Abstract
The mitochondrial genomes of a wide variety of species contain an insufficient number of functional tRNA genes, and translation of mitochondrial mRNAs is sustained by import of nucleus-encoded tRNAs. In Leishmania, transfer of tRNAs across the inner membrane can be regulated by positive and negative interactions between them. To define the factors involved in such interactions, a large multisubunit complex (molecular mass, ∼640 kDa) from the inner mitochondrial membrane of the kinetoplastid protozoon Leishmania, consisting of ∼130-Å particles, was isolated. The complex, when incorporated into phospholipid vesicles, induced specific, ATP- and proton motive force-dependent transfer of Leishmania tRNATyr as well as of oligoribonucleotides containing the import signal YGGYAGAGC. Moreover, allosteric interactions between tRNATyr and tRNAIle were observed in the RNA import complex-reconstituted system, indicating the presence of primary and secondary tRNA binding sites within the complex. By a combination of antibody inhibition, photochemical cross-linking, and immunoprecipitation, it was shown that binding of tRNAIle to a 21-kDa component of the complex is dependent upon tRNATyr, while binding of tRNATyr to a 45-kDa component is inhibited by tRNAIle. This “ping-pong” mechanism may be an effective means to maintain a balanced tRNA pool for mitochondrial translation.
There is remarkable diversity in the scope and mechanism of mitochondrial tRNA import (reviewed in reference 18). Human mitochondria do not import tRNA, but a number of neuromuscular degenerative and metabolic diseases are caused by mutations in mitochondrial tRNA genes (21). In yeast, a single tRNA is imported, apparently through protein import channels and requiring at least two soluble factors, including the mitochondrial form of the cognate aminoacyl-tRNA synthetase (8). By contrast, in kinetoplastid protozoa (Leishmania and trypanosomes), import of a whole spectrum of tRNAs is necessitated by the complete lack of mitochondrial tRNA genes (5, 19). In this system, membrane-bound tRNA binding proteins recognize specific structural motifs (import signals) on tRNA, soluble factors are not required, and the translocation pathway appears to be distinct from that for protein import (11, 14, 17). Moreover, the sequence and bioenergetic requirements for outer and inner membrane transfer are nonidentical (2), indicating the presence of a distinct transport machinery (the RNA import complex [RIC]) at the inner membrane, a situation similar to the TOM and TIM complexes for protein import (15). A 15-kDa polypeptide has been shown to be required for import into Leishmania mitochondria (1); otherwise, the import machinery remains undefined.
Using an in vitro evolution protocol, it was recently shown that Leishmania mitochondria recognize a number of short sequence motifs homologous to multiple domains in tRNAs, suggesting the presence of several import signals (3). Moreover, novel positive and negative allosteric interactions between these aptamers, as well as between intact tRNAs, at the inner membrane were described (3). The RNAs could be classified into two types: type I RNAs are efficiently transferred through the inner membrane but are inhibited by type II. In contrast, type II RNAs have poor inner membrane transfer efficiencies and are stimulated by type I. For example, tRNATyr(GUA) is a type I RNA containing the conserved motif UAGAGC in the D domain, while tRNAIle(UAU) is type II with the sequence UCGCGGGUU in the variable loop-T domain (V-T) region (3). The mechanism of these allosteric interactions is unknown, but there are several possibilities. A single conformationally flexible dimeric or multimeric receptor could bind to either a type I or a type II motif. Alternatively, distinct type I and type II receptors may interact directly or indirectly through a third subunit. A related issue is whether the effector and substrate binding subunits for either RNA are identical or different.
To begin to define the molecular components of the inner membrane import apparatus involved in such allosteric interactions, we have isolated a protein complex from mitochondrial inner membrane that is functional for import and probed the role of individual subunits of the complex using a combination of chemical cross-linking and immunochemical approaches. The results reveal specific interactions between distinct type I and type II receptors at the inner membrane.
MATERIALS AND METHODS
Isolation of mitochondria.
Mitochondria were prepared from Leishmania tropica strain UR6 and purified by Percoll gradient centrifugation (11). The outer membrane was solubilized by digitonin treatment, and the resultant mitoplasts were subjected to freeze-thaw cycles to separate particulate (inner membrane) and soluble (matrix) fractions (13).
Purification of RIC.
Purified Leishmania mitochondria (20 mg of protein) were extracted with 320 μM digitonin as above, and mitoplasts were separated by centrifugation at 3,450 × g. Inner membranes were then prepared by three freeze-thaw cycles, suspended at 20 mg of protein/ml in sucrose-Tris-EDTA (STE) containing 0.5% sodium deoxycholate (DOC), 0.4 M KCl, 0.1 mM phenylmethylsulfonyl fluoride, and 1 mM EGTA, and incubated on ice for 30 min with gentle agitation. The DOC extract was clarified by centrifugation at 16,000 × g for 10 min. An RNA affinity column was prepared by immobilizing 500 pmol of RNA containing the D arm of tRNATyr (see Fig. 3) and a 3′-terminal poly(A) tail on 0.1 ml of oligo(dT) cellulose and equilibrated with buffer DB (1) containing 0.1 M KCl. The DOC extract was diluted to 0.1 M in KCl with DB and loaded on the column, which was then washed sequentially with DB containing 0.125% DOC and 0.1, 0.25, 0.5, and 1.0 M KCl. The 0.5 and 1.0 M KCl fractions were concentrated by dialysis against dry Sephadex (1) or by centrifugal ultrafiltration. RNA binding activity was assayed by incubating the fractions with 32P-labeled tRNATyr in binding buffer (1) for 30 min on ice and observing the electrophoretic mobility shift of the RNA-protein complex in 6% polyacrylamide gel (60:1) in the presence of 5 mM MgAc2. Polyclonal antibodies against individual bands resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were raised in BALB/c mice. The RIC21p gel band was trypsinized, and the digest was analyzed by liquid chromatography/mass spectrometry (MS) and MS/MS in a Finnigan LCQ ion trap mass spectrometer system. BLAST analysis was performed against the L. major sequence database (http://www.sanger.ac.uk/Projects/L_major/).
FIG. 3.
RIC-mediated import of tRNA into phospholipid vesicles. (A) Import of Leishmania tRNATyr. Import was assayed by RNase protection. Lane 1, BSA-containing vesicles. Lanes 2 to 6 and 9 to 11, RIC-reconstituted vesicles. The complete system (lanes 2 and 10) contained 100 fmol of tRNATyr and 4 mM ATP. Lanes 3 and 9, ATP omitted. Lane 4, ATP replaced by AMPPCP. Lane 5, 0.5% DOC added after import incubation. Lane 6, tRNAGln(CUG) replacing tRNATyr. Lane 11, 50 μM carbonylcyanide m-chlorophenylhydrazone added. Lanes 7 and 8, input tRNATyr (1 fmol) and tRNAGln (2 fmol), respectively. (B) Left, structure of tRNATyr D arm oligoribonucleotide. Positions are numbered as in the intact tRNA sequence (10). The conserved motif is shown in bold. Right, import of D arm derivatives. Lanes 1 and 2, wild type. Lanes 3 to 6 contained mutants G22:C; G22:C, C13:G; A23:U; and A23:U, U12:A, respectively. Lane 1, BSA-containing vesicles. Lanes 2 to 6, RIC-containing vesicles. (C) Interactions between tRNATyr and tRNAIle. RIC-containing vesicles were incubated with high-specific-activity substrate (50 fmol) with or without low-specific-activity effector (5 fmol), and uptake was assayed. Lanes 1 to 3, tRNATyr substrate; lanes 4 to 6, tRNAIle substrate. Lanes 1 and 4, no effector; lane 2, tRNAIle effector; lanes 3 and 6, tRNAGln(CUG) effector; lane 5, tRNATyr effector. Lanes 7 and 8, input tRNAtyr and tRNAIle, respectively (1 fmol).
Reconstitution in phospholipid vesicles.
Phosphatidylcholine (0.5 mg), dispersed in 150 μl of liposome suspension buffer (50 mM HEPES-KOH [pH 7.5], 10 mM MgAc2, 2 mM dithiothreitol [DTT], 10% glycerol, 0.25% DOC), was mixed with purified RIC or bovine serum albumin (BSA) (5 μg in 50 μl of DB-DOC-1 M KCl) and dialyzed against a mixture of 5 mM HEPES-KOH [pH 7.5], 5 mM MgAc2, and 20 mM KCl for 18 h at 4°C (4). Aggregated material was removed by centrifugation at 2,300 × g, and the supernatant containing mainly unilamellar vesicles was used. Intactness of the vesicles was checked by observing the release of entrapped carboxyfluorescein in the presence of 1% Triton X-100. Washed proteoliposomes were analyzed by SDS-PAGE to reveal incorporation of RIC subunits (∼36 μg of protein/ml).
Import substrates.
32P-labeled Leishmania tRNATyr(GUA), tRNAIle(UAU), tRNAGln(CUG) and D arm oligoribonucleotides were prepared by T7 RNA polymerase transcription (1-3, 12). tRNAIle [42-66], covering positions 42 to 66 of the V-T region, was transcribed from a clone containing the internal HpaII fragment of the tRNAIle(UAU) gene in plasmid pGCN8 (3). High- and low-specific-activity substrates and effectors for allosteric interaction assays were prepared as described previously (3). 5-Bromouridine (5-BrU)-labeled tRNA was synthesized in transcription reactions containing 10 μM [α-32P]UTP and 20 μM 5-BrU triphosphate.
Import assays.
Import into intact mitochondria or mitoplasts was assayed by incubating the vesicles (100 μg of protein) with 32P-labeled RNA (100 fmol) in 20 μl of import buffer (10 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 1 mM DTT, and 4 mM ATP) at 37°C for 15 min; then, RNases A and T1 were added to final concentrations of 2.5 μg/ml and 50 units/ml, respectively, and RNase digestion continued at 37°C for 15 min. The mitochondria were washed with STE by centrifugation, and imported RNA was isolated by guanidinium isothiocyanate extraction and ethanol precipitation (12). Proteoliposomes (∼0.7 μg of RIC) were incubated with 100 fmol of substrate RNA as above and treated with RNase; then, 3 ml of liposome washing buffer (5 mM HEPES-KOH [pH 7.5], 2 mM DTT, 5 mM EDTA) was added, and the vesicles were recovered by centrifugation at 120,000 × g for 30 min at 4°C for RNA analysis. For allosteric interaction assays, proteoliposomes were incubated with a mixture of high-specific-activity substrate (50 fmol) and low-specific-activity effector (5 fmol) as indicated.
Electrophoretic and sedimentation analyses.
Native RIC was electrophoresed on 4 to 15% gradient polyacrylamide (40:1) containing 50 mM Tris-HCl [pH 8.9], with 25 mM Tris-glycine as running buffer, at 8 V/cm. For Western blots, the gel was equilibrated with Tris-glycine-SDS buffer before transfer to a polyvinylpyrrolidone membrane. The blots were probed with mouse polyclonal anti-subunit serum at 1:100 dilution and developed with horseradish peroxidase-conjugated secondary immunoglobulin G. Sedimentation analysis was performed in 15 to 30% glycerol gradients in buffer DB at 150,000 × g for 4.5 h at 4°C.
Electron microscopy.
Freshly purified RIC (nondialyzed 1 M KCl fraction) was diluted to ∼3 μg/ml with buffer DB, and 10 μl was applied on carbon-coated grids for 2 min; the grids were washed twice with deionized water and stained with 1% uranyl acetate for 2 min before observation at 30,000×. Photomicrographs were scanned on a Bio-Rad G710 densitometer. Particle size measurements were performed using Quantity One software.
Photochemical cross-linking and immunoprecipitation.
Mitoplasts were incubated with 5-BrU-labeled tRNA (1 nM) for 30 min on ice, washed, and irradiated with 313-nm UV on a transilluminator for 30 min with cooling to form RNA-protein cross-links (20). SDS was then added to 0.2%, and the proteins were diluted 20-fold into 1× TETN buffer (250 mM Tris-HCl [pH 7.5], 5 mM EDTA, 250 mM NaCl, 1% Triton X-100, 2 mg of BSA/ml) containing a 1:50 dilution of polyclonal mouse antiserum. Immunoprecipitation was performed with protein A-Sepharose. Immune complexes were dissociated in SDS-PAGE sample buffer and resolved by 8 M urea-6% PAGE in the presence of 0.05% SDS. Antibody-inhibition experiments were performed on mitoplasts as described previously (1).
RESULTS
A large complex from the inner membrane.
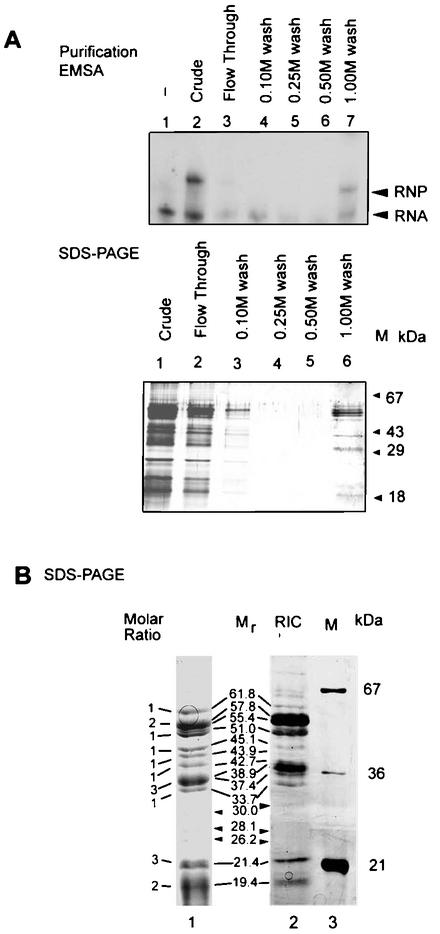
To purify the RIC, we exploited the observation that the D domain of Leishmania tRNATyr(GUA) contains a necessary and sufficient import signal (12). A DOC extract of inner membrane preparations was chromatographed on an affinity column containing the D arm oligoribonucleotide (see below) as an affinity ligand. By gel-shift assay using the crude extract, a specific ribonucleoprotein complex containing tRNATyr could be discerned (Fig. 1). The activity was retained by the affinity column and was specifically eluted with 1 M K+. This step resulted in a 40-fold enrichment of the protein in greater-than-80% yield (Table 1).
FIG. 1.
Purification of RIC from Leishmania mitochondria. (A) Upper panel, electrophoretic mobility shift of Leishmania tRNATyr incubated without protein (lane 1), with crude DOC extract of inner membrane (lane 2), or with the indicated fractions eluting from a D arm affinity column (lanes 3 to 7). Lower panel, SDS-PAGE (silver stain) of the same fractions. M, size markers. (B) SDS-PAGE of RIC. Lanes: 1, Coomassie stain; 2, silver stain; 3, markers. Mole ratios (determined from lane 1) and molecular sizes are shown adjacent to the bands. Substoichiometric bands are indicated by arrowheads.
TABLE 1.
Enrichment of RIC during purification
| Fraction | Protein (mg/ml) | Activity (U/ml)a | Total activity (U) | Specific activity (U/mg)b | Purifi- cation (fold)c | Yield (%)d |
|---|---|---|---|---|---|---|
| Inner membrane extract | 1.2 | 73 | 58.4 | 60 | —e | 100 |
| Affinity 1 M K+ fraction | 0.21 | 510 | 51 | 2,380 | 40 | 87 |
| Glycerol gradient peak fraction | 0.19 | 700 | 28 | 3,684 | 61 | 48 |
One unit of activity corresponds to the binding of 1 fmol of tRNATyr in the electrophoretic mobility shift assay (Fig. 1).
Specific activity is expressed in terms of units per milligram of protein.
Increase in specific activity over the crude inner membrane extract.
Yield is the percentage of the total activity in the crude extract recovered in each fraction.
—, not applicable.
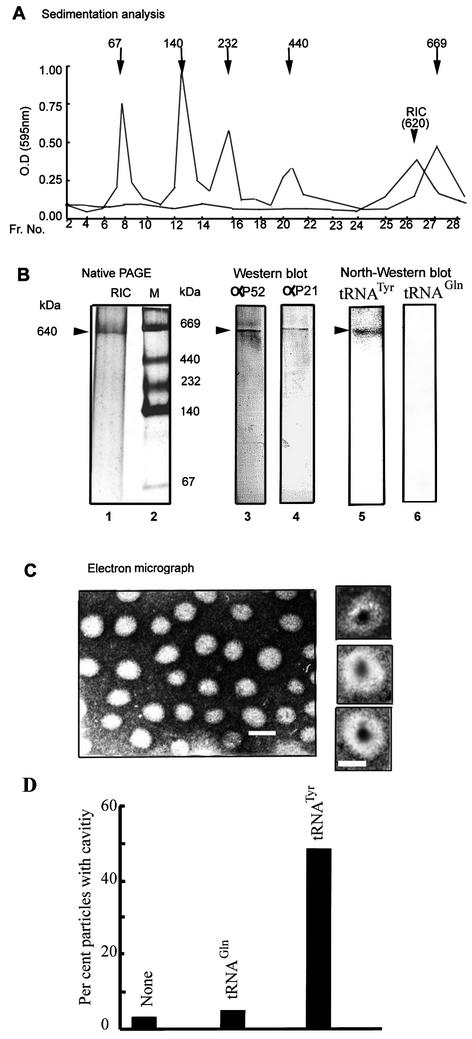
SDS-PAGE analysis of the 1 M K+ fraction showed the presence of 10 major bands ranging from 19 to 62 kDa, present in the apparent stoichiometry of 2:3:1:3:1:1:1:1:2:1, respectively, with an aggregate molecular mass of ∼620 kDa (Fig. 1). The profile, though complex, is reproducible, with correspondence between the Coomassie- and silver-stained bands (Fig. 1). A number of substoichiometric bands were also present; these could be contaminants or proteolytic degradation products. Native gel electrophoresis of the affinity-purified fraction revealed the presence of a single protein species migrating with an apparent molecular mass of 640 kDa (Fig. 2). Sedimentation analysis yielded a value of ∼620 kDa (Fig. 2). Glycerol gradient centrifugation resulted in a 50% increase of specific activity over the affinity fraction but a relatively low yield (Table 1), probably due to inactivation during sedimentation. The peak protein fraction in the glycerol gradient was sufficient to induce import, and no other fraction contained activity (data not shown), indicating the absence of smaller essential components; however, the possibility of multiple large complexes of identical size cannot be presently excluded. Western blot analyses demonstrated the presence of several of the subunits, including the 52- and the 21-kDa species, as well as one or more proteins binding specifically to tRNATyr, but not to tRNAGln(CUG), which is not imported, in the native 640-kDa complex (Fig. 2).
FIG. 2.
Size and morphology of RIC. (A) Glycerol gradient ultracentrifugation of purified RIC or markers run in parallel. Marker sizes (kDa) are shown at the top. Each fraction was assayed by the Bradford method, and the absorbance at 595 nm was recorded. (B) Left, native PAGE of affinity-purified RIC, silver stain. Middle, Western blot of native complex. Probes: anti-RIC51p (strip 3) or anti-RIC21p (strip 4). Right, Northwestern blot (1) of native RIC. Probe: strip 5, 32P-tRNATyr; strip 6, 32P-tRNAGln. The 640-kDa band is indicated. (C) Electron micrograph (negative stain). Bar, 160 Å. Individual particles with fluid-filled cavities are shown at the right. Bar, 140 Å. (D) Fraction of particles with cavities observed by preincubating RIC alone (bar 1) or with tRNAGln (bar 2) or tRNATyr (bar 3).
Electron microscopy of negatively stained preparations of RIC revealed the presence of particles with cross-sections of 130 ± 32 Å (n = 200). About 3% of the particles had fluid-filled cavities with diameters of 45 ± 9 Å (Fig. 2). It is not known for certain whether these cavities represent the tRNA transport pore. Interestingly, the import substrate tRNATyr, but not the nonimported tRNAGln(CUG), induced a significant increase in the number of cavities (Fig. 2), suggesting these to be RNA-gated channels.
Preliminary mass spectrometric peptide sequencing (data not shown) indicated that the 21-kDa band of RIC contains two major proteins, in keeping with the stoichiometry of two to three copies per RIC (Fig. 1). These include a homologue of assembly subunit 6 (UCR_14kDa) (7) of the ubiquinol-cytochrome c reductase complex (respiratory complex III) of the inner mitochondrial membrane and an unknown protein with no database matches. The present data are not sufficient to define a precise function of each of these proteins in import but may indicate a close relationship between RIC and the mitochondrial electron transport machinery.
The complex is necessary and sufficient for import.
To examine whether the RIC is functional for RNA import, it was incorporated into phospholipid vesicles. RIC-containing vesicles were incubated with radiolabeled tRNATyr and then treated with RNase and reisolated. In the presence of ATP, protection of the tRNA from RNase was observed, indicating uptake into the vesicles (Fig. 3A). No protection was observed with RIC-free vesicles. Disruption of the vesicles with DOC after the incubation rendered the RNA nuclease-sensitive, showing that intactness of the membrane is essential for the protection and that the RIC itself does not protect the RNA. The apparent Km for RIC-mediated import of tRNATyr into phospholipid vesicles was 1.16 nM (Table 2). tRNAGln(CUG), which is not imported in vivo (9), was not translocated by RIC (Fig. 3A); the tRNA was recovered intact from the postincubation supernatant (data not shown), arguing against specific degradation of tRNAGln as the reason for the lack of its import.
TABLE 2.
Kinetic characteristics of RIC-mediated import
| Experimental variable | Parameter | Value |
|---|---|---|
| tRNATyr | Kma | 1.16 nM |
| ATP | Kma | 1.35 mM |
| tRNATyr (effector) on tRNAIle (substrate) | EC50b | 0.033 nM |
| tRNAIle (effector) on tRNATyr (substrate) | IC50c | 0.018 nM |
Concentration of tRNATyr or ATP for half-maximal rate of import into RIC-reconstituted phosphatidylcholine vesicles.
Concentration of tRNATyr effector required for half-maximal stimulation of binding of tRNAIle (2 nM).
Concentration of tRNAIle effector for half-maximal inhibition of binding of tRNATyr (2 nM).
Uptake into RIC-reconstituted vesicles was dependent upon ATP; the nonhydrolyzable β-γ imido analogue of ATP was unable to substitute for ATP, indicating the requirement for ATP hydrolysis (Fig. 3A). The apparent Km for ATP was 1.35 mM; this is comparable to the value of 1.52 mM for import into intact mitochondria (2). It has been shown previously that in intact mitochondria or mitoplasts, transfer of tRNA to the matrix requires a transmembrane proton motive force probably generated by the oligomycin-sensitive F1F0-ATPase's pumping protons out of the matrix at the expense of ATP hydrolysis (13). In a similar manner, RIC-mediated transfer was sensitive to the protonophore uncoupler carbonylcyanide m-chlorophenylhydrazone, which dissipates proton gradients (Fig. 3A), indicating that both ATP hydrolytic and proton pumping activities are contained within the RIC.
Signal recognition by RIC is sensitive to mutations within the conserved motif.
In addition to intact tRNAs, Leishmania mitochondria import short oligoribonucleotides containing the import signal YGGYAGAGC in the D arm of tRNATyr (2, 3). RIC-dependent transfer of a 27-mer oligonucleotide containing the wild-type sequence was observed (Fig. 3B). The mutation G22:C in the conserved import motif abolished import, whereas restoration of the loop-closing base pair of the hairpin in the double mutant G22:C, C13:G restored import to about two-thirds of the wild-type level. The mutation A23:U in the next position of the motif also abolished importability, but restoration of the base pair in A23:U, U12:A failed to regenerate activity. These results indicate that the stability of the hairpin is critical for signal recognition by RIC and, further, that the identity of A23 is important. The behavior of these mutants in the RIC-reconstituted system is identical to that in mitoplasts and different from that in intact mitochondria (2). Thus, RIC retains the RNA recognition specificity of the inner membrane.
Allosteric interactions within RIC.
A recently described property of inner membrane import is that of cooperative and antagonistic interactions between two types of RNA ligand (3). Type I ligands, of which tRNAs (including tRNATyr) containing the conserved D arm domain are an example, can be transferred independently of other ligands, whereas type II ligands (such as tRNAIle) are dependent on type I ligands for transfer. Moreover, type II ligands inhibit transfer of type I ligands. To examine whether such interactions occur within the isolated complex, RIC-reconstituted phospholipid vesicles were assayed for import of tRNATyr in the presence of low concentrations of tRNAIle and vice versa. Indeed, the uptake of tRNATyr was inhibited by tRNAIle, while tRNATyr stimulated uptake of tRNAIle (Fig. 3C). The 50% effective concentration for tRNATyr effector was 0.033 nM, while the 50% inhibitory concentration for tRNAIle was 0.016 nM (Table 2). Thus, RIC contains receptors for both tRNAs, namely, the primary receptor (that for tRNATyr), which binds directly to its ligand, and the secondary receptor (that for tRNAIle), whose binding to its ligand depends upon conformational changes transmitted within the native complex subsequent to loading of tRNATyr.
Regulated interaction of tRNATyr with RIC45p.
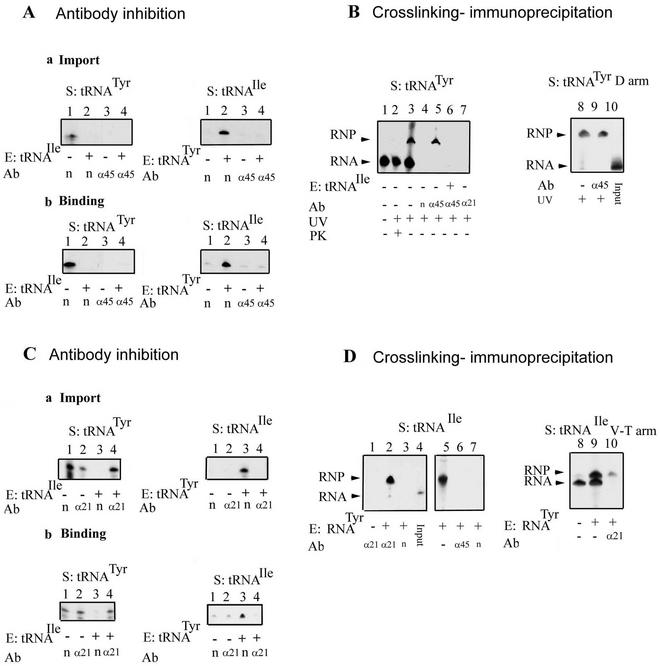
To detect these allosteric interactions at the levels of individual components of RIC, subunit-specific antibodies were employed. A preliminary Northwestern blot analysis of RIC revealed the presence of several tRNA binding proteins. Polyclonal mouse antibodies against these bands were prepared, and their specificity was verified by Western blots (data not shown). Antibody against the 45-kDa subunit (RIC45p) (Fig. 1) inhibited binding as well as import of tRNATyr (Fig. 4A). Moreover, in the presence of the antibody, tRNATyr was unable to stimulate the import of tRNAIle, implying either that RIC45p directly binds tRNATyr or that it has some other role unrelated to tRNA binding.
FIG. 4.
Role of RIC45p and RIC21p in import. (A) Inhibition by anti-RIC45p antibody. Mitoplasts were incubated with the indicated antiserum (1:50), washed, and assayed for import (a) or binding (b) of the indicated 32P-labeled tRNA substrate (S) in the presence or absence of the indicated effector (E). Ab, antibody; n, nonimmune serum; α, anti-RIC 45p serum. (B) Immunoprecipitation of cross-linked tRNA. Left, 5-BrU-, 32P-labeled tRNATyr (20 fmol) was incubated with mitoplasts in the presence (lane 6) or absence (lanes 1 to 5 and 7) of low-specific-activity tRNAIle (2 fmol) under binding conditions. In lanes 1 to 3, the mitoplasts were solubilized and directly loaded; lanes 4 to 7 show immunoprecipitates. RNA-protein complexes were UV cross-linked (except lane 1, a no-UV control) and immunoprecipitated with anti-RIC45p (lanes 5 and 6), anti-RIC21p (lane 7), or nonimmune serum (lane 4). Right, photo-cross-linking of 5-BrU-labeled D arm oligonucleotide (Fig. 3). Lane 10, input RNA. Lane 8, cross-linked product. Lane 9, anti-RIC45p immunoprecipitate. (C) Inhibition by anti-RIC21p antibody. Mitoplasts were incubated with nonimmune or anti-RIC21p antibody and assayed for import (a) or binding (b) of the indicated substrate in the absence or presence of effector. (D) Cross-linking of RIC21p to intact tRNAIle (left) or tRNAIle [42-66] (right). Mitoplasts were incubated with the indicated substrates and effectors before UV irradiation. Lane 4, input tRNAIle. Lane 5, cross-linked product. Lanes 1 to 3, 6, and 7, immunoprecipitates with anti-RIC21p (lanes 1 and 2), nonimmune serum (lanes 3 and 7), or anti-45p antibody (lane 6). Right, tRNAIle [42-66] was incubated with mitoplasts in the absence (lane 8) or presence (lanes 9 and 10) of tRNATyr effector and then irradiated. Lane 10, anti-21p immunoprecipitate.
To detect a direct interaction between RIC45p and tRNA, a photochemical cross-linking experiment was performed using RNA containing the photoactivable analogue 5-BrU triphosphate (20). Radiolabeled tRNATyr containing 5-BrU residues was incubated with mitoplasts and then UV cross-linked to the membrane; after dissociation of protein complexes with detergent, immunoprecipitation was performed with anti-RIC45p antibody. A major cross-linked product, migrating behind free tRNATyr and sensitive to protease, was observed (Fig. 4B). This complex was precipitated by anti-RIC45p antibody, but not by normal serum or by antibody against the 21-kDa subunit. Moreover, the yield of the complex was reduced in the presence of tRNAIle effector in the initial incubation step (Fig. 4B). Thus, RIC45p directly contacts tRNATyr, and this binding is inhibited by tRNAIle, i.e., it has the properties of a type I receptor.
The type I receptor is expected to interact with the corresponding import signal present in the D domain of tRNATyr. To examine whether this is the case for RIC45p, a cross-linking experiment was carried out with 5-BrU-labeled D arm oligonucleotide. A specific RNA-protein complex immunoprecipitable with anti-RIC45p antibody was observed (Fig. 4B), showing direct contacts between RIC45p and the D arm signal.
tRNATyr-dependent interaction of tRNAIle with RIC21p.
Information was obtained on type II interactions by similar immunochemical experiments. Antibody against the 21-kDa band (RIC21p) had no effect on the binding of tRNATyr to the inner membrane and had only a two- to threefold effect on its import (Fig. 4). In contrast, the antibody totally inhibited binding as well as import of tRNAIle in the presence of tRNATyr (Fig. 4). It also antagonized the inhibition of binding and import of tRNATyr by tRNAIle. These results indicate that either RIC21p is the binding site for tRNAIle or that it somehow relays the conformational changes between type I and type II receptors. With 5-BrU-labeled tRNAIle, a tRNA-RIC21p complex which migrates slower than the free RNA and which is sensitive to protease (data not shown) was recovered using anti-RIC21p, but not anti-RIC45p, antibody when tRNAIle was incubated with mitoplasts in the presence of tRNATyr, but not in its absence (Fig. 4C). Moreover, the 5-BrU-labeled V-T segment of tRNAIle, which contains a type II signal, was specifically cross-linked to form a complex immunoprecipitable by anti-RIC21p antibody (Fig. 4D). We conclude that a component of RIC21p has the properties of a type II receptor for tRNAIle.
DISCUSSION
In this study, progress has been made towards the molecular definition of the inner membrane import machinery of Leishmania mitochondria. Affinity chromatography using a type I import signal (i.e., the D arm of tRNATyr) was a particularly effective way to separate the functional import complex from the host of respiratory complexes on the inner membrane (Fig. 1). Subsequent resolution of the complex into its constituent polypeptides, followed by a band-by-band immunochemical analysis of the individual components for import-related activities, resulted in the identification of two factors with tRNA receptor function. This biochemical approach, coupled with sequence analysis and genome database searches, should be of general applicability to those systems, e.g., plants and protozoa, in which import mutants are not available.
The complex appears to be composed of roughly spherical particles of uniform size (Fig. 2). A solid protein sphere of 130 Å diameter (as determined by electron microscopy) would have a mass of ∼950 kDa, assuming a partial specific volume of 0.73 cm3 g−1 (9). However, the subunit composition, native gel migration, and sedimentation properties support a mass of 620 to 640 kDa (Fig. 1 and 2). This difference can be accounted for by assuming a hollow sphere: if one assumes a cylindrical pore of ∼45 Å diameter, the calculated mass of ∼750 kDa comes to within 20% of the determined value. The reason for only ∼3% of the particles showing fluid-filled cavities may be that the channels are gated by tRNA. This is supported by the observation that in the presence of tRNATyr, there is an increase in the fraction of particles showing pores (Fig. 2). Thus, one consequence of the binding of tRNA to its receptor on the RIC would be to induce the opening of the import channel. Assembly of proton translocation channels on the endoplasmic reticulum membrane induced by ribosome “ligand” has been previously observed (6).
The diameter of the putative translocation channel (∼45 Å) is considerably larger than the 20-Å protein transport channels on the mitochondrial outer membrane (9) or endoplasmic reticulum (6), which can accommodate unfolded polypeptide chains. The canonical tRNA structure is L shaped, with two 20-Å helical arms at an angle of ∼90° and a distance of 80 Å between the two extremities of the L (16). This means that in order to pass through the 45-Å pore, the tRNA structure has to be distorted by reducing or increasing the hinge angle. It may not be a coincidence that both the type I (D arm) and type II (V-T region) are located at or close to the hinge; interaction with the cognate receptors could lead to conformational changes within the tRNA appropriate for translocation.
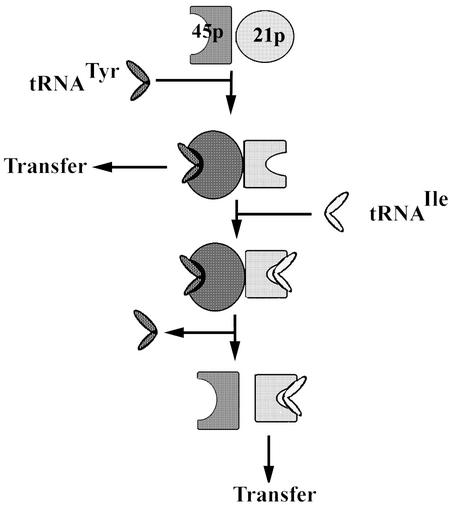
By antibody inhibition, photochemical cross-linking, and immunoprecipitation, two components of the import complex were identified as proximal contact sites for tRNATyr and tRNAIle, respectively (Fig. 4). The RIC45p-tRNATyr complex is inhibited by tRNAIle, whereas the RIC21p-tRNAIle complex is stimulated by tRNATyr, as expected of type I and type II receptors, respectively. These complexes are specific for their respective tRNAs, i.e., the receptors are distinct. Additionally, blocking of RIC21p by antibody is sufficient to prevent inhibition of tRNATyr binding by tRNAIle (Fig. 4), indicating that a separate tRNAIle binding protein is not required to negatively regulate the type I tRNA. These findings can be accommodated by a simple two-receptor model (Fig. 5). RIC45p binds tRNATyr to form a translocation-competent complex from which the tRNA can be transferred to the import pore. The tRNATyr-bound complex contacts RIC21p, activating the tRNAIle binding site of the latter. Subsequent loading of tRNAIle on RIC21p results in a second conformational change that is transmitted back to RIC45p, leading to the dissociation of tRNATyr, loss of contact between the two receptors, and formation of a translocation-competent RIC21p-tRNAIle complex. Such feed-forward and feedback interactions involving transient contacts between the respective receptors explain how the type I and type II tRNAs can be transported through the same import pore without directly competing with one another, while maintaining a properly balanced pool within the mitochondrial matrix.
FIG. 5.
The “ping-pong” model of allosteric interactions. Heavily and lightly shaded objects represent RIC45p and RIC21p, respectively. Different shapes indicate different conformations. For details, see the text.
More work is necessary on the identities of these two proteins and on the details of their interactions with tRNAs. Peptide analysis of the 21-kDa component indicated the presence of two or more proteins. It is not obvious which of these is the tRNAIle binding protein detected by photochemical cross-linking (Fig. 4), as neither contains a known RNA binding domain. The complex III homologue contains an extra N-terminal domain that could be involved in tRNA binding; alternatively, the unknown protein could perform this function. In mitochondria, electron transfer through the respiratory chain results in vectorial proton transport coupled to a number of inner membrane transport processes as well as ATP synthesis. The sensitivity of RIC-mediated tRNA import to uncouplers (Fig. 3) implies a similar electron transfer-coupled proton pumping by the complex and, by inference, the presence of respiratory chain components within RIC. Thus the tRNA import complex may be viewed as a specialized molecular motor combining elements of mitochondrial energy transduction with RNA binding and transport.
Acknowledgments
This work was supported by a grant from the Department of Science and Technology, Government of India. S.N.B. and S.G. received research fellowships from the Council of Scientific and Industrial Research, and S.C. received a research fellowship from the University Grants Commission.
We thank Nirmalendu Das for help with the liposome preparation and the W. M. Keck Biomedical Mass Spectrometry Laboratory, University of Virginia, for peptide analysis.
REFERENCES
- 1.Adhya, S., T. Ghosh, A. Das, S. K. Bera, and S. Mahapatra. 1997. Role of an RNA binding protein in import of tRNA into Leishmania mitochondria. J. Biol. Chem. 272:21396-21402. [DOI] [PubMed] [Google Scholar]
- 2.Bhattacharyya, S. N., S. Mukherjee, and S. Adhya. 2000. Mutations in a tRNA import signal define distinct receptors at the two membranes of Leishmania mitochondria. Mol. Cell. Biol. 20:7410-7417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattacharyya, S. N., S. Chatterjee, and S. Adhya. 2002. Mitochondrial RNA import in Leishmania: aptamers homologous to multiple tRNA domains that interact cooperatively or antagonistically at the inner membrane. Mol. Cell. Biol. 22:4372-4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darley-Usmar, V. M., R. A. Capaldi, S. Takamiya, F. Millett, M. T. Wilson, F. Malatesta, and P. Sarti. 1987. Reconstitution and molecular analysis of the respiratory chain, p. 113-152. In V. M. Darley-Usmar, D. Rickwood, and M. T. Wilson (ed.), Mitochondria, a practical approach. IRL Press, Oxford, United Kingdom.
- 5.Hancock, K., and S. L. Hajduk. 1990. The mitochondrial tRNAs of Trypanosoma brucei are nuclear encoded. J. Biol. Chem. 265:19203-19215. [PubMed] [Google Scholar]
- 6.Hanein, D., K. E. S. Matlack, B. Jungnickel, K. Plath, K.-U. Kalies, K. R. Miller, T. A. Rapoport, and C. W. Akey. 1996. Oligomeric rings of the Sec 61p complex induced by ligands required for protein translocation. Cell 87:721-732. [DOI] [PubMed] [Google Scholar]
- 7.Iwata, S., J. W. Lee, K. Okada, J. K. Lee, M. Iwata, B. Rasmussen, T. A. Link, S. Ramaswamy, and B. K. Jap. 1998. Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science 281:64-71. [DOI] [PubMed] [Google Scholar]
- 8.Kolesnikova, O. A., N. S. Entelis, H. Mireau, T. D. Fox, R. P. Martin, and I. A. Tarassov. 2000. Suppression of mutations in mitochondrial DNA by tRNAs imported from the cytoplasm. Science 289:1931-1933. [DOI] [PubMed] [Google Scholar]
- 9.Künkele, K.-P., S. Heins, M. Dembowski, F. E. Nargang, R. Benz, M. Thieffry, J. Walz, R. Lill, S. Nussberger, and W. Neupert. 1998. The preprotein translocation channel of the outer membrane of mitochondria. Cell 93:1009-1019. [DOI] [PubMed] [Google Scholar]
- 10.Lye, L.-F., D.-H. T. Chen, and Y. Suyama. 1993. Selective import of nuclear-encoded tRNAs into mitochondria of the protozoan Leishmania tarentolae. Mol. Biochem. Parasitol. 58:233-246. [DOI] [PubMed] [Google Scholar]
- 11.Mahapatra, S., and S. Adhya. 1996. Import of RNA into Leishmania mitochondria occurs through direct interaction with membrane bound receptors. J. Biol. Chem. 271:20432-20437. [DOI] [PubMed] [Google Scholar]
- 12.Mahapatra, S., S. Ghosh, S. K. Bera, T. Ghosh, A. Das, and S. Adhya. 1998. The D arm of tRNATyr is necessary and sufficient for import into Leishmania mitochondria in vitro. Nucleic Acids Res. 26:2037-2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mukherjee, S., S. N. Bhattacharyya, and S. Adhya. 1999. Stepwise transfer of tRNA through the double membrane of Leishmania mitochondria. J. Biol. Chem. 274:31249-31255. [DOI] [PubMed] [Google Scholar]
- 14.Nabholz, C. E., E. K. Horn, and A. Schneider. 1999. tRNAs and proteins are imported into Leishmania mitochondria of Trypanosoma brucei by two distinct mechanisms. Mol. Biol. Cell 10:2547-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neupert, W. 1997. Protein import into mitochondria. Annu. Rev. Biochem. 66:863-917. [DOI] [PubMed] [Google Scholar]
- 16.Saenger, W. 1984. Principles of nucleic acid structure, p. 331-349. Springer-Verlag, New York, N.Y.
- 17.Schleyer, M., B. Schmidt, and W. Neupert. 1982. Requirement of a membrane potential for the posttranslational transfer of proteins into mitochondria. Eur. J. Biochem. 125:109-116. [DOI] [PubMed] [Google Scholar]
- 18.Schneider, A., and L. Marechal-Drouard. 2000. Mitochondrial tRNA import: are there distinct mechanisms? Trends Cell. Biol. 10:509-513. [DOI] [PubMed] [Google Scholar]
- 19.Simpson, A. M., Y. Suyama, H. Dewes, D. Campbell, and L. Simpson. 1989. Kinetoplastid mitochondria contain functional tRNAs which are encoded in nuclear DNA and also contain small minicircle and maxicircle transcripts of unknown function. Nucleic Acids Res. 17:5427-5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanner, N. K., M. M. Hanna, J. Abelson. 1988. Binding interactions between yeast tRNA ligase and a precursor transfer ribonucleic acid containing two photoreactive uridine analogues. Biochemistry 27:8852-8861. [DOI] [PubMed] [Google Scholar]
- 21.Wallace, D. C. 1999. Mitochondrial diseases in man and mouse. Science 283:1482-1487. [DOI] [PubMed] [Google Scholar]