/T160 (Mouse Genome Database [4] accession number 107912), the POB3 counterpart in the mammalian FACT complex, is a member of the high mobility group (HMG) of chromatin-associated proteins (2, 10). While similar to SSRP1, POB3 lacks an HMG box domain and relies instead on an HMG box protein, Nhp6a or Nhp6b, to recruit the yeast CP complex to chromatin (5, 15). Thus, two proteins apparently function as orthologs of SSRP1 in yeast: POB3, which lacks an HMG box, and the genetically redundant Nhp6a/Nhp6b proteins that consist of little more than HMG boxes.
SSRP1/T160 was initially identified in screens for proteins that interact with immunoglobulin V(D)J recombination sites and cisplatin DNA adducts (8, 38). However, these binding activities are shared by other HMG proteins and could reflect the general ability of HMG proteins to bind distorted DNA conformations (18, 41). Subsequent studies have implicated SSRP1 in transcription control, for example, as a sequence-specific transcription factor of the embryonic ɛ globin gene (13) and as a coactivator working in concert with serum response factor (39) and the p53-related p63 protein (49). The Drosophila SSRP1 protein also localizes to transcriptionally active chromatin on polytene chromosomes (26).
Whether SSRP1 can function independently of the FACT complex is not known; however, there is growing evidence that SSRP1 and/or FACT interacts with a larger network of proteins. Proteins reported to interact with SSRP1 in mammalian cells include the PU.1 and serum response factor transcription factors (33, 39) and CHD1, a chromodomain-, ATPase-helicase-containing protein implicated in chromatin remodeling (26). Finally, components of the yeast CP complex have been reported to interact with three distinct complexes: (i) histones, (ii) Chd1 and casein kinase 2, and (iii) Rtf1, Paf1, Ctr9, Cdc73, and a previously uncharacterized protein, Leo1 (27).
In summary, SSRP1 has been implicated in transcriptional initiation and elongation and in DNA replication and repair—apparent manifestations of its ability to influence chromatin structure. However, efforts to understand the biochemical functions of mammalian SSRP1 are complicated by the fact that by altering DNA and chromatin structure, this highly abundant protein may have pleiotropic effects that are not directly related to its function.
To assist in the functional analysis of SSRP1, the Ssrp1 gene was disrupted in embryonic stem (ES) cells, and the targeted mutation was introduced into the murine germ line. Mice homozygous for the mutation die soon after implantation, and preimplantation blastocysts are defective for cell outgrowth and/or survival in vitro. Thus, like yeast POB3, Ssrp1 appears to encode essential and nonredundant functions necessary for cell viability.
MATERIALS AND METHODS
Targeted disruption of Ssrp1.
A phage clone containing the complete Ssrp1 gene was isolated from a mouse 129/Sv genomic library and subcloned into the NotI site of pBlueScript II KS(−) (Strategene). A targeting vector was constructed that consisted of a phosphoglycerol kinase (PGK)-Neo cassette flanked by sequences homologous to the Ssrp1 gene: an 8.9-kb NotI-KpnI fragment and a 1.3-kb XhoI-BglII fragment. After homologous recombination, the vector replaces 3.9 kb of genomic sequences (located between KpnI and XhoI sites) that contain exons 14 to 17 with the PGK-Neo cassette. The vector also contained a PGK-thymidine kinase (TK) gene to select against nonhomologous inserts. Twenty micrograms of vector DNA was linearized with XhoI and electroporated into 2 × 107 J1 ES cells. ES cell colonies were cultured on irradiated mouse embryonic fibroblast (MEF) cells in the presence of 0.4 mg of G418/ml and 1 mM gancyclovir. Correctly targeted clones were identified by Southern blot hybridization and used for microinjection into C57BL/6 blastocysts.
Genotype analysis.
The Ssrp1 genotypes of ES cells and mice were assessed by Southern blot hybridization to a [32P]dCTP-labeled, 0.7-kb BglII/BamHI probe derived from genomic sequences located downstream of the 3′ homologous region in the targeting vector, as described previously (45).
The Ssrp1 genotype of embryonic day 6.5 (E6.5) to E8.5 embryos was assessed by PCR, using a mixture of three primers: a (5′CCGGCCCAGTAGGTATTTTC), b (5′CAGACTGCCTTGGGAAAAGC), and c (5′TCCCTCCAAGGAGCTATGTG). Each 50-μl reaction mixture contained 10 mM Tris-HCl (pH 8.3), 5 mM KCl, 1.5 mM MgCl2, 200 μM (each) deoxyribonucleoside triphosphate, each primer at 2 μM, and 2.5 U of Amplitaq (Roche). Reactions involved 30 cycles of denaturation (94°C; 1 min), primer annealing (59°C; 1 min), and primer extension (72°C; 2 min).
The Ssrp1 genotypes of E3.5 blastocysts and blastocyst-derived colonies were assessed by nested PCR. The first reaction used a mixture of three primers: 1 (5′AGGCTGGCTGTGACTTAGTG), 2 (5′ACTTGTGTAGCGCCAAGTG), and 3 (5′CATCCGTGAGGGCTTACT), as described above but for 20 cycles of denaturation (94°C; 1 min), primer annealing (55°C; 1 min), and primer extension (72°C;2 min). A second round of PCR used primers a, b, and c for 30 cycles as described above.
p53 genotypes were assessed by PCR. The wild-type allele was detected with a pair of primers: X7 (5′TATACTCAGAGCCGGCCT) and X6.5 (5′ACAGCGTGGTGGTACCTTAT). The null allele (28) was detected using primers X7 and Neo19 (5′CTATCAGGACATAGCGTTGG). Reactions involved 30 cycles of denaturation (94°C; 1 min), primer annealing (55°C; 1 min), and primer extension (72°C; 2 min) as described above.
Analysis of Ssrp1 mutant cells and embryos.
The expression of SSRP1 protein in ES cell clones was assessed by Western blot analysis. Cell lysates were fractioned by electrophoresis on a sodium dodecyl sulfate-8% polyacrylamide gel and transferred onto a polyvinylidene difluoride membrane (NEN Life Science), and SSRP1 proteins were detected by using rabbit polyclonal antibodies raised against the amino terminus of the protein, as described elsewhere (20).
E3.5 blastocysts were isolated and used to derive ES cell lines as described earlier (45). For studies of blastocyst outgrowth, E3.5 embryos were cultured, either with or without irradiated MEF feeder layers, in ES cell medium (Dulbecco modified Eagle medium [Mediatech] supplemented with 15% fetal bovine serum [heat inactivated at 55°C for 30 min], 0.1 mM 2-mercaptoethanol, 100 mM nonessential amino acids, and 100 U of penicillin-streptomycin [Gibco BRL]/ml).
The morphologies of postimplantation embryos were assessed from serial sections (7 μm thick) of paraffin-embedded deciduas stained with hematoxylin and eosin (36). Apoptosis in preimplantation embryos was detected with an in situ cell death detection kit (Roche). Blastocysts were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 15 min and then permeablized for 30 min with 0.3% Triton X-100 and 1.5% bovine serum albumin in PBS. Blastocysts treated with 0.5 mg of DNase I/ml for 10 min provided positive controls for DNA fragmentation. Embryos were incubated with the terminal deoxynucleotidyl transferase biotin-dUTP nick end labeling (TUNEL) reaction mixture for 1 h at 37°C, and cell nuclei were stained with DAPI (1 mg/ml in PBS) for 10 min. The embryos were viewed and photographed using a fluorescence microscope and then genotyped by nested PCR.
RESULTS
Targeted disruption of Ssrp1.
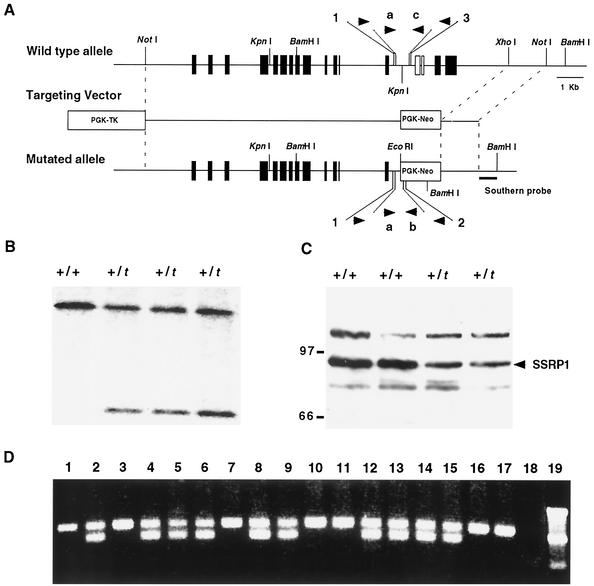
To assess the function of Ssrp1 in cell development and growth, a targeted mutation was generated via homologous recombination in mouse ES cells. The targeting vector replaced Ssrp1 sequences containing exons 14 to 17 (located in a 3.9-kb KpnI-XhoI fragment) with a PGK-Neo cassette (Fig. 1A). The mutant allele (designated Ssrp1t) thus deletes the HMG box domain (exons 14 and 15) along with 87 amino acids from the C terminus. Targeted ES cells were identified by Southern blot hybridization to a flanking sequence probe (Fig. 1B).
FIG. 1.
Targeted disruption of the Ssrp1 gene. (A) Schematic diagram of the genomic organization of the Ssrp1 gene, the targeting vector, and the mutated allele after homologous recombination. The locations of sequences used as probes for Southern blot hybridization and PCR primers (1, 2, 3, a, b, and c) are indicated. The targeting vector contained a neomycin resistance gene (Neo) expressed from the PGK gene promoter for positive selection. The Neo gene replaced sequences of the Ssrp1 gene located between the KpnI and XhoI sites. The targeting vector also contained a herpes simplex virus TK gene for negative selection. (B) Southern blot analysis showing correct targeting of the Ssrp1 gene. Genomic DNAs from ES cell clones isolated following positive-negative selection were digested with BamHI and analyzed by Southern blot hybridization using the 3′ flanking probe shown in Fig. 1A. The 9.5-kb band represents the wild type (+) allele, and the 2.5-kb band corresponds to the correctly targeted allele (t). (C) Western blot analysis of SSRP1 expression in wild-type and heterozygous targeted ES cells. Whole-cell extracts from Ssrp1+/t and Ssrp1+/t ES cells were analyzed with antisera against the amino-terminal region of SSRP1. The native, 86-kDa SSRP1 protein was detected (arrow) in both cell types, while no truncated proteins were detected in Ssrp1+/t cells. The mobilities of the 97- and 66-kDa molecular mass standards are shown. Comparable amounts of protein were loaded for all samples based on Coomassie staining (not shown). (D) PCR genotyping of wild-type and heterozygous mutant mice. Primers a, b, and c (Fig. 1A) were mixed and used to amplify sequences corresponding to the wild-type (374-nucleotide) and targeted (250-nucleotide) alleles. Tail DNA from 3-week-old offspring produced by crossing Ssrp1+/t heterozygotes (lanes 1 to 17) and a sample lacking DNA (lane 18) were amplified by PCR and fractionated on a 1.5% agarose gel, together with molecular weight markers (lane 19). Seven of the animals analyzed were wild type, and 10 were heterozygous, while none was homozygous for the targeted allele.
Since SSRP1 is expressed in many cell types, including ES cells, the effects of the targeted mutation on SSRP1 expression could be assessed by Western blot analysis. As shown in Fig. 1C, clones containing the targeted allele expressed approximately half as much SSRP1 as wild-type cells. Moreover, truncated proteins potentially encoded by the targeted allele were not detected.
Four Ssrp1+/t ES clones independently injected into C57BL/6 blastocysts gave rise to germ line chimeras. Agouti offspring inheriting the targeted allele were identified by either Southern blot or PCR analysis (data not shown), and the mutation was bred into a 129sv or C57BL/6 background for three generations before being intercrossed.
Ssrp1 is required for early embryonic development.
Ssrp1+/t mice were intercrossed in an attempt to generate homozygous mutant mice. However, no Ssrp1t/t mice were detected among 220 offspring analyzed (Fig. 1D and Table 1). To determine the stage at which the homozygous mutant mice die, embryos from timed matings were genotyped after different times of gestation (Table 1). Again, none of the embryos that could be dissected from decidua at E6.5 to E8.5 was homozygous for the targeted allele. However, embryos in 16 out of 60 decidua examined were nearly or completely resorbed and could not be genotyped. This proportion (26.7%) was sufficient to account for the absence of homozygous mutants. Embryos were also examined at E5.5 in serial sections cut through intact decidua. Once again, the presumptive homozygous mutant embryos (4 out of 16) were almost completely resorbed (Fig. 2).
TABLE 1.
Genotypes of offspring and embryos produced by Ssrp1+/t intercrossesa
| Age | No. of mice
|
|||
|---|---|---|---|---|
| +/+ | +/t | t/t | Total | |
| 3 wk | 75 | 145 | 0 | 220 |
| E7.5-8.5 | 13 | 31 | 0 | 44b |
| E3.5 | 32 | 54 | 29 | 115 |
Mice heterozygous for the targeted mutation in the Ssrp1 gene were mated, and embryos and progeny of the indicated ages were genotyped.
This total does not include 16 empty deciduas that could not be genotyped.
FIG. 2.
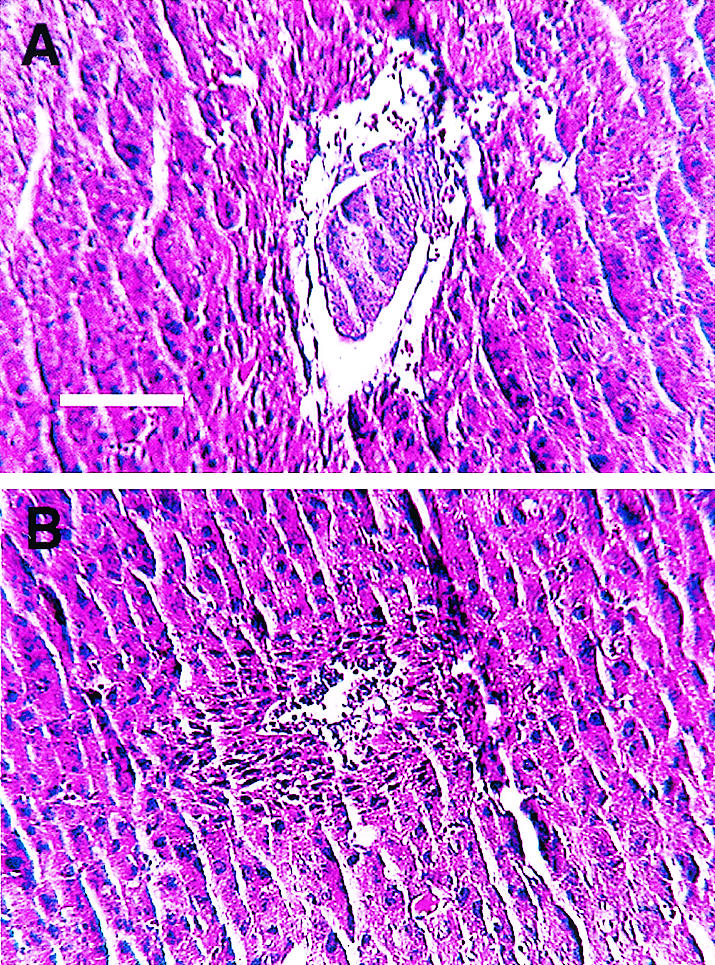
Histological sections of E5.5 embryos. Decidua containing E5.5 embryos from Ssrp1+/t intercrosses were sectioned and stained with hematoxylin and eosin. Representative normal (A) and presumptive homozygous (B) mutant embryos are shown. The panels are reproduced at the same magnification (bar, 100 μm).
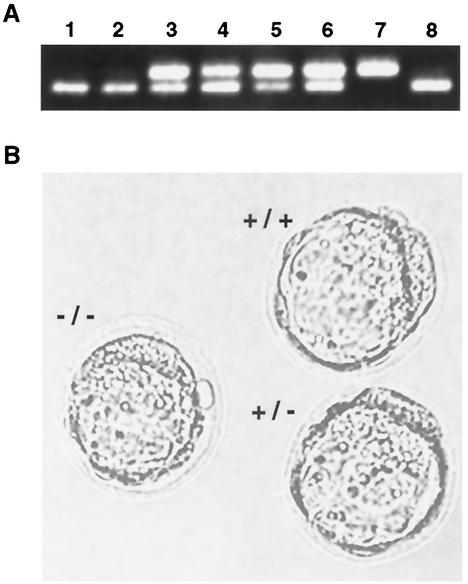
The observed decidual reactions suggested that the mutant embryos die after implantation. To test this idea further, 115 preimplantation, blastocyst stage embryos (E3.5) were genotyped (Fig. 3A) by a nested-PCR strategy (Fig. 1A). Of these, 29 were homozygous for the mutant allele, 32 were wild type, and 54 were heterozygous for the Ssrp1t mutation—consistent with a Mendelian distribution (Table 1). The mutant blastocysts appeared morphologically normal (Fig. 3B). Taken together, these results suggest that Ssrp1t/t embryos die between implantation and E5.5.
FIG. 3.
PCR genotyping of E3.5 blastocysts. (A) Blastocysts from Ssrp1 heterozygous intercrosses were analyzed by nested PCR, using primers 1, 2, and 3 and a, b, and c (Fig. 1A) in the first and second rounds of amplification, respectively. (B) Phase-contrast photomicrographs of E3.5 blastocysts of the indicated genotypes. Ssrp1t/t blastocysts appear morphologically normal.
Defective outgrowth of ICM cells from Ssrp1t/t blastocysts.
ES cell lines are readily derived from the inner cell mass (ICM) of E3.5 blastocysts if the cells are maintained under conditions that suppress their differentiation. Cell lines homozygous for early embryonic lethal mutations can also be derived when the affected genes are not required for the growth or viability of ICM cells. For example, we have isolated ES cells deficient in hnRNP C or the protein arginine methyltransferase 1, even though the mutant embryos die at or before E6.5 (36, 45). Ssrp1-deficient ES cells could provide a valuable system to study the biochemical functions of SSRP1. However, when blastocysts from Ssrp1+/t intercrosses were cultured in vitro, none of the 12 resulting ES cell lines was homozygous for the targeted allele (data not shown). By contrast, eight lines were heterozygous for the mutant allele and four were wild type. These numbers, while relatively small, are consistent with a Mendelian distribution, assuming homozygous mutant cells are selectively lost during cultivation. The probability (chi square) of recovering no homozygous mutants out of 12 cell lines by chance alone is 0.08.
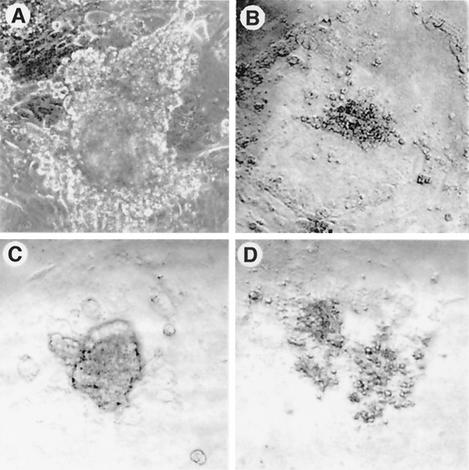
The failure to recover homozygous mutant ES cell lines raised questions about whether SSRP1 is required for the growth or viability of ICM cells. To address this issue, 32 blastocysts from Ssrp1+/t intercrosses were individually cultured on MEF feeder cells and examined daily. During the first 24 h, all of the embryos attached and hatched from the zona pellucida. After 4 days of culture, the ICMs from 23 blastocysts formed well-delineated colonies on top of the more adherent trophoblast giant cells (Fig. 4A). ICM cells from the remaining nine blastocysts, after limited proliferation, appeared to degenerate and/or detach from the dish (Fig. 4B). Cells from the 23 ICM-derived colonies were collected into drawn glass capillaries and were analyzed by PCR to determine their genotypes. As summarized in Table 2, eight of the colonies were wild type, 15 were heterozygous for the targeted allele, and none was homozygous for the mutation.
FIG. 4.
Defective outgrowth of Ssrp1 homologous mutant blastocysts. E3.5 blastocysts from Ssrp1 heterozygous intercrosses were cultured in vitro for 4 days, both with (A and B) and without (C and D) MEF feeder layers. All embryos attached within 24 h and hatched from their zonae pellucidae. The ICM of Ssrp1+/t embryos produced adherent colonies of proliferating cells (A and C), whereas ICM cells from Ssrp1t/t embryos died and/or detached from the culture vessel (B and D).
TABLE 2.
Genotypes of blastocyst-derived colonies cultured with or without MEF feeder layersa
| Type | No. of mice
|
||||
|---|---|---|---|---|---|
| +/+ | +/t | t/t | Unknown | Total | |
| With MEFs | |||||
| Normal ICM colonies | 8 | 15 | 0 | 0 | 23 |
| Aberrant ICM colonies | 0 | 0 | 0 | 9b | 0 |
| Without MEFs | |||||
| Normal ICM colonies | 13 | 26 | 0 | 0 | 39 |
| Aberrant ICM colonies | 0 | 0 | 8 | 3 | 11 |
E3.5 embryos produced by mating mice heterozygous for the targeted mutation in the Ssrp1 gene were cultured with and without MEF feeder layers and were genotyped by nested PCR after 4 days.
These colonies had degenerated and could not be genotyped.
Fifty additional blastocysts were cultured without MEFs to eliminate the potential for contamination by wild-type feeder cells that might interfere with genotyping. After 3 to 4 days of culture, the ICMs from 39 blastocysts formed colonies, although these were smaller than those produced on MEF feeder cells (Fig. 4C). ICM cells from the remaining 11 blastocysts degenerated and/or detached from the culture vessel instead of forming distinct colonies (Fig. 4D). Three of the defective outgrowths could not be genotyped, but the other eight were confirmed as Ssrp1t/t. The genotypes of the 39 normal blastocyst-derived colonies included 13 wild type, 26 heterozygotes, and no homozygous mutants (Table 2). Taken together, these results indicate that the murine Ssrp1 gene is necessary for the growth and/or survival of the ICM.
Apoptotic cells in Ssrp1-deficient blastocysts.
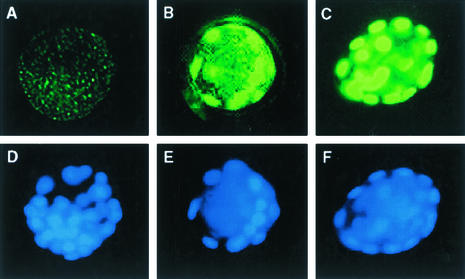
The SSRP1 protein has been reported to bind V(D)J recombination sequences and cisplatin adducts, suggesting a possible role in DNA repair and/or recombination (31, 38). DNA repair defects associated with the Ssrp1 mutation could result in cell death by apoptosis and contribute to the rapidity with which Ssrp1-deficient cells lose viability both in vivo and in vitro. While Ssrp1-deficient blastocysts appear normal, freshly isolated E3.5 embryos were analyzed for the presence of apoptotic cells by TUNEL. Seventy-three blastocysts, of which 24 were wild type and 20 were homozygous for the Ssrp1 mutation, were stained, photographed, and genotyped, and the apoptotic cells were counted blind from coded photographs. Wild-type, heterozygous, and mutant embryos averaged 1.2 ± 1.2, 1.7 ± 1.7, and 2.6 ± 3.4 TUNEL-positive cells per blastocyst, respectively (Fig. 5). Although statistically significant (P < 0.04), the difference between wild-type and mutant embryos was relatively small; thus, apoptosis in E3.5 embryos does not appear to account for the survival defects observed in Ssrp1-deficient blastocysts.
FIG. 5.
TUNEL staining of wild-type and Ssrp1-deficient blastocysts. E3.5 blastocysts from Ssrp1+/t intercrosses were fixed, permeabilized, and then stained with TUNEL mixture (A to C) and with DAPI (D to F). The blastocysts were genotyped as Ssrp1+/+ (A and D) and Ssrp1t/t (B and E). A DNase I-treated Ssrp1+/+ blastocyst (C and F) provided a positive a control for DNA fragmentation.
p53 deficiency does not rescue embryonic lethality caused by the Ssrp1t mutation.
As an important regulator of the cellular responses to DNA damage, the p53 tumor suppressor could interact with SSRP1. Several genes involved in the cellular response to DNA damage, including BRCA1, Lig4, and XRCC4, are required for embryonic development. Death of BRCA1, Lig4, and XRCC4 mutant embryos occurs at or before E7.5, E17.5, and E16.5, respectively, and requires active participation by the p53 tumor suppressor, which promotes apoptosis in DNA repair-deficient cells (16, 17, 29). As a consequence, inactivating mutations in p53 delay or prevent embryonic lethal phenotypes associated with these DNA repair deficiencies.
To assess the role of p53 in the death of Ssrp1-deficent embryos, mice with mutations in Ssrp1 and p53 were intercrossed. Since the Ssrp1 and p53 genes are unlinked, 1 in 16 offspring from mice doubly heterozygous for inactivating Ssrp1 and p53 mutations is expected to be homozygous for both mutations if p53 deficiency rescues Ssrp1t/t embryos from embryonic death. However, none of the 106 pups analyzed, including 21 p53−/− offspring, was homozygous for the Ssrp1 mutation. We also analyzed 65 embryos from E7.5 to E12.5, but none was homozygous for the targeted allele. Finally, p53 status had no discernible effect on the outgrowth of Ssrp1t/t blastocysts. Specifically, of 25 blastocysts produced by intercrossing Ssrpt/t heterozygotes in a p53-null background, 8 Ssrp+/+ and 12 Ssrp+/t blastocysts gave rise to normal ICM-derived colonies. The remaining five blastocyts produced defective colonies, including three that were Ssrpt/t and two that could not be genotyped. These results indicate that p53 status does not influence the growth and/or survival defects of Ssrp1 mutant embryos, either in vivo or ex vivo.
DISCUSSION
The present study shows that murine Ssrp1 is essential for the growth and survival of early embryonic cells both in vivo and ex vivo. Thus, while a number of HMG chromatin-associated proteins are encoded by the mouse genome (2), none of these HMG box family members appears to compensate for the loss of Ssrp1 function. Ssrp1 is required for the survival of postimplantation blastocysts and for the viability of cells of the ICM—phenotypes associated with genes essential for cell viability (12, 19, 32). These results are consistent with studies linking Ssrp1 expression and cell proliferation (20) and with antisense ablation experiments implicating Ssrp1 in the proliferation of murine fibroblasts (21). However, we cannot exclude the possibility that the gene is dispensable in some cell types (9).
The targeted mutation deleted 87 amino acids from the carboxyl terminus of SSRP1, including the HMG box. Since the HMG box is required for DNA binding, the mutation is expected to inactivate SSRP1 function. The mutation was inherited as a simple, recessive embryonic lethal trait; however, due to the severity of the growth and/or survival defects exhibited by Ssrp1-deficient embryos, we were unable to determine if the targeted mutation is a null allele. ES cells heterozygous for the mutation expressed approximately half as much SSRP1 protein as wild-type cells and did not appear to express truncated forms of the protein. Moreover, heterozygous mice displayed no phenotypes that might result from the trans-dominant activity of a truncated protein.
While SSRP1 shares features in common with other HMG box proteins, it also has several unique features that distinguish it from other HMG family members. SSRP1 is most similar to HMG-1 within the HMG domain, but unlike HMG-1, SSRP1 is capable of binding DNA in a sequence-specific manner (13). SSRP1 also contains only a single HMG domain, like the sequence-specific HMG box proteins SRY and LEF-1/TCF-1a. Because of these and other features, SSRP1 was classified into a distinct subfamily of HMG domain proteins (2). Since targeted mutations in other HMG family members have thus far resulted in far more restricted phenotypes (3, 7, 11, 23, 40, 42, 43), the present study provides further evidence that Ssrp1 encodes unique, nonredundant functions.
As a component of the phylogenetically conserved FACT/DUF/SPT16-POB3 complex, SSRP1 appears to assist in chromatin remodeling during transcription initiation and elongation and DNA replication (22, 34, 35, 44, 46, 47). In S. cerevisiae, orthologous functions of SSRP1 appear to be supplied by the combined actions of POB3, which is similar to SSRP1 but lacks the HMG box domain, and Nhp6a/Nhp6b, which are genetically redundant proteins consisting of little more than HMG boxes (5, 15). The phenotype of the Ssrp1t mutation in murine embryos provides additional evidence for phylogenetic conservation between SSRP1 and POB3, as POB3 is required for cell viability in S. cerevisiae.
Several observations suggest that SSRP1 may play a role in DNA repair and recombination. SSRP1 binds cisplatin DNA adducts and V(D)J recombination sites (31, 38), and SSRP1 has been reported to enhance the activities of the p53 tumor suppressor (24, 25) and the p53-related protein p63 (49). As a component of the FACT complex, SSRP1 would be positioned to activate p53-dependent responses to DNA damage at the sites of transcription or DNA replication. However, phenotypes caused by deficiencies in several DNA repair genes (e.g., BRCA1, Lig4, and XRCC4) are less severe than those caused by loss of Ssrp1, and phenotypes resulting from these DNA repair defects, unlike Ssrp1 deficiency, are reduced in severity in the absence of p53 (16, 17, 29). Thus, the phenotype of the Ssrp1t mutation, and its apparent lack of genetic interaction with p53, suggest that SSRP1 does not function primarily in DNA repair and recombination.
A protein complex containing SSRP1 and SPT16 (FACT) has been reported to influence CK2-dependent phosphorylation and activity of p53 (25). Since FACT enhances transcription elongation on chromatin templates in vitro, the complex has been postulated to activate p53 as part of a transcription-dependent mechanism for recognizing DNA damage. However, since embryos tolerate the loss of p53 and not SSRP1, SSRP1 apparently does not function primarily as an upstream activator of p53.
In summary, while genetic inferences are necessarily indirect, Ssrp1 appears to encode nonredundant functions that play essential roles in cellular metabolism. As direct functional studies are currently hampered by the lack of Ssrp1-deficient cells, future studies will benefit from the development of conditional systems to regulate Ssrp1 function in mammalian cells.
Acknowledgments
We thank Fred Alt (Harvard University), in whose laboratory the initial work on this project was conducted, and Jin Chen for assistance with fluorescence microscopy.
This work was supported by Public Health Service Grants to E.M.O. and H.E.R. (P01HL68744) and H.S. (R37AI18790). Additional support was provided by a Cancer Center Support grant (P30CA68485) for the Vanderbilt-Ingram Cancer Center.
REFERENCES
- 1.Aalfs, J. D., and R. E. Kingston. 2000. What does ‘chromatin remodeling’ mean? Trends Biochem. Sci. 25:548-555. [DOI] [PubMed] [Google Scholar]
- 2.Baxevanis, A. D., S. H. Bryant, and D. Landsman. 1995. Homology model building of the HMG-1 box structural domain. Nucleic Acids Res. 23:1019-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bianchi, M. E., and M. Beltrame. 2000. Upwardly mobile proteins. Workshop: the role of HMG proteins in chromatin structure, gene expression and neoplasia. EMBO Rep. 1:109-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blake, J. A., J. E. Richardson, C. J. Bult, J. A. Kadin, and J. T. Eppig. 2002. The Mouse Genome Database (MGD): the model organism database for the laboratory mouse. Nucleic Acids Res. 30:113-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brewster, N. K., G. C. Johnston, and R. A. Singer. 2001. A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol. Cell. Biol. 21:3491-3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brewster, N. K., G. C. Johnston, and R. A. Singer. 1998. Characterization of the CP complex, an abundant dimer of Cdc68 and Pob3 proteins that regulates yeast transcriptional activation and chromatin repression. J. Biol. Chem. 273:21972-21979. [DOI] [PubMed] [Google Scholar]
- 7.Britsch, S., D. E. Goerich, D. Riethmacher, R. I. Peirano, M. Rossner, K. A. Nave, C. Birchmeier, and M. Wegner. 2001. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev. 15:66-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruhn, S. L., P. M. Pil, J. M. Essigmann, D. E. Housman, and S. J. Lippard. 1992. Isolation and characterization of human cDNA clones encoding a high mobility group box protein that recognizes structural distortions to DNA caused by binding of the anticancer agent cisplatin. Proc. Natl. Acad. Sci. USA 89:2307-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bultman, S., T. Gebuhr, D. Yee, C. La Mantia, J. Nicholson, A. Gilliam, F. Randazzo, D. Metzger, P. Chambon, G. Crabtree, and T. Magnuson. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6:1287-1295. [DOI] [PubMed] [Google Scholar]
- 10.Bustin, M. 1999. Regulation of DNA-dependent activities by the functional motifs of the high-mobility-group chromosomal proteins. Mol. Cell. Biol. 19:5237-5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calogero, S., F. Grassi, A. Aguzzi, T. Voigtlander, P. Ferrier, S. Ferrari, and M. E. Bianchi. 1999. The lack of chromosomal protein Hmg1 does not disrupt cell growth but causes lethal hypoglycaemia in newborn mice. Nat. Genet. 22:276-280. [DOI] [PubMed] [Google Scholar]
- 12.Carlone, D. L., and D. G. Skalnik. 2001. CpG binding protein is crucial for early embryonic development. Mol. Cell. Biol. 21:7601-7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dyer, M. A., P. J. Hayes, and M. H. Baron. 1998. The HMG domain protein SSRP1/PREIIBF is involved in activation of the human embryonic beta-like globin gene. Mol. Cell. Biol. 18:2617-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Evans, D. R., N. K. Brewster, Q. Xu, A. Rowley, B. A. Altheim, G. C. Johnston, and R. A. Singer. 1998. The yeast protein complex containing cdc68 and pob3 mediates core-promoter repression through the cdc68 N-terminal domain. Genetics 150:1393-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Formosa, T., P. Eriksson, J. Wittmeyer, J. Ginn, Y. Yu, and D. J. Stillman. 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20:3506-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frank, K. M., N. E. Sharpless, Y. Gao, J. M. Sekiguchi, D. O. Ferguson, C. Zhu, J. P. Manis, J. Horner, R. A. DePinho, and F. W. Alt. 2000. DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol. Cell 5:993-1002. [DOI] [PubMed] [Google Scholar]
- 17.Gao, Y., D. O. Ferguson, W. Xie, J. P. Manis, J. Sekiguchi, K. M. Frank, J. Chaudhuri, J. Horner, R. A. DePinho, and F. W. Alt. 2000. Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature 404:897-900. [DOI] [PubMed] [Google Scholar]
- 18.Gariglio, M., G. G. Ying, L. Hertel, M. Gaboli, R. G. Clerc, and S. Landolfo. 1997. The high-mobility group protein T160 binds to both linear and cruciform DNA and mediates DNA bending as determined by ring closure. Exp. Cell Res. 236:472-481. [DOI] [PubMed] [Google Scholar]
- 19.Herceg, Z., W. Hulla, D. Gell, C. Cuenin, M. Lleonart, S. Jackson, and Z. Q. Wang. 2001. Disruption of Trrap causes early embryonic lethality and defects in cell cycle progression. Nat. Genet. 29:206-211. [DOI] [PubMed] [Google Scholar]
- 20.Hertel, L., M. De Andrea, G. Bellomo, P. Santoro, S. Landolfo, and M. Gariglio. 1999. The HMG protein T160 colocalizes with DNA replication foci and is down-regulated during cell differentiation. Exp. Cell Res. 250:313-328. [DOI] [PubMed] [Google Scholar]
- 21.Hertel, L., P. Foresta, G. Barbiero, G. G. Ying, G. Bonelli, F. M. Baccino, S. Landolfo, and M. Gariglio. 1997. Decreased expression of the high-mobility group protein T160 by antisense RNA impairs the growth of mouse fibroblasts. Biochimie 79:717-723. [DOI] [PubMed] [Google Scholar]
- 22.John, S., L. Howe, S. T. Tafrov, P. A. Grant, R. Sternglanz, and J. L. Workman. 2000. The something about silencing protein, Sas3, is the catalytic subunit of NuA3, a yTAF(II)30-containing HAT complex that interacts with the Spt16 subunit of the yeast CP (Cdc68/Pob3)-FACT complex. Genes Dev. 14:1196-1208. [PMC free article] [PubMed] [Google Scholar]
- 23.Kanai-Azuma, M., Y. Kanai, J. M. Gad, Y. Tajima, C. Taya, M. Kurohmaru, Y. Sanai, H. Yonekawa, K. Yazaki, P. P. Tam, and Y. Hayashi. 2002. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development 129:2367-2379. [DOI] [PubMed] [Google Scholar]
- 24.Keller, D. M., and H. Lu. 2002. p53 serine 392 phosphorylation increases after UV through induction of the assembly of the CK2-hSPT16-SSRP1 complex. J. Biol. Chem. 277:50206-50213. [DOI] [PubMed] [Google Scholar]
- 25.Keller, D. M., X. Zeng, Y. Wang, Q. H. Zhang, M. Kapoor, H. Shu, R. Goodman, G. Lozano, Y. Zhao, and H. Lu. 2001. A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol. Cell 7:283-292. [DOI] [PubMed] [Google Scholar]
- 26.Kelley, D. E., D. G. Stokes, and R. P. Perry. 1999. CHD1 interacts with SSRP1 and depends on both its chromodomain and its ATPase/helicase-like domain for proper association with chromatin. Chromosoma 108:10-25. [DOI] [PubMed] [Google Scholar]
- 27.Krogan, N. J., M. Kim, S. H. Ahn, G. Zhong, M. S. Kobor, G. Cagney, A. Emili, A. Shilatifard, S. Buratowski, and J. F. Greenblatt. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol. Cell. Biol. 22:6979-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowe, S. W., E. M. Schmitt, S. W. Smith, B. A. Osborne, and T. Jacks. 1993. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 362:847-849. [DOI] [PubMed] [Google Scholar]
- 29.Ludwig, T., D. L. Chapman, V. E. Papaioannou, and A. Efstratiadis. 1997. Targeted mutations of breast cancer susceptibility gene homologs in mice: lethal phenotypes of Brca1, Brca2, Brca1/Brca2, Brca1/p53, and Brca2/p53 nullizygous embryos. Genes Dev. 11:1226-1241. [DOI] [PubMed] [Google Scholar]
- 30.Lycan, D., G. Mikesell, M. Bunger, and L. Breeden. 1994. Differential effects of Cdc68 on cell cycle-regulated promoters in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:7455-7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malone, E. A., C. D. Clark, A. Chiang, and F. Winston. 1991. Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:5710-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Medghalchi, S. M., P. A. Frischmeyer, J. T. Mendell, A. G. Kelly, A. M. Lawler, and H. C. Dietz. 2001. Rent1, a trans-effector of nonsense-mediated mRNA decay, is essential for mammalian embryonic viability. Hum. Mol. Genet. 10:99-105. [DOI] [PubMed] [Google Scholar]
- 33.Nagulapalli, S., J. M. Pongubala, and M. L. Atchison. 1995. Multiple proteins physically interact with PU.1. Transcriptional synergy with NF-IL6 beta (C/EBP delta, CRP3). J. Immunol. 155:4330-4338. [PubMed] [Google Scholar]
- 34.Okuhara, K., K. Ohta, H. Seo, M. Shioda, T. Yamada, Y. Tanaka, N. Dohmae, Y. Seyama, T. Shibata, and H. Murofushi. 1999. A DNA unwinding factor involved in DNA replication in cell-free extracts of Xenopus eggs. Curr. Biol. 9:341-350. [DOI] [PubMed] [Google Scholar]
- 35.Orphanides, G., W. H. Wu, W. S. Lane, M. Hampsey, and D. Reinberg. 1999. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400:284-288. [DOI] [PubMed] [Google Scholar]
- 36.Pawlak, M. R., C. A. Scherer, J. Chen, M. J. Roshon, and H. E. Ruley. 2000. Arginine N-methyltransferase 1 is required for early postimplantation mouse development, but cells deficient in the enzyme are viable. Mol. Cell. Biol. 20:4859-4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prendergast, J. A., L. E. Murray, A. Rowley, D. R. Carruthers, R. A. Singer, and G. C. Johnston. 1990. Size selection identifies new genes that regulate Saccharomyces cerevisiae cell proliferation. Genetics 124:81-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shirakata, M., K. Huppi, S. Usuda, K. Okazaki, K. Yoshida, and H. Sakano. 1991. HMG1-related DNA-binding protein isolated with V-(D)-J recombination signal probes. Mol. Cell. Biol. 11:4528-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spencer, J. A., M. H. Baron, and E. N. Olson. 1999. Cooperative transcriptional activation by serum response factor and the high mobility group protein SSRP1. J. Biol. Chem. 274:15686-15693. [DOI] [PubMed] [Google Scholar]
- 40.Staal, F. J., and H. Clevers. 2000. Tcf/Lef transcription factors during T-cell development: unique and overlapping functions. Hematol. J. 1:3-6. [DOI] [PubMed] [Google Scholar]
- 41.Travers, A. 2000. Recognition of distorted DNA structures by HMG domains. Curr. Opin. Struct. Biol. 10:102-109. [DOI] [PubMed] [Google Scholar]
- 42.Vasseur, S., A. Hoffmeister, A. Garcia-Montero, G. V. Mallo, R. Feil, S. Kuhbandner, J. C. Dagorn, and J. L. Iovanna. 2002. p8-deficient fibroblasts grow more rapidly and are more resistant to adriamycin-induced apoptosis. Oncogene 21:1685-1694. [DOI] [PubMed] [Google Scholar]
- 43.Verbeek, S., D. Izon, F. Hofhuis, E. Robanus-Maandag, H. te Riele, M. van de Wetering, M. Oosterwegel, A. Wilson, H. R. MacDonald, and H. Clevers. 1995. An HMG-box-containing T-cell factor required for thymocyte differentiation. Nature 374:70-74. [DOI] [PubMed] [Google Scholar]
- 44.Wada, T., G. Orphanides, J. Hasegawa, D. K. Kim, D. Shima, Y. Yamaguchi, A. Fukuda, K. Hisatake, S. Oh, D. Reinberg, and H. Handa. 2000. FACT relieves DSIF/NELF-mediated inhibition of transcriptional elongation and reveals functional differences between P-TEFb and TFIIH. Mol. Cell 5:1067-1072. [DOI] [PubMed] [Google Scholar]
- 45.Williamson, D. J., S. Banik-Maiti, J. DeGregori, and H. E. Ruley. 2000. hnRNP C is required for postimplantation mouse development but is dispensable for cell viability. Mol. Cell. Biol. 20:4094-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wittmeyer, J., and T. Formosa. 1997. The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol. Cell. Biol. 17:4178-4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wittmeyer, J., L. Joss, and T. Formosa. 1999. Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry 38:8961-8971. [DOI] [PubMed] [Google Scholar]
- 48.Yarnell, A. T., S. Oh, D. Reinberg, and S. J. Lippard. 2001. Interaction of FACT, SSRP1, and the high mobility group (HMG) domain of SSRP1 with DNA damaged by the anticancer drug cisplatin. J. Biol. Chem. 276:25736-25741. [DOI] [PubMed] [Google Scholar]
- 49.Zeng, S. X., M. S. Dai, D. M. Keller, and H. Lu. 2002. SSRP1 functions as a co-activator of the transcriptional activator p63. EMBO J. 21:5487-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]