Abstract
Expression of the recombinase proteins RAG-1 and RAG-2 is discordant: while RAG-1 is relatively long lived, RAG-2 is degraded periodically at the G1-S transition. Destruction of RAG-2 is mediated by a conserved interval in the recombination-dispensable region. The need for RAG-2 to reaccumulate in the nucleus at each cell division suggested the existence of an intrinsic RAG-2 nuclear localization signal (NLS). RAG-1 or RAG-2, expressed individually, is a nuclear protein. A screen for proteins that bind the recombination-dispensable region of RAG-2 identified the nuclear transport protein Importin 5. Mutation of residues 499 to 508 in RAG-2 abolished Importin 5 binding, nuclear accumulation, and periodic degradation of RAG-2. The Importin 5 binding site overlaps an NLS, defined by mutagenesis. RAG-1 rescued the localization of degradation-defective, RAG-2 NLS mutants; this required an intact RAG-1 NLS. Mutations in RAG-2 that abolish intrinsic nuclear accumulation but spare periodic degradation impaired recombination in cycling cells; induction of quiescence restored recombination to wild-type levels. Recombination defects were correlated with a cell cycle-dependent defect in the ability of RAG-1 to rescue localization of the RAG-2 mutants. These results suggest that the intrinsic RAG-2 NLS functions in the nuclear uptake of RAG-2 following its reexpression in cycling cells.
The antigen receptor genes of lymphocytes are encoded in separate DNA segments that are brought together during lymphoid development by V(D)J recombination, the only form of site-specific DNA recombination known in vertebrates and the central process by which immunologic diversity is generated (11). RAG-1 and RAG-2, the sole lymphoid-specific components of the recombinase machinery (37, 43), initiate V(D)J recombination by cleaving participating gene segments at specific recombination signal sequences, producing two signal ends, terminating in flush, double-stranded breaks, and two coding ends, terminating in hairpin structures (32). Recombination is then completed by components of the general cellular machinery for nonhomologous DNA end joining (14, 36).
The full-length RAG-1 and RAG-2 proteins are 1040 and 527 amino acid residues long, respectively, but core fragments comprising residues 384 through 1008 of RAG-1 and 1 through 387 of RAG-2 are sufficient to support recombination of transfected, extrachromosomal substrates (6, 22, 33, 40-42, 46, 48). Since the core fragments are more readily isolated than the full-length proteins, they have been used to define the initial chemical steps of V(D)J recombination in vitro (8, 25, 32, 39, 51, 52). The core RAG-1 fragment, which contains the catalytic site for DNA nicking and transesterification, mediates binding to recombination signal sequences and contacts the coding flank near the scissile bond (10, 20, 23). Although RAG-2 contains no known catalytic residues and has no intrinsic DNA binding activity, it is essential for V(D)J recombination. The core RAG-2 fragment stabilizes and extends interactions of RAG-1 with the recombination signal sequence; its presence in RAG-DNA complexes is essential for helical distortion near the scissile bond, a possible prerequisite for transesterification (3, 16, 35, 49, 50). Accordingly, mutations that perturb DNA binding and transesterification have been identified in core RAG-2 (9, 38).
Residues 387 through 527 of RAG-2, while phylogenetically conserved, are dispensable for recombination of extrachromosomal substrates in vivo and for DNA cleavage by the RAG proteins in vitro. Nonetheless, removal of the noncore region has been reported to reduce the efficiency of recombination within extrachromosomal substrates (6, 22, 33, 40-42, 46, 48), to increase the production of hybrid joints at the expense of coding and signal joints (44), and to impede VH-to-DJH joining at endogenous loci (2, 21, 28), although the mechanisms underlying these effects are not well understood. The noncore region of RAG-2 couples V(D)J recombination to the cell cycle by supporting the periodic destruction of RAG-2 protein, which accumulates preferentially in G0/G1 cells and is degraded at the G1-S transition in dividing cells (27, 30, 31). Two determinants of RAG-2 degradation have been mapped to the noncore region: a consensus phosphorylation site for cyclin-dependent kinases (CDKs) surrounding Thr490 and a cationic interval spanning residues 499 to 508. Destruction of RAG-2 is triggered upon phosphorylation of Thr490 by cyclin A/CDK2 (24); the contribution of the cationic region to degradation is less clear. By constraining initiation of V(D)J recombination to the G0 and G1 cell cycle phases, the periodic destruction of RAG-2 may serve to coordinate RAG-mediated DNA cleavage with the completion of V(D)J recombination by nonhomologous end joining (24).
The amount of RAG-1, in contrast to that of RAG-2, does not fluctuate greatly during cell division (31). Consequently, RAG-1 and RAG-2 are expressed discordantly in dividing cells, implying a requirement for reincorporation of newly synthesized RAG-2 into a correctly localized recombinase once every cell cycle. The RAG proteins are diffusely nuclear when coexpressed from their endogenous loci in lymphoid cells or when coexpressed in transfected fibroblasts (47). When expressed individually, RAG-1 and RAG-2 are also localized to the nucleus (47). Sites governing the nuclear localization of RAG-1 reside near the carboxyl terminus at two intervals spanning residues 826 through 840 and 969 through 973; mutation of either region results in redistribution of RAG-1 to the cytoplasm (47). Both regions bind importin-α, consistent with the interpretation that they function as classical nuclear localization signals (NLSs).
The potential function of an independent import strategy for RAG-2 has remained unexplored. We now show that a site within the noncore region of RAG-2 is bound by the nuclear import receptor Importin 5. The Importin 5 binding site is contained within a nonclassical NLS which, in turn, overlaps residues required for periodic destruction of RAG-2. By systematic mutagenesis of the RAG-2 recombination-dispensable region, the overlapping, noncore functions of cell cycle periodicity and nuclear localization were separated; this allowed us to assess the relative contributions of RAG-1 and RAG-2 nuclear import signals to localization and activity of the recombinase in quiescent and dividing cells. Our results suggest a model in which the intrinsic import signal of RAG-2 supports nuclear reaccumulation as dividing cells reenter the G1 cell cycle phase.
MATERIALS AND METHODS
DNA constructs.
The RAG expression plasmids pcRAG-1 and pcRAG-2 and RAG-2 alanine scanning mutants have been described previously (27, 31). Point mutations in RAG-2 and RAG-1 were generated by site-directed mutagenesis (Chameleon mutagenesis kit; Stratagene). RAG-2 truncation mutants were generated by the PCR using appropriate primers. For localization assays, RAG-1, RAG-2, and RAG mutants were subcloned into pEGFP-C vectors (Clontech). For colocalization assays, RAG-1 or its mutants were subcloned into pEYFP-C1, and RAG-2 or its mutants were subcloned into pECFPC-1. The cDNA segment specifying amino acid residues 439 through 527 of RAG-2 was amplified by the PCR and subcloned between the BlpI and NotI restriction sites of pGFPLacZ (a gift of J. Han and J. Boeke). For binding assays, cDNA spanning codons 439 to 527 of wild-type RAG-2 or the 499/508A10 mutant were amplified by the PCR and subcloned between the BamHI and EcoRI sites of pGEX2-T (Pharmacia). The plasmids pJH200 and pJH290, used in recombination assays, have been described (15, 29).
Cell lines and transfection.
The human embryonic kidney cell line 293 was propagated in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. NIH 3T3 cells were propagated in Dulbecco's modified Eagle medium supplemented with 10% calf serum. The RAG-2-deficient, pro-B-cell line 63-12 (45) and the mature B-cell line A20 were maintained in RPMI-1640 supplemented with 10% calf serum and 50 μM beta-mercaptoethanol.
NIH 3T3 cells were transfected using Lipofectamine 2000 (Invitrogen). Unless otherwise stated, NIH 3T3 cells were 60% confluent at the time of transfection. B-lymphoid cells were transfected in log phase by electroporation. Cells were washed and resuspended in growth medium containing 7.5% calf serum at a density of 4 × 107/ml. A cell suspension of 300 μl was mixed with 30 μg of plasmid DNA in a 4-mM gap electroporation cuvette and allowed to stand for 15 min at room temperature before being subjected to a single pulse from a BMX ECM 830 square-wave electroporator at 300 V for 10 ms. Cells were allowed to stand for an additional 15 min following the pulse and then were transferred to complete medium and incubated for 48 h at 37°C.
Immunofluorescence.
Transfected cells were washed, fixed in 3.7% paraformaldehyde for 10 min at room temperature, and mounted (Vectashield; Vector Laboratories); nuclei were stained with DAPI. For localization of single proteins in fibroblastoid and B-lymphoid cells, analysis was performed using a Zeiss LSM510 confocal microscope and LSM510 software (Zeiss). For colocalization studies, cells were visualized using a Zeiss Axiovert S100 TV microscope and Metamorph 4.5.6 software (Universal Imaging). In both cases, at least 100 cells in total were scored from two or more independent transfections. Localization of RAG protein(s) was classified by visual inspection as nuclear; more nuclear than cytoplasmic (nuclear > cytoplasmic); diffuse in the cell (nuclear = cytoplasmic); or cytoplasmic (www.bs.jhmi.edu/mbg/RossEtAlAppendix.pdf).
Binding assays.
Plasmids encoding glutathione S-transferase (GST) fusion proteins were introduced into Escherichia coli BL21. Bacterial cells were cultured overnight at 37°C in Luria broth supplemented with 50 μg of ampicillin/ml. Overnight cultures were diluted 1:10 into fresh medium and grown at 37°C to an optical density at 595 nm of 0.6 to 1.0; expression of fusion proteins was then induced by addition of isopropyl-β-d-thiogalactopyranoside to a 0.4 mM concentration. After 4 h, bacteria were pelleted and resuspended in ice-cold phosphate-buffered saline (PBS) supplemented with protease inhibitors and Triton X-100 to a final concentration of 1%. The mixtures were incubated for 30 min on ice, and lysis was completed by three cycles of sonication at 10 pulses each with a Branson microtip sonifier (5 mM diameter tip, power output 5, duty cycle 30%). Lysates were clarified by centrifugation at 10,000 × g for 15 min at 4°C. Supernatants were mixed with 1 ml of a 50% slurry of glutathione-agarose beads and incubated for 20 min at room temperature. Beads were washed four times with PBS; the amount of bound protein was estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Coomassie staining against a dilution series of bovine serum albumin. To prepare affinity matrices for use in binding assays, the amount of bacterial lysate used in each adsorption was adjusted to produce beads of similar substituent density.
Log-phase 293 fibroblasts, 63-12 pro-B, or A20 mature B cells were harvested by centrifugation and resuspended in lysis buffer (20 mM Tris Cl [pH 7.6], 150 mM NaCl, 20 mM NaF, 5 mM KCl, 5 mM EDTA, 1 mM dithiothreitol, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 0.25% Tween 20, 10 μg of leupeptin/ml, 10 μg of aprotinin/ml, and 5 μg of pepstatin/ml). Cells were disrupted on ice by 30 strokes of a Dounce homogenizer fitted with a type A pestle. The lysate was clarified by centrifugation at 21,000 × g for 10 min. The protein concentration was adjusted to 0.75 mg/ml with lysis buffer. The lysate was precleared by addition of glutathione-Sepharose (50:50 slurry) at a ratio of 37.5 μl of beads to each milliliter of lysate. The mixture was tumbled for 1 h at 4°C, and beads were removed by centrifugation at 2,300 × g for 1 min. Agarose bead-bound GST fusion protein (100 μg in the form of a 50:50 slurry in lysis buffer at 1 mg/ml) was then added to 1.6 ml of precleared lysate, and the mixture was tumbled for 3 h at 4°C. Following binding, beads were recovered by centrifugation at 2,300 × g for 1 min. Protein was eluted sequentially with lysis buffer containing 150, 250, 350, or 450 mM NaCl; three washes of 1 ml each were performed at each salt concentration. For each wash, beads were tumbled in buffer for 5 min at 4°C and then collected by centrifugation at 2,300 × g for 1 min. Eluted protein was precipitated by trichloroacetic acid using insulin or deoxycholate as a carrier and fractionated by SDS-PAGE.
Protein identification.
Proteins were excised from preparative, Coomassie-stained polyacrylamide gels. Polypeptides of interest were digested with trypsin, and peptide sequences were determined by liquid chromatography and tandem mass spectroscopy and Edman degradation at the Wistar Proteomics Facility. The identity of Importin 5 (karyopherin β3) was confirmed by immunoblotting with a karyopherin β3 (H-300) rabbit polyclonal antibody (Santa Cruz Biotechnology).
Recombination assays.
NIH 3T3 cells were transfected with pJH200 or pJH290 (18 μg), pcRAG-1 (5 μg), pcRAG-2 (5 μg), and pRSV-T (2 μg) using Lipofectamine 2000 (Invitrogen). The plasmid pcDNA1 was added when required to keep the total amount of DNA constant. Cells were harvested at 36 h after transfection, and V(D)J recombination was assayed as described (15).
Cell cycle analysis.
For experiments involving cell cycle manipulation, cells were transfected at 20 or 90% confluence. At the time of transfection and at 36 h after transfection, a sample of cells was harvested and DNA was stained by incubation on ice for 30 min in PBS containing 0.025% NP-40, 100 μg of propidium iodide/ml, and 10 μg of RNase/ml. Stained cells were analyzed using a FACScan (Becton Dickinson). After collection of 10,000 events, the percentage of cells with 2C DNA content was estimated using CELLQuest software (Becton Dickinson).
RESULTS
Specific interaction of a nuclear import protein with residues in the noncore region of RAG-2.
Earlier work established that one function of the RAG-2 noncore region—the coupling of RAG-2 degradation to the cell cycle—could be conferred on other proteins by transfer of the C-terminal 89 amino acid residues of RAG-2 (31). To identify proteins that interact with this region of RAG-2, we established a biochemical screen. Residues 439 through 527 of RAG-2 were fused to the C terminus of GST, and the resulting protein, termed GST-RAG-2(439-527), was expressed in bacteria. A similar GST fusion, termed GST-RAG-2(439-527mt), was made to residues 439 through 527 of the degradation-deficient mutant RAG-2(499/508A10). Accumulation of GST-RAG-2(439-527), relative to GST-RAG-2(439-527mt), was reduced in asynchronous, transfected mammalian cells (data not shown), suggesting that the C-terminal 89 residues of RAG-2, when fused to GST, retain at least one biochemical function, the ability to target degradation.
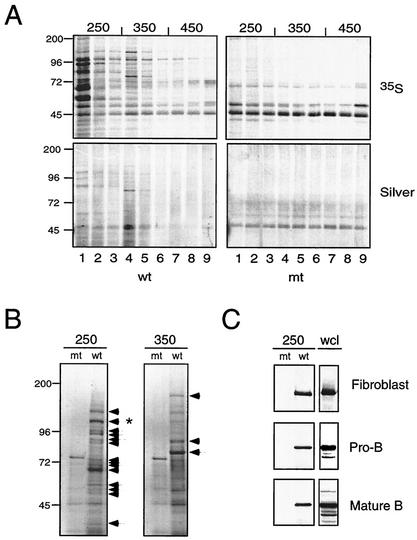
GST-RAG-2(439-527), GST-RAG-2(439-527mt), or GST alone, affixed to glutathione-Sepharose beads, was incubated with a whole-cell lysate derived from 293 cells that had been labeled metabolically with [35S]methionine and [35S]cysteine. Bound protein was eluted sequentially with buffers containing NaCl at 150, 250, 350, and 450 mM concentrations. Elution profiles for GST and GST-RAG-2(439-527mt) were similar (data not shown). Beginning at 250 mM NaCl, a number of species were recovered from GST-RAG-2(439-527) that were not bound to GST-RAG-2(439-527mt); most of these proteins were eluted in the 250 and 350 mM washes (Fig. 1A).
FIG. 1.
Specific binding of the nuclear transport receptor Importin 5 to a site in the recombination-dispensable region of RAG-2. Agarose bead-bound GST-RAG-2(439-527) or GST-RAG-2(439-527mt) fusions were incubated with whole-cell lysates from 293 fibroblastoid, 63-12 pro-B, or A20 mature B cells, and bound proteins were eluted by sequential washes at increasing ionic strength. (A) Elution profiles of proteins retained by wild-type (wt) or mutant (mt) RAG-2 beads. Prior to lysis, 293 cells were labeled metabolically with [35S]methionine-cysteine. Sequential fractions eluting at 250 mM (lanes 1 to 3), 350 mM (lanes 4 to 6), and 450 mM (lanes 7 to 9) NaCl were fractionated by SDS-PAGE, and protein was detected using a phosphorimager (upper panel) or by silver staining (lower panel). Positions and sizes (in kilodaltons) of molecular markers are indicated at left. (B) Preparative gel electrophoresis of proteins from 293 cells eluting from mutant (mt) or wild-type (wt) RAG-2 beads at 250 mM (left) and 350 mM (right) NaCl. Protein was visualized with Coomassie blue. Arrows indicate species that were subjected to microsequence analysis (www.bs.jhmi.edu/mbg/RossEtAlAppendix.pdf). The species identified by sequence as Importin 5 is marked by an asterisk. Positions and sizes (in kilodaltons) of molecular markers are indicated at left. (C) Identification of Importin 5 among proteins from fibroblastoid and lymphoid cells that specifically bound to RAG-2. Proteins eluting from mutant (mt) or wild-type (wt) RAG-2 beads at 250 mM salt (250, left) or a sample of whole-cell lysate (wcl, right) were fractionated by SDS-PAGE, and Importin 5 was detected by immunoblotting.
Proteins bound specifically to GST-RAG-2(439-527) were isolated by preparative gel electrophoresis and identified by microsequencing (Fig. 1B; www.bs.jhmi.edu/mbg/RossEtAlAppendix.pdf). Among these was Importin 5, whose presence in the 250 mM eluate from GST-RAG-2(439-527) was confirmed by immunoblotting (Fig. 1C). Similar experiments were conducted with lysates derived from a pro-B-cell line (63-12) or from a mature B-cell line (A20). Elution profiles were similar for lysates from pro-B cells and mature B cells and resembled those for lysates derived from 293 cells (data not shown). Immunoblotting confirmed the presence of Importin 5 in pro-B and mature B cells and its specific binding to GST-RAG-2(439-527) (Fig. 1C). Importin 5 (karyopherin β3, RanBP5) is a Ran-binding protein related to importin β and functions as a mediator of nucleocytoplasmic transport (7, 18, 54). Identification of this protein suggested that the noncore region of RAG-2 contains a nuclear import signal and that this signal overlaps the interval mutated in GST-RAG-2(439-527mt).
A nuclear targeting signal near the C terminus of RAG-2.
RAG-2 localizes to the nucleus whether coexpressed with RAG-1 or expressed alone (47). While NLSs have been identified within RAG-1, the RAG-2 protein lacks a consensus NLS (47). To test whether the C-terminal region of RAG-2 could function as a nuclear targeting signal, chimeric polypeptides consisting of green-fluorescent protein (GFP) fused to the N terminus of full-length wild-type or mutant RAG-2 were constructed. GFP-fused RAG-1 and RAG-2 exhibited recombinational activities similar to their unfused counterparts (data not shown). Results obtained with GFP-fused RAG-2 were similar to those obtained by immunofluorescence using unfused RAG-2 and anti-RAG-2 antibodies (data not shown).
Expression of the full-length chimera, GFP-RAG-2(1-527), in NIH 3T3 cells confirmed that RAG-2 is localized to the nucleus, even in the absence of RAG-1, and that RAG-2, when overexpressed in fibroblastoid cells, is excluded from nucleoli (Fig. 2A, a to c; Table 1; also data not shown) (47). In contrast, deletion of the C-terminal 140 amino acid residues of RAG-2 [GFP-RAG-2(1-387)] or a smaller deletion of the C-terminal 29 amino acids [GFP-RAG-2(1-498)] resulted in partitioning of the chimera between the nucleus and the cytoplasm (Fig. 2A, g to l; Table 1). Removal of the C-terminal 10 residues of RAG-2 [GFP-RAG-2(1-517)] had a smaller debilitating effect on nuclear localization of the chimera (Fig. 2A, d to f; Table 1). Transfer of the C-terminal 89 amino acid residues of RAG-2 to GFP [GFP-RAG-2(439-527)] recapitulated the localization phenotype of the full-length chimera, suggesting that this domain is sufficient for nuclear import and retention (Fig. 2A, m to o; Table 1). These observations were confirmed for the mature B-cell line A20 and the RAG-2-deficient pro-B-cell line 63-12. In either cell line, the subcellular distribution of GFP-RAG-2(1-527), GFP-RAG-2(1-498), or GFP-RAG-2(439-527) was similar to that seen in NIH 3T3 cells (Fig. 2B and C; Tables 2 and 3).
FIG. 2.
Identification of an NLS in the RAG-2 recombination-dispensable region. (A) Fluorescence confocal microscopy of NIH 3T3 fibroblasts transiently transfected with constructs expressing GFP alone (panels p to r) or GFP fusions to portions of RAG-2 (panels a to o). RAG-2 amino acid residues contained within each fusion protein are indicated at right. A20 mature B cells (B) and 63-12 pro-B cells (C) are shown. (D) NIH 3T3 cells transfected with GFP-LacZ (147 kDa) or GFP-LacZ fused to the 89 carboxy-terminal amino acids of the RAG-2 protein. In all cases DAPI is used as a counterstain for the nucleus, and representative cells are shown. Numbers indicate the percentage of cells demonstrating localization similar to those shown and correspond to Tables 1 to 3.
TABLE 1.
Intracellular localization of wild-type and mutant RAG-2 proteins expressed in the absence of RAG-1 in NIH 3T3 cellsa
| Protein, segment, or mutation | Localization (% of cells)
|
|||
|---|---|---|---|---|
| N | N > C | N = C | C | |
| GFP | 0 | 11 | 89 | 0 |
| RAG-2 (wild-type) | 73 | 25 | 2 | 0 |
| 1-517 | 0 | 80 | 19 | 1 |
| 1-498 | 0 | 7 | 93 | 0 |
| 1-387 | 1 | 0 | 96 | 3 |
| 439-527 | 81 | 14 | 5 | 0 |
| 439/448A10 | 14 | 70 | 15 | 2 |
| 449/458A10 | 32 | 58 | 8 | 2 |
| 459/468A10 | 62 | 22 | 16 | 0 |
| 469/478A10 | 31 | 57 | 11 | 1 |
| 479/488A10 | 35 | 56 | 8 | 1 |
| 489/498A10 | 0 | 16 | 84 | 0 |
| 499/508A10 | 0 | 4 | 93 | 2 |
| 509/518A10 | 0 | 7 | 83 | 9 |
| 519/528A10 | 0 | 0 | 27 | 73 |
| 489/508A10 | 0 | 1 | 95 | 4 |
| K499A | 55 | 43 | 2 | 0 |
| M502A | 0 | 51 | 38 | 11 |
| K503A | 0 | 19 | 80 | 2 |
| KK507/508A2 | 0 | 20 | 68 | 13 |
| K499A; K503A; KK507/508A2 | 0 | 23 | 73 | 4 |
| KPPM499/502A4; SLH504/506A3 | 0 | 29 | 62 | 10 |
| K499A; K503R; KK507/508R2 | 0 | 38 | 54 | 8 |
| T490A | 84 | 15 | 1 | 0 |
| K492A | 40 | 51 | 9 | 0 |
| R493A | 21 | 72 | 7 | 0 |
| KR492/493A2 | 18 | 66 | 16 | 0 |
| NPPLQ494/498A5 | 0 | 63 | 30 | 7 |
| KRNNPLQ492/498A7 | 0 | 5 | 93 | 2 |
| K512A | 9 | 74 | 17 | 0 |
| K518A | 29 | 53 | 16 | 2 |
| K519A | 19 | 65 | 13 | 2 |
| KK518/519A2 | 0 | 59 | 34 | 7 |
| R523A | 1 | 79 | 20 | 0 |
| R524A | 50 | 43 | 7 | 0 |
| RR523/524A2 | 0 | 56 | 43 | 1 |
| KR492/493A2; K512A | 0 | 37 | 39 | 25 |
| KR492/493A2; K523A | 0 | 19 | 81 | 0 |
| LacZ | 0 | 0 | 18 | 82 |
| LacZ-RAG-2(439-527) | 67 | 29 | 4 | 0 |
N, nuclear; N > C, more nuclear than cytoplasmic; N = C, diffuse in the cell; C, cytoplasmic.
TABLE 2.
Intracellular localization of wild-type and mutant RAG-2 proteins expressed in 63-12 pro-B cellsa
| Protein or segment | Localization (% of cells)
|
|||
|---|---|---|---|---|
| N | N > C | N = C | C | |
| GFP | 0 | 15 | 85 | 0 |
| RAG-2 (wild type) | 87 | 11 | 2 | 0 |
| 1-498 | 0 | 20 | 80 | 0 |
| 439-527 | 94 | 6 | 0 | 0 |
See Table 1, footnote a, for explanation of categories.
TABLE 3.
Intracellular localization of wild-type and mutant RAG-2 proteins expressed in A20 mature B cellsa
| Protein or segment | Localization (% of cells)
|
|||
|---|---|---|---|---|
| N | N > C | N = C | C | |
| GFP | 0 | 13 | 87 | 0 |
| RAG-2 (wild type) | 89 | 11 | 0 | 0 |
| 1-498 | 0 | 22 | 77 | 1 |
| 439-527 | 95 | 5 | 0 | 0 |
See Table 1, footnote a, for explanation of categories.
The diffuse localization of GFP-RAG-2(1-387) and GFP-RAG-2(1-498) in the steady state could indicate inefficient import resulting in cytoplasmic accumulation, an imbalance in the nuclear shuttling of RAG-2 to favor cytoplasmic accumulation, or a combination of the two. Accordingly, the localization of RAG-2 and RAG-2 mutants was examined in the presence of the nuclear export inhibitor leptomycin B (53). Even at high concentrations of the drug (up to 10 ng/ml) and over extended periods of time (up to 10 h), no appreciable differences were seen in the localization of the full-length chimera or any of the GFP-tagged mutants, despite the nuclear accumulation of a positive control after 2 h at 5 ng/ml (data not shown). While these data indicate that RAG-2 is not exported via the CRM1 pathway in NIH 3T3 cells, export by another mechanism cannot be excluded.
Although residues 439 through 527 of RAG-2 appeared sufficient to support nuclear accumulation of GFP, it remained possible that the small size (37 kDa) of the GFP-RAG-2(439-527) protein had allowed passive diffusion into the nucleus, where it could have been retained (4). To confirm that the 439-527 segment of RAG-2 is capable of mediating active import, it was fused to β-galactosidase (LacZ), a large heterologous protein, and the fusion was tagged at the N terminus with GFP. The overall size of the GFP-LacZ segment is 147 kDa. While GFP-LacZ alone accumulates in the cytoplasm of NIH 3T3 cells, the addition of residues 439 through 527 of RAG-2 resulted in nuclear accumulation of the protein (Fig. 2D; Table 1).
Definition of a complex nuclear localization signal within RAG-2.
To delineate the region responsible for nuclear targeting of RAG-2, a series of clustered alanine mutations, scanning residues 439 through 527, was introduced into full-length GFP-RAG-2 fusion proteins and tested for the mutations' effects on subcellular localization (Fig. 3). Mutant proteins carrying alanine substitutions over the intervals 489 to 498, 499 to 508, and 509 to 518 were distributed between the nucleus and the cytoplasm, as was observed for GFP-RAG-2(1-387) and GFP-RAG-2(1-498) (Fig. 3f, g, and h). Mutation of the C-terminal nine residues, however, resulted in reduced expression and exclusion of the protein from the nucleus, consistent with instability due to misfolding (Fig. 3i).
FIG. 3.
Identification of a RAG-2 nuclear localization sequence by clustered alanine scanning mutagenesis. Full-length GFP-RAG-2 fusion proteins bearing the indicated alanine scanning mutations were transfected into NIH 3T3 cells and localized by fluorescence microscopy. The wild-type (wt) RAG-2 amino acid sequence is given at top for residues 439 through 527; for each mutant protein, designated at left by an index letter (a through j), the positions of alanine substitutions are indicated (upper panel). The distributions of DAPI, GFP, or merged DAPI and GFP fluorescence (overlay) are shown in the lower panel. Representative cells are shown. Letters (a to j) correspond to individual mutant proteins as defined in the upper panel. Numbers in upper right corners of the overlay series indicate the percentages of cells with localization patterns similar to those shown.
The nuclear targeting signal was defined further by point mutagenesis (Table 1). Because the 499-508 region of RAG-2 bound Importin 5, and because basic residues are commonly found in nuclear targeting signals, we began by mutating positively charged residues in this region. Residues K503 and K507 or K508 are necessary for normal localization, with K507 and K508 being redundant (Table 1 and data not shown). Mutation of the phylogenetically conserved residue M502 had a partial effect on localization, suggesting that structure as well as charge density in this region may be important for proper nuclear targeting. In accordance with this, mutation of all residues in the 499-508 interval except for the essential lysines resulted in diffuse localization. Mutations in the 489-498 region of the RAG-2 protein also uncovered requirements for charge and structure, since the mutants of KR492/493AA and NPPLQ494/498A5 were partially redistributed to the cytoplasm (Table 1).
RAG-1 and RAG-2 can cooperate in localization of the V(D)J recombinase.
The RAG-1 and RAG-2 proteins associate in vitro in the absence of accessory factors (49) and can be coimmunoprecipitated from cell lysates (1, 12, 26, 34, 47). Thus, during the G0 and G1 cell cycle phases, when both proteins are present, RAG-1 and RAG-2 are expected to exist as a complex (24, 27, 30, 31). Because RAG-1 is localized to the nucleus even in the absence of RAG-2 (47), we tested the ability of wild-type RAG-1 to override the nuclear localization defects of the RAG-2 mutants described above. Conversely, we asked whether wild-type RAG-2 could support the nuclear accumulation of RAG-1 mutants defective in nuclear localization.
Wild-type or mutant RAG-1 proteins were tagged with yellow fluorescent protein (YFP) and coexpressed in NIH 3T3 cells with wild-type or mutant RAG-2 proteins, tagged with cyan fluorescent protein (CFP). In agreement with previous results (47), wild-type RAG-1 in the absence of RAG-2 was predominantly nuclear and concentrated in the nucleoli (Fig. 4A). When RAG-1 was coexpressed with wild-type RAG-2 or with RAG-2 mutants defective in nuclear localization, RAG-1 remained mostly nuclear, albeit slightly more cytoplasmic in the presence of RAG-2 NLS mutants, and was diffusely distributed in the nucleus. Moreover, in the presence of RAG-1, the RAG-2 mutants accumulated in the nucleus (Fig. 4A; www.bs.jhmi.edu/mbg/RossEtAlAppendix.pdf).
FIG.4.
RAG-1 and RAG-2 cooperate in localizing the V(D)J recombinase. NIH 3T3 cells were transfected with CFP-RAG-2 constructs (green pseudo-color) and YFP-RAG-1 constructs (red pseudo-color) and assayed by fluorescence microscopy. Representative cells are shown. (A) Wild-type YFP-RAG-1 was transfected either alone or in combination with CFP-fused RAG-2 proteins. DAPI, RAG-1 (YFP), or merged DAPI and YFP signals are displayed for transfection with RAG-1 alone (top panel). Coexpression patterns of YFP-RAG-1 and CFP fusions containing full-length RAG-2(1-527) or RAG-2(499/508A10) are displayed in the lower panel. In each case, RAG-2, RAG-1, and merged signals are shown. Numbers indicate the percentages of cells positive for RAG-1 and RAG-2 and demonstrating RAG-2 localization patterns similar to those shown. The graph at right displays the localization pattern of RAG-2(499/508A10) in the presence (solid bars) or absence (hatched bars) of wild-type RAG-1. (B) Transfection of YFP-RAG-1(BV) alone (top panel) or with CFP-RAG-2 (lower panel), displayed as in panel A. Numbers indicate the percentages of cells positive for RAG-1 and RAG-2 and demonstrating RAG-2 localization patterns similar to those shown. The graph at right displays localization pattern of RAG-2(499/508A10) in the presence (solid bars) or absence (hatched bars) of RAG-1(BV). (C) Transfection of YFP-RAG-1(1/2BIV) either alone (top panel) or with CFP fused to wild-type RAG-2 (lower panel), displayed as in panel A. Numbers indicate the percentages of cells positive for RAG-1 and RAG-2 and demonstrating RAG-1 localization patterns similar to those shown. The graph at right displays localization pattern of RAG-1(1/2 BIV) in the presence (solid bars) or absence (hatched bars) of wild-type RAG-2.
Several RAG-1 mutants defective in nuclear localization have been described (47); among these are 1/2 BIV (K835I, R838L, K839I, R840L, and K844I) and BV (RR969/970IL and RK972/973LI). As previously reported (47), these mutants, when expressed in the absence of RAG-2, exhibited nuclear localization defects (Fig. 4B and C; www.bs.jhmi.edu/mbg/RossEtAlAppendix.pdf). The BV mutation impaired the ability of RAG-1 to support nuclear accumulation of RAG-2 localization mutants (Fig. 4B and Table A2), while coexpression of wild-type RAG-2 with RAG-1 BV resulted in relocalization of the mutant RAG-1 protein to the nucleus (Fig. 4B; www.bs.jhmi.edu/mbg/RossEtAlAppendix.pdf). Interestingly, RAG-2 nuclear localization mutants were also able to support relocalization of RAG-1 BV to the nucleus, suggesting that this property is independent of the nuclear localization sequence that we had mapped to the RAG-2 carboxy-terminal region (Fig. 4B; www.bs.jhmi.edu/mbg/RossEtAlAppendix.pdf). In contrast, wild-type RAG-2 failed to support nuclear redistribution of the RAG-1 1/2 BIV mutant (Fig. 4C; www.bs.jhmi.edu/mbg/RossEtAlAppendix.pdf), and coexpression of localization-defective RAG-2 mutants with RAG-1 1/2 BIV had no effect on the distribution of either protein (data not shown), suggesting that association of RAG-1 with RAG-2 may be impaired by the 1/2 BIV mutation.
Cell cycle-dependent impairment of V(D)J recombination by selective mutation of the RAG-2 nuclear localization sequence.
Experiments described above identified a nuclear localization signal within the C-terminal 40 amino acid residues of RAG-2. Mutations that abolish cell cycle-specific regulation of RAG-2 accumulation (27) or that impair endogenous VH to DJH joining (2, 21, 28) have also been mapped to this region, as have mutations that reduce the overall efficiency of rearrangement in assays employing extrachromosomal substrates (48). The RAG-2 degradation and localization phenotypes are, however, separable: the RAG-2(K503A) and RAG-2(492/498)A7 mutants, for example, exhibit defective localization but normal degradation at the G1-S boundary (Table 1) (27). This allowed us to assess the potential contribution of the RAG-2 NLS to the efficiency of V(D)J recombination independently of any effect on periodic degradation. The recombination activities of RAG-2(K503A) and RAG-2(492/498)A7 were compared to those of wild-type RAG-2 or of the RAG-2(499/508)A10 and RAG-2(489/498)A10 mutants, which are defective for both nuclear localization and degradation (Fig. 5). Recombination frequencies were measured in transfected NIH 3T3 cells, whose cell cycle distribution is responsive to altered cell density. In subconfluent, cycling cells, RAG-2(499/508)A10 and RAG-2(489/498)A10 supported normal levels of recombination with signal or coding joint substrates (Fig. 5A and C and data not shown), as was expected from the ability of RAG-1 to direct nuclear localization of RAG-2 NLS mutants. In contrast, recombination frequencies were reproducibly decreased by about twofold in cycling cells expressing RAG-2(K503A) or RAG-2(492/498)A7 (Fig. 5A and C and data not shown). Notably, the decrease in recombination frequency was positively correlated with diminished ability of RAG-1 to rescue the nuclear localization of the RAG-2(K503A) and RAG-2(492/498)A7 mutants (Fig. 6B and D).
FIG. 5.
Cell cycle-dependent impairment of V(D)J recombination by selective mutation of the RAG-2 nuclear localization sequence. Cells were transfected at either 20% confluency (A and C) or 90% confluency (B and D) with wild-type RAG-1, the indicated RAG-2 constructs, and the extrachromosomal substrate pJH200. At 36 h after transfection, cells were collected and assayed for rearrangement (A and B) or for DNA content by fluorescence-activated cell sorting (C and D). Complete data for recombination assays are listed at www.bs.jhmi.edu/mbg/RossEtAlAppendix.pdf. Representative fluorescence-activated cell sorter profiles are shown. Numbers indicate the percentages of cells with 2C DNA content.
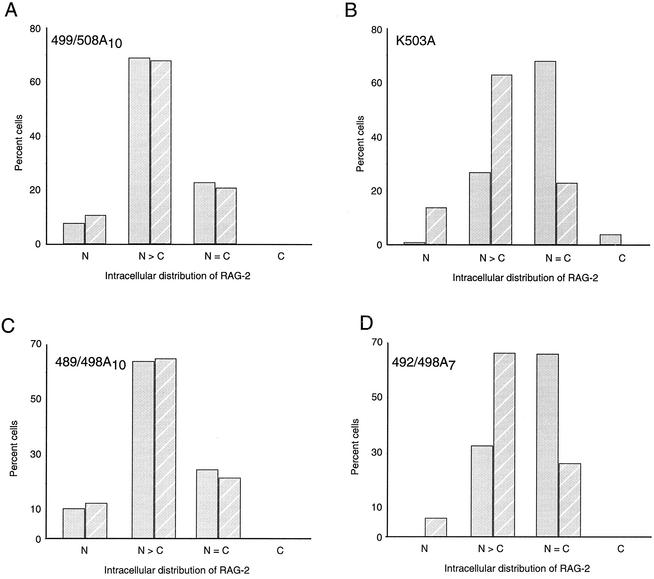
FIG. 6.
Incomplete rescue of RAG-2 localization by RAG-1 in cycling cells. NIH 3T3 cells were cotransfected at either 20% (solid bars) or 90% (hatched bars) confluency with wild-type YFP-RAG-1 and CFP-RAG-2(499/508A10) (A), CFP-RAG-2(K503A) (B), CFP-RAG-2(489/498A10) (C), or CFP-RAG-2(492/498A7) (D). Cells expressing both RAG-1 and RAG-2 were scored by fluorescence microscopy for the intracellular distribution of RAG-2 protein.
The behavior of RAG-2(K503A) and RAG-2(492/498)A7, in contrast to that of RAG-2 (499/508)A10 and RAG-2(489/498)A10, suggested that the NLS that we had identified in the noncore region of RAG-2 might play a specific role in reconcentrating the protein in the nucleus during its reaccumulation in the G1 cell cycle phase. We reasoned that if this were correct, then the debilitating effects of the K503A or (492/498)A7 mutation would exhibit cell cycle dependence. Indeed, under conditions of contact inhibition, when the G0/G1 fraction is markedly increased, RAG-2(K503A), RAG-2(492/498)A7, and wild-type RAG-2 supported similar frequencies of recombination (Fig. 5B and D). Moreover, under these conditions the ability of RAG-1 to rescue nuclear localization of RAG-2(K503A) or RAG-2(492/498)A7 was similar to that seen with the RAG-2(499/508)A10 and RAG-2(489/498)A10 mutants (Fig. 6B and D). In accordance with its constitutive accumulation in the cell cycle, no cell cycle-dependent change in recombination frequency compared to wild-type (Fig. 5) or nuclear localization (Fig. 6A and C) was observed for the RAG-2(499/508)A10 or RAG-2(489/498)A10 mutant.
DISCUSSION
The V(D)J recombinase components RAG-1 and RAG-2 are discordantly regulated at the posttranscriptional level. While the amount of RAG-1 protein varies little as a function of cell cycle phase, RAG-2 is destroyed and resynthesized at each cell division. The periodic destruction of RAG-2 is initiated at the G1-to-S transition by cyclin A/CDK2, which phosphorylates a conserved sequence motif in the noncore region of the protein, thereby triggering proteolysis (24, 27, 30, 31). Consequently, RAG activity is restricted to the G0 and G1 cell cycle phases.
We now describe a second functional site within the noncore region of RAG-2, overlapping the interval required for regulated degradation, which binds the nuclear transport protein Importin 5 and which serves as a RAG-1-independent nuclear import signal. The presence of a separate, intrinsic NLS may ensure the efficient nuclear uptake of RAG-2 as this protein reaccumulates in G1. The distinct behaviors of two classes of RAG-2 NLS mutants are consistent with this proposal. Mutations that simultaneously disrupt nuclear import and periodic degradation exhibit no reduction in the frequency of V(D)J recombination. In contrast, ablation of the RAG-2 NLS in the context of an intact degradation signal impairs V(D)J recombination in cycling cells but not in cells that have been induced to enter quiescence.
An intrinsic nuclear import signal in the noncore region of RAG-2.
Importin 5 is a member of a family of nuclear import receptors that interact with complex NLSs (13). Specific binding of Importin 5 to a site near the carboxyl terminus of RAG-2 suggested the presence of a nuclear import signal in this region. This was confirmed by a combination of confocal fluorescence microscopy and mutational analysis, which localized an NLS to an interval spanning amino acid residues 492 to 524 of RAG-2. While this region is rich in basic residues, positive charge was not sufficient to direct nuclear localization, as evidenced by the debilitating effects of nonconservative mutations at uncharged positions (M502A, KPPM499/502A4; SLH504/506A3, and NPPQL494/498A5) or of conservative mutations at basic positions (K499A, K503R, and KK507/508R2).
When expressed in the absence of RAG-1, RAG-2 mutants lacking the carboxy-terminal NLS are not excluded from the nucleus but rather are distributed between the nucleus and the cytoplasm. This suggests that the core region of RAG-2 may contain an additional, weakly functional NLS or mediate association with an imported protein other than RAG-1. During the course of the studies we describe here, others reported that the carboxy-terminal 37 amino acids of RAG-2 could support its nuclear accumulation, although the determinants of nuclear localization were not defined (5). Furthermore, while removal of this region was reported to result in nuclear exclusion, this phenomenon seems likely to be peculiar to the 293 cell line. Although we noted exclusion of RAG-2 NLS mutants from the nuclei of 293 cells (data not shown), we observed partitioning of these mutants between the nucleus and the cytoplasm in B lymphoid cell lines, as well as in HeLa and NIH 3T3 cells.
The specific binding of Importin 5 to residues in the C-terminal NLS of RAG-2 is consistent with a role for this interaction in nuclear import. We note, in addition, that nuclear import receptors can also act as chaperones, preventing cytoplasmic aggregation of nuclear proteins (19). In accordance with this possibility, we observed cytoplasmic aggregation of RAG-2 nuclear localization mutants in 293 cells (data not shown).
Although additional proteins were also associated, directly or indirectly, with the RAG-2 C-terminal region (www.bs.jhmi.edu/mbg/RossEtAlAppendix.pdf), we chose to focus our initial attention on Importin 5 because its identity suggested hypotheses that were immediately testable. Nonetheless, the other associated proteins, some of which have been suggested to play roles in chromatin reorganization and transcriptional regulation, are interesting in light of recent findings concerning potential regulatory functions of the RAG-2 noncore region at the immunoglobulin heavy chain and T-cell receptor β loci (2, 21, 28).
Contributions of RAG-1 and RAG-2 to localization of V(D)J recombinase.
The presence of distinct nuclear localization signals in RAG-1 and RAG-2 suggested the existence of two pathways for nuclear import of the V(D)J recombinase, involving interaction of RAG-1 with the importin α/β receptor or of RAG-2 with Importin 5.
We observed that RAG-2 mutants defective in intrinsic nuclear import can be directed to the nucleus when expressed together with RAG-1. This property of RAG-1 was abolished by mutation of its NLS and therefore appears to be mediated in trans by the NLS of RAG-1 in complex with RAG-2. In contrast, both wild-type RAG-2 and RAG-2 nuclear localization mutants supported partial rescue of the RAG-1 nuclear localization mutant BV. The inability of RAG-2 to rescue the RAG-1 localization mutant 1/2BIV is consistent with impaired association of RAG-1(1/2BIV) with RAG-2. In a recent report, mutation of RAG-1 at residues R838, K839, and R840 reduced the formation of signal ends, signal joints, and particularly coding joints in vivo but had relatively little effect on DNA binding, DNA cleavage, and hairpin formation in vitro (17). On this basis, as well as on the basis of the ability of certain substrate conformations to suppress the coding joint defect, it was proposed that mutation of R838, K839, and R840 destabilized a postcleavage complex between RAG proteins and DNA (17). Alternatively or in addition, our results suggest that this mutation may weaken the specific association of RAG-1 with RAG-2 in vivo, thereby contributing to the observed decreases in DNA cleavage and rejoining.
A role for an intrinsic RAG-2 NLS during the periodic accumulation of RAG-2 protein.
We were unable to determine the contribution, if any, of the intrinsic RAG-1 NLS to V(D)J recombination activity in vivo because of functional overlap between sequence intervals that mediate nuclear localization, specific DNA binding and, possibly, association with RAG-2 (data not shown) and (17). In contrast, the determinants of RAG-2 nuclear localization and periodic degradation do not overlap regions of the protein essential for recombination. A lysine-to-alanine mutation of RAG-2 at position 503 inactivated the NLS without perturbing cell cycle-dependent degradation as did an alanine scanning mutation in the region spanning residues 492 through 498 of RAG-2, which spared the cyclin A/CDK2 phosphorylation site. In cycling cells, the RAG-2(K503A) and RAG-2(492/498)A7 mutations were associated with a decrease in recombination frequency as assessed using extrachromosomal substrates. The conservative nature of these mutations, and the ability of quiescence to rescue their recombination defects, indicated that the K503A and (492/498)A7 phenotypes do not result from an overall structural defect such as might result from misfolding. Decreased recombination was positively correlated with reduced ability of RAG-1 to support nuclear localization of RAG-2(K503A) or RAG-2(492/498)A7. This suggests that cotransport with RAG-1 is insufficient to support nuclear reaccumulation of RAG-2 under conditions of periodic destruction.
Our observations suggest a simple model. In quiescent cells the amount of RAG-2 protein does not fluctuate and RAG-2 is cotransported into the nucleus with RAG-1. In cycling cells, RAG-2 is degraded at the G1-S boundary and reaccumulates as cells reenter G1. We propose that under these conditions the nuclear import of RAG-1 may proceed at a rate that is insufficient to accommodate the rapid reaccumulation of RAG-2. Import of RAG-2 by means of its intrinsic NLS, either alone or in association with RAG-1, would permit nuclear import of RAG-2 at a rate greater than that supported by the NLS of RAG-1. Under this model, the RAG-2(K503A) and RAG-2(492/498)A7 phenotypes can be rationalized as follows. In cycling cells, as these mutants reaccumulate, their rate of nuclear import is limited by that of RAG-1; inefficient nuclear import is associated with a relative decrease in recombination frequency. In quiescent cells, RAG-2(K503A) and RAG-2(492/498)A7, like wild-type RAG-2, are expected to accumulate at an approximately constant rate; we propose that their nuclear import under these conditions can be accommodated by cotransport with RAG-1. The extensive overlap between determinants of cell cycle-dependent degradation and nuclear localization in the noncore region of RAG-2 provokes the speculation that these two functions were acquired together during evolution of the V(D)J recombinase.
Acknowledgments
We thank Michael Cooper and Joshua Mendell for technical advice; Nini Guo and Claire Letourneux for technical assistance; Jeffrey Han, Jef Boeke, Joshua Mendell, Hal Dietz, and Eugene Oltz for reagents; the staff of the Johns Hopkins Biopolymers Facility for synthesis of oligonucleotides; and our colleagues in the Department of Molecular Biology and Genetics for stimulating discussions.
This work was supported by the Howard Hughes Medical Institute and by grant CA16519 from the National Cancer Institute. A.E.R. is a predoctoral fellow of the Medical Scientist Training Program of the NIH.
REFERENCES
- 1.Aidinis, V., D. C. Dias, C. A. Gomez, D. Bhattacharyya, E. Spanopoulou, and S. Santagata. 2000. Definition of minimal domains of interaction within the recombination-activating genes 1 and 2 recombinase complex. J. Immunol. 164:5826-5832. [DOI] [PubMed]
- 2.Akamatsu, Y., R. Monroe, D. D. Dudley, S. K. Elkin, F. Gartner, S. R. Talukder, Y. Takahama, F. W. Alt, C. H. Bassing, and M. A. Oettinger. 2003. Deletion of the RAG2 C terminus leads to impaired lymphoid development in mice. Proc. Natl. Acad. Sci. USA 100:1209-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akamatsu, Y., and M. S. Oettinger. 1998. Distinct roles of RAG1 and RAG2 in binding the V(D)J recombination signal sequences. Mol. Cell. Biol. 18:4670-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonner, W. M. 1978. Protein migration and accumulation in nuclei, p. 97-148. In H. Busch (ed.), The cell nucleus, vol. 6. Academic, New York, N.Y.
- 5.Corneo, B., A. Benmerah, and J. P. de Villartay. 2002. A short peptide at the C terminus is responsible for the nuclear localization of RAG2. Eur. J. Immunol. 32:2068-2073. [DOI] [PubMed] [Google Scholar]
- 6.Cuomo, C. A., and M. A. Oettinger. 1994. Analysis of regions of RAG-2 important for V(D)J recombination. Nucleic Acids Res. 22:1810-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deane, R., W. Schafer, H. P. Zimmermann, L. Mueller, D. Gorlich, S. Prehn, H. Ponstingl, and F. R. Bischoff. 1997. Ran-binding protein 5 (RanBP5) is related to the nuclear transport factor importin-beta but interacts differently with RanBP1. Mol. Cell. Biol. 17:5087-5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eastman, Q. M., T. M. Leu, and D. G. Schatz. 1996. Initiation of V(D)J recombination in vitro obeying the 12/23 rule. Nature 380:85-88. [DOI] [PubMed] [Google Scholar]
- 9.Fugmann, S. D., and D. G. Schatz. 2001. Identification of basic residues in RAG2 critical for DNA binding by the RAG1-RAG2 complex. Mol. Cell 8:899-910. [DOI] [PubMed] [Google Scholar]
- 10.Fugmann, S. D., I. J. Villey, L. M. Ptaszek, and D. G. Schatz. 2000. Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol. Cell 5:97-107. [DOI] [PubMed] [Google Scholar]
- 11.Gellert, M. 2002. V(D)J recombination: RAG proteins, repair factors and regulation. Annu. Rev. Biochem. 71:101-132. [DOI] [PubMed] [Google Scholar]
- 12.Gomez, C. A., L. M. Ptaszek, A. Villa, F. Bozzi, C. Sobacchi, E. G. Brooks, L. D. Notarangelo, E. Spanopoulou, Z. Q. Pan, P. Vezzoni, P. Cortes, and S. Santagata. 2000. Mutations in conserved regions of the predicted RAG2 kelch repeats block initiation of V(D)J recombination and result in primary immunodeficiencies. Mol. Cell. Biol. 20:5653-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorlich, D., and U. Kutay. 1999. Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol. 15:607-660. [DOI] [PubMed] [Google Scholar]
- 14.Grawunder, U., R. B. West, and M. R. Lieber. 1998. Antigen receptor gene rearrangement. Curr. Opin. Immunol. 10:172-180. [DOI] [PubMed] [Google Scholar]
- 15.Hesse, J. E., M. R. Lieber, M. Gellert, and K. Mizuuchi. 1987. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D)-J joining signals. Cell 49:775-783. [DOI] [PubMed] [Google Scholar]
- 16.Hiom, K., and M. Gellert. 1997. A stable RAG-1-RAG2-DNA complex that is active in V(D)J cleavage. Cell 88:65-72. [DOI] [PubMed] [Google Scholar]
- 17.Huye, L. E., M. M. Purugganan, M. M. Jiang, and D. B. Roth. 2002. Mutational analysis of all conserved basic amino acids in RAG-1 reveals catalytic, step arrest, and joining-deficient mutants in the V(D)J recombinase. Mol. Cell. Biol. 22:3460-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jakel, S., and D. Gorlich. 1998. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 17:4491-4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jakel, S., J. M. Mingot, P. Schwarzmaier, E. Hartmann, and D. Gorlich. 2002. Importins fulfill a dual function as nuclear import receptors and cytoplasmic chaperones for exposed basic domains. EMBO J. 21:377-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, D. R., Y. Dai, C. L. Mundy, W. Yang, and M. A. Oettinger. 1999. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 13:3070-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirch, S. A., G. A. Rathbun, and M. A. Oettinger. 1998. Dual role of RAG2 in V(D)J recombination: catalysis and regulation of ordered Ig gene assembly. EMBO J. 17:4881-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirch, S. A., P. Sudarsanam, and M. A. Oettinger. 1996. Regions of RAG1 protein critical for V(D)J recombination. Eur. J. Immunol. 26:886-891. [DOI] [PubMed] [Google Scholar]
- 23.Landree, M. A., J. A. Wibbenmeyer, and D. B. Roth. 1999. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 13:3059-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee, J., and S. Desiderio. 1999. CyclinA/CDK2 regulates V(D)J recombination by coordinating RAG-2 accumulation and DNA repair. Immunity 11:771-781. [DOI] [PubMed] [Google Scholar]
- 25.Leu, T. M., Q. M. Eastman, and D. G. Schatz. 1997. Coding joint formation in a cell-free V(D)J recombination system. Immunity 7:303-314. [DOI] [PubMed] [Google Scholar]
- 26.Leu, T. M., and D. G. Schatz. 1995. rag-1 and rag-2 are components of a high-molecular-weight complex, and association of rag-2 with this complex is rag-1 dependent. Mol. Cell. Biol. 15:5657-5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li, Z., D. I. Dordai, J. Lee, and S. Desiderio. 1996. A conserved degradation signal regulates RAG-2 accumulation during cell division and links V(D)J recombination to the cell cycle. Immunity 5:575-589. [DOI] [PubMed] [Google Scholar]
- 28.Liang, H. E., L. Y. Hsu, D. Cado, L. G. Cowell, G. Kelsoe, and M. S. Schlissel. 2002. The “dispensable” portion of RAG2 is necessary for efficient V-to-DJ rearrangement during B and T cell development. Immunity 17:639-651. [DOI] [PubMed] [Google Scholar]
- 29.Lieber, M. R., J. E. Hesse, S. Lewis, G. C. Bosma, N. Rosenberg, K. Mizuuchi, M. J. Bosma, and M. Gellert. 1988. The defect in murine severe combined immune deficiency: joining of signal sequences but not coding segments in V(D)J recombination. Cell 55:7-16. [DOI] [PubMed] [Google Scholar]
- 30.Lin, W. C., and S. Desiderio. 1994. Cell cycle regulation of V(D)J recombination-activating protein RAG-2. Proc. Natl. Acad. Sci. USA 91:2733-2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, W. C., and S. Desiderio. 1993. Regulation of V(D)J recombination activator protein RAG-2 by phosphorylation. Science 260:953-959. [DOI] [PubMed] [Google Scholar]
- 32.McBlane, J. F., D. C. van Gent, D. A. Ramsden, C. Romeo, C. A. Cuomo, M. Gellert, and M. A. Oettinger. 1995. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell 83:387-395. [DOI] [PubMed] [Google Scholar]
- 33.McMahan, C. J., M. J. Difilippantonio, N. Rao, E. Spanopoulou, and D. G. Schatz. 1997. A basic motif in the N-terminal region of RAG1 enhances V(D)J recombination activity. Mol. Cell. Biol. 17:4544-4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMahan, C. J., M. J. Sadofsky, and D. G. Schatz. 1997. Definition of a large region of RAG1 that is important for coimmunoprecipitation of RAG2. J. Immunol. 158:2202-2210. [PubMed] [Google Scholar]
- 35.Mo, X., T. Bailin, and M. J. Sadofsky. 1999. RAG1 and RAG2 cooperate in specific binding to the recombination signal sequence in vitro. J. Biol. Chem. 274:7025-7031. [DOI] [PubMed] [Google Scholar]
- 36.Moshous, D., I. Callebaut, R. de Chasseval, B. Corneo, M. Cavazzana-Calvo, F. Le Deist, I. Tezcan, O. Sanal, Y. Bertrand, N. Philippe, A. Fischer, and J. P. de Villartay. 2001. Artemis, a novel DNA double-strand break repair/V(D)J recombination protein, is mutated in human severe combined immune deficiency. Cell 105:177-186. [DOI] [PubMed] [Google Scholar]
- 37.Oettinger, M. A., D. G. Schatz, C. Gorka, and D. Baltimore. 1990. RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science 248:1517-1523. [DOI] [PubMed] [Google Scholar]
- 38.Qiu, J. X., S. B. Kale, H. Yarnell Schultz,, and D. B. Ross. 2001. Separation-of-function mutants reveal critical roles for RAG2 in both the cleavage and joining steps of V(D)J recombination. Mol. Cell 7:77-87. [DOI] [PubMed] [Google Scholar]
- 39.Ramsden, D. A., T. T. Paull, and M. Gellert. 1997. Cell-free V(D)J recombination. Nature 388:488-491. [DOI] [PubMed] [Google Scholar]
- 40.Roman, C. A., S. R. Cherry, and D. Baltimore. 1997. Complementation of V(D)J recombination deficiency in RAG-1(−/−) B cells reveals a requirement for novel elements in the N-terminus of RAG-1. Immunity 7:13-24. [DOI] [PubMed] [Google Scholar]
- 41.Sadofsky, M. J., J. E. Hesse, and M. Gellert. 1994. Definition of a core region of RAG-2 that is functional in V(D)J recombination. Nucleic Acids Res. 22:1805-1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sadofsky, M. J., J. E. Hesse, J. F. McBlane, and M. Gellert. 1993. Expression and V(D)J recombination activity of mutated RAG-1 proteins. Nucleic Acids Res. 21:5644-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schatz, D. G., M. A. Oettinger, and D. Baltimore. 1989. The V(D)J recombination activating gene, RAG-1. Cell 59:1035-1048. [DOI] [PubMed] [Google Scholar]
- 44.Sekiguchi, J. A., S. Whitlow, and F. W. Alt. 2001. Increased accumulation of hybrid V(D)J joints in cells expressing truncated versus full-length RAGs. Mol. Cell 8:1383-1390. [DOI] [PubMed] [Google Scholar]
- 45.Shinkai, Y., G. Rathbun, K. P. Lam, E. M. Oltz, V. Stewart, M. Mendelsohn, J. Charron, M. Datta, F. Young, A. M. Stall, and F. W. Alt. 1992. RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell 68:855-867. [DOI] [PubMed] [Google Scholar]
- 46.Silver, D. P., E. Spanopoulou, R. C. Mulligan, and D. Baltimore. 1993. Dispensable sequence motifs in the RAG-1 and RAG-2 genes for plasmid V(D)J recombination. Proc. Natl. Acad. Sci. USA 90:6100-6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spanopoulou, E., P. Cortes, C. Shih, C. M. Huang, D. P. Silver, P. Svec, and D. Baltimore. 1995. Localization, interaction, and RNA binding properties of the V(D)J recombination-activating proteins RAG1 and RAG2. Immunity 3:715-726. [DOI] [PubMed] [Google Scholar]
- 48.Steen, S. B., J. O. Han, C. Mundy, M. A. Oettinger, and D. B. Roth. 1999. Roles of the “dispensable” portions of RAG-1 and RAG-2 in V(D)J recombination. Mol. Cell. Biol. 19:3010-3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swanson, P. C., and S. Desiderio. 1999. RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol. Cell. Biol. 19:3674-3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swanson, P. C., and S. Desiderio. 1998. V(D)J recombination signal recognition: distinct, overlapping DNA-protein contacts in complexes containing RAG1 with and without RAG2. Immunity 9:115-125. [DOI] [PubMed] [Google Scholar]
- 51.van Gent, D. C., J. F. McBlane, D. A. Ramsden, M. J. Sadofsky, J. E. Hesse, and M. Gellert. 1995. Initiation of V(D)J recombination in a cell-free system. Cell 81:925-934. [DOI] [PubMed] [Google Scholar]
- 52.van Gent, D. C., D. A. Ramsden, and M. Gellert. 1996. The RAG1 and RAG2 proteins establish the 12/23 rule in V(D)J recombination. Cell 85:107-113. [DOI] [PubMed] [Google Scholar]
- 53.Wolff, B., J. J. Sanglier, and Y. Wang. 1997. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem. Biol. 4:139-147. [DOI] [PubMed] [Google Scholar]
- 54.Yaseen, N. R., and G. Blobel. 1997. Cloning and characterization of human karyopherin beta3. Proc. Natl. Acad. Sci. USA 94:4451-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]