Abstract
The association between histone acetylation and replacement observed during spermatogenesis prompted us to consider the testis as a source for potential factors capable of remodelling acetylated chromatin. A systematic search of data banks for open reading frames encoding testis-specific bromodomain-containing proteins focused our attention on BRDT, a testis-specific protein of unknown function containing two bromodomains. BRDT specifically binds hyperacetylated histone H4 tail depending on the integrity of both bromodomains. Moreover, in somatic cells, the ectopic expression of BRDT triggered a dramatic reorganization of the chromatin only after induction of histone hyperacetylation by trichostatin A (TSA). We then defined critical domains of BRDT involved in its activity. Both bromodomains of BRDT, as well as flanking regions, were found indispensable for its histone acetylation-dependent remodelling activity. Interestingly, we also observed that recombinant BRDT was capable of inducing reorganization of the chromatin of isolated nuclei in vitro only when the nuclei were from TSA-treated cells. This assay also allowed us to show that the action of BRDT was ATP independent, suggesting a structural role for the protein in the remodelling of acetylated chromatin. This is the first demonstration of a large-scale reorganization of acetylated chromatin induced by a specific factor.
The histone code hypothesis postulating a role for posttranslational modifications of histones in the control of chromatin structure and function (39, 41) is now supported by a large body of data. The milestone of this hypothesis is the existence of protein domains capable of specifically recognizing particular posttranslational histone modifications (22). To date, at least two distinct domains have been shown to interact with modified histones: bromodomains, recognizing acetylated lysines (43), and some chromodomains, interacting with methylated lysine 9 present in the histone H3 N-terminal tail (2, 19, 20, 25, 40).
Bromodomains are conserved modules present in many chromatin- and transcription-related proteins, including histone acetyltransferases and chromatin remodelling factors (11). The first bromodomain shown to specifically interact with an acetylated peptide, corresponding to the histone H4 N terminus, was that of P/CAF (9). Further investigations showed that bromodomains can also recognize acetylated lysines in nonhistone proteins and play an essential role in establishing new acetylation-dependent functions (10, 33, 37). Bromodomain-containing proteins are therefore likely to interpret signals generated by protein acetylation and, more specifically in the case of chromatin, mediate functions depending on histone acetylation (1, 16, 26, 31).
Interestingly, there is at least one physiological situation where histone acetylation can unambiguously be linked to chromatin remodelling. In many species, including the fly, trout, rooster, rat, mouse, and human, male germ cell nuclear maturation is characterized by the hyperacetylation of histones followed by their replacement by testis-specific basic proteins (4, 5, 15, 17, 32, 34). Since the hyperacetylation of histones is frequently observed just before or simultaneously with their replacement, it has been proposed that it could serve as a signal for histone replacement. We therefore considered the testis as a source of factors capable of modifying the structure of chromatin in an acetylation-dependent manner. Accordingly, bromodomain-containing proteins specifically expressed in the testis were the first candidates. The search of databases for testis-specific bromodomain-containing proteins allowed the identification of BRDT (named for bromodomain testis specific), a human testis-specific protein of unknown function containing two canonical bromodomains. We cloned the murine cDNA encoding this protein and confirmed its testis-specific expression. Very interestingly, in somatic cells, although it had no dramatic effect on the organization of chromatin, the protein was capable of inducing, in vivo and in vitro, spectacular chromatin remodelling when histone hyperacetylation was induced. This acetylation-dependent chromatin remodelling and the capacity of the protein to interact with the acetylated N-terminal tail of histone H4 were dependent on the integrity of both bromodomains and their flanking regions. Therefore, this work shows for the first time the ability of a specific bromodomain-containing protein to induce a large-scale acetylation-dependent chromatin reorganization.
MATERIALS AND METHODS
Search for bromodomain-containing testis-specific proteins.
dbEST and dbEST Cumulative Updates were searched using the bromodomain region of GCN5 as a query. In order to rapidly identify interesting expressed sequence tags (ESTs) (those presenting the best homology to the query), we designed a computer program to treat the raw data and to set up a schematic representation of homologous ESTs considering all homology parameters (see below). For each EST, both the homology score and the number of homologous subfragments (n) and p(n) (see BLAST server) were considered. When n was greater than 1, union of the fragment showing the highest score (the highest scoring segment pair) and other segment pairs (lower scoring pair) was performed. To obtain the most significant possible representation, ESTs having a high score of greater than 100 were listed. Finally, the ESTs were sorted according to their origin, and only ESTs present in libraries from testis but absent from other libraries were retained.
Cloning of murine BRDT cDNA.
Several primer pairs, designed from murine ESTs homologous to the human BRDT sequence (National Center for Biotechnology Information accession number NM_001726) were used to amplify three overlapping mouse BRDT fragments from a Marathon-Ready testis cDNA library (Clontech). All PCR products were cloned in pCR2.1-TOPO vector (Invitrogen) by TA cloning. New PCRs were performed to fuse the fragments using Pfu DNA polymerase (Stratagene). The 2.05-kb DNA fragment, encoding the short version of BRDT (sBRDT) open reading frame, was digested with EcoRI and NotI (present in the 5′ and 3′ primers) and cloned into pcDNA3.1/HA C vector (Invitrogen).
Plasmid constructs.
Deletion and point mutations were generated by PCR and confirmed by DNA sequencing. The δC-sBRDT mutant, corresponding to the N-terminus-containing double-bromodomain region (amino acids 1 to 443), was obtained during the first amplification from the testis library using primers 5′-CGTCGAATTCCAGGATCCGAAGTGGTTGAGAATG-3′ (sense) and 5′-ATAGTTTAGCGGCCGCGAATGGAACTTGGGACAGAACTTGCAGC-3′(antisense). For subsequent mutants, pcDNA-HA-δC-sBRDT was used as the template. Mutant m5 was constructed by deleting amino acids 392 to 443, mutant m6 was constructed by deleting amino acids 1 to 13, and mutant m1 was constructed by deleting amino acids 1 to 13 and 392 to 443. Mutant Brd1 included the first bromodomain and corresponded to amino acids 14 to 141, and mutant Brd2 included the second bromodomain and corresponded to amino acids 254 to 383. Mutant Brd1-2 corresponded to Brd1 and Brd2 fused together.
Site-directed mutations were introduced with the QuikChange site-directed mutagenesis kit (Stratagene), using δC-sBRDT DNA as a template. Conserved amino acids present in the two bromodomains were mutated. Specifically, P50, F51, and V55 in the first bromodomain were mutated (Brd1pfv mutant), and P293, F294, and V298 in the second bromodomain were mutated (Brd2pfv mutant). Mutations were then confirmed by DNA sequencing.
Peptide pull-down assays.
Cos7 cells were transiently transfected by the following constructions: HA-δC-sBRDT, HA-Brd1pfv, and HA-Brd2pfv. Twenty-four hours posttransfection, cells were harvested and disrupted by incubation in lysis buffer (20% glycerol, 3 mM MgCl2, 50 mM HEPES [pH 7.9], 500 mM KCl, 0.1% NP-40, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride [PMSF]) during 30 min on ice. After centrifugation, the supernatant was recovered, and the extracts were incubated with biotinylated peptides corresponding to the N-terminal tail of histone H4, either not modified or tetra-acetylated (Upstate) and bound to streptavidin-agarose beads, in incubation buffer (20% glycerol, 3 mM MgCl2, 50 mM HEPES [pH 7.9], 150 mM KCl, 0,1% NP-40, 1 mM DTT, 1 mM PMSF) for 1 h, and the beads were washed in incubation buffer. The proteins were then eluted from the agarose beads and analyzed by Western blotting using antihemagglutinin (anti-HA) antibody and horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G (IgG).
Spermatogenic cell fractionation.
Our spermatogenic cell fractionation technique was derived from the method of Bellvé (3). After euthanasia, the testes of two adult mice were dissected, and the albuginea were removed. The seminiferous tubules were released by a 10-min incubation at 37°C in 10 ml of a collagenase solution (Sigma) (1 mg/ml in phosphate-buffered saline [PBS]) in a conic 50-ml tube. The tubules were then allowed to sediment at the bottom of the tube, and the collagenase solution was replaced by 2 ml of the sedimentation medium (Ham F-12 medium or Dulbecco's modified Eagle medium [DMEM] [GIBCO]) containing 0.5% bovine serum albumin (BSA) (Sigma) and DNase (2 μg/ml) (Sigma). The germinal and Sertoli cells were then released from the tubules by pipetting for approximately 10 min with a glass Pasteur pipette until a homogenous cell suspension was obtained, which was then filtered twice using 100-μm-pore-size filters (Becton Dickinson). The cells were resuspended in 18 ml of sedimentation solution and laid on a 360-ml BSA gradient (2 to 4%) in an airtight sedimentation chamber at 4°C. The gradient was obtained by gently filling the chamber with two 180-ml solutions of 2 and 4% BSA in Ham F-12 medium or DMEM, using a tap system which enables the two solutions to mix before they enter the chamber, avoiding air bubbles. The cells were allowed to sediment at 4°C for 70 min. Ten-milliliter fractions were then collected on ice and centrifuged (134 × g for 10 min at 4°C), the cells present in each collected fraction were identified by observation under a phase-contrast microscope, and the fractions were pooled in order to obtain cell suspensions enriched in pachytene spermatocytes, round and elongating spermatids, and condensing spermatids. Each cell suspension was then washed again in PBS at 4°C (10 min at 134 × g) and subjected to RNA extraction for Northern blots. Three sedimentation procedures had to be performed (and the corresponding fractions kept at −20°C before being pooled together) in order to prepare one Northern blot membrane.
Northern blot analysis.
Total RNA was extracted, using Tri reagent (Sigma), either from several mouse tissues or from pools of cell fractions obtained from three spermatogenic cell fractionations. Total RNA, 10 μg for each fraction, was subjected to electrophoresis in a denaturing 1% agarose gel containing 2 M formaldehyde, transferred to a nitrocellulose membrane (Hybond N+; Amersham), and incubated with 32P-labeled probe at 60°C in sodium phosphate buffer (pH 7) and 8% sodium dodecyl sulfate (SDS). The RNA blot was then washed at 60°C in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% SDS. Ethidium bromide staining of the 18S and 28S RNA was used to indicate that equal loads of the samples were used.
δC-sBRDT production and purification.
δC-sBRDT-encoding cDNA was amplified by PCR using primers 5′-CGTCGAATTCCAGGATCCGAAGTGGTTGAGAATG-3′ (sense) and 5′-ATAGTTTAGCGGCCGCGAATGGAACTTGGGACAGAACTTGCAGC-3′ (antisense). The product was digested with EcoRI and NotI and cloned into the cleaved pET-28a+ (Novagen) expression vector. This construct was used to transform Escherichia coli BL21(DE3) cells. Cells were grown in Luria-Bertani (LB) medium, induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (until an optical density at 600 nm of 0.6), and incubated for 3 h. Cells were then harvested and resuspended in lysis buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 10 mM imidazole, 1 mM PMSF) and then disrupted by sonication. The cell extract was clarified by centrifugation, and the supernatant was mixed with 50% Ni2+-nitrilotriacetic acid (Ni-NTA; Qiagen) resin equilibrated with lysis buffer. The suspension was mixed gently for 1 h at 4°C, and after centrifugation (2 min at 2,000 rpm), the resin was washed four times in wash buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 20 mM imidazole, 1 mM PMSF). Bound proteins were then eluted with the same buffer containing 250 mM imidazole. The purity of the preparation was checked by SDS-polyacrylamide gel electrophoresis and Coomassie blue staining. The purified His-δC-sBRDT was then dialyzed against buffer D (60 mM KCl, 15 mM NaCl, 15 mM Tris-HCl [pH 8], 0.34 M sucrose, 0.65 mM spermidine, 1 mM DTT, 0.5 mM PMSF, 10% glycerol) before being used for in vitro remodelling.
In vitro remodelling.
The in vitro remodelling activity of BRDT was assessed using nuclei isolated from murine erythroleukemia (MEL) cells (Friend cells) or BALB/c 3T3 cells. Cells in culture were either not treated or treated with trichostatin A (TSA) (100 ng/ml) for 12 h, harvested, and washed with PBS. Nuclei were purified as follows: approximately 5 × 106 cells were lysed by incubation in lysis buffer (0.05% Triton X-100, 60 mM KCl, 15 mM NaCl, 15 mM Tris-HCl [pH 8], 0.34 M sucrose, 2 mM EDTA, 0.5 mM EGTA, 0.65 mM spermidine, 1 mM DTT, 1 mM PMSF, 10% glycerol) for 5 min on ice. After centrifugation, the pellet (containing the nuclei) was recovered. For each remodelling assay, 105 control or TSA-treated nuclei were incubated with 5 μg of recombinant δC-sBRDT for 1 h at room temperature. Incubation was performed in the presence of either 1 mM ATP (Amersham) or 5 U of apyrase (Sigma). The mixture was then placed on glass slides, fixed with 4% paraformaldehyde (Sigma), and examined under a microscope after 4′,6′-diamidino-2-phenylindole (DAPI) staining (250 ng/ml).
Cell lines, transfection, and cell treatment.
Cos7 cells were cultured in monolayers in DMEM (GIBCO) supplemented with penicillin, streptomycin, and 10% fetal calf serum (FCS) (GIBCO). Cells at 50% confluence in one-well coverslips were transfected with 2 μg of DNA using the FuGENE 6 reagent (Roche). Twelve hours after transfection, cells were treated with TSA (100 ng/ml) for 12 h. Twenty-four hours after transfection, cells were washed twice with PBS, and immunofluorescence staining was performed.
MEL cells (Friend cells) were cultured in suspension in Eagle minimum essential medium (BioWhittaker), supplemented with 10% FCS, 5% glutamine, and 1% penicillin-streptomycin. BALB/c 3T3 cells were cultured in monolayers in RPMI 1640 medium (BioWhittaker), supplemented with 10% FCS, 10% glutamine, and 1% penicillin-streptomycin.
Immunofluorescence staining.
Transfected cells (Cos7) grown on coverslips were fixed in 4% paraformaldehyde for 1 min at room temperature and then permeabilized in 4% paraformaldehyde-0.1% Triton X-100 for 1 min at room temperature. Following incubation with blocking buffer (PBS containing 5% skim milk) for 30 min, rabbit antibody against the HA tag (Santa Cruz) diluted 1:1,000 (in blocking buffer) was added to the coverslips, which were incubated for 1 h at 37°C. The cells were then washed in PBS and further incubated with Alexa 488-conjugated anti-rabbit IgG (Molecular Probes) for 30 min at 37°C. After three washes in PBS, the cells were counterstained with Hoechst stain (250 ng/ml) and examined under a fluorescence microscope.
Microscopy.
Cell preparations were first analyzed with an epifluorescence microscope (Axiophot; Zeiss) using a 63× oil immersion lens objective. For three-dimensional analysis, a confocal laser scanning microscope (LSM 510-NLO; Zeiss), equipped with a 40× oil immersion lens objective (numerical aperture of 1.3), was used. Following excitation at the appropriate wavelengths (488 nm with an argon ion laser, 543 nm with a HeNe laser, and 750 nm with a Ti-Saphir laser [SpectraPhysics]) of the fluorescein isothiocyanate (FITC), tetramethyl rhodamine isothiocyanate (TRITC), and Hoechst signals, stacks of 128 × 128 pixel size images (with 0.1 μm between the images) (with a zoom of 3.9×, one pixel = 0.1 μm) were recorded. Images were averaged eight times. The optical sections were then transferred to a Unix workstation (Silicon Graphics) where three-dimensional images were obtained and analyzed using the appropriate software (Edit3D [36]).
Western blots.
Samples were subjected to electrophoresis on an SDS-8% polyacrylamide gel and transferred to nitrocellulose membranes (Hybond C+; Amersham) by standard procedures. Following a 30-min incubation in PBS containing 10% skim milk and 0.3% Tween (blocking solution), the blots were incubated for 1 h at room temperature with primary antibody diluted in PBS containing 10% FCS and 0.2% Tween. After three 10-min washes at room temperature, the blots were incubated with horseradish peroxidase-conjugated secondary antibody (goat anti-rabbit IgG; Amersham) diluted 1/5,000 in PBS containing 1% dry milk. The final detection was performed using the ECL+ kit (Amersham).
RESULTS
Identification and cloning of testis-specific bromodomain-containing proteins.
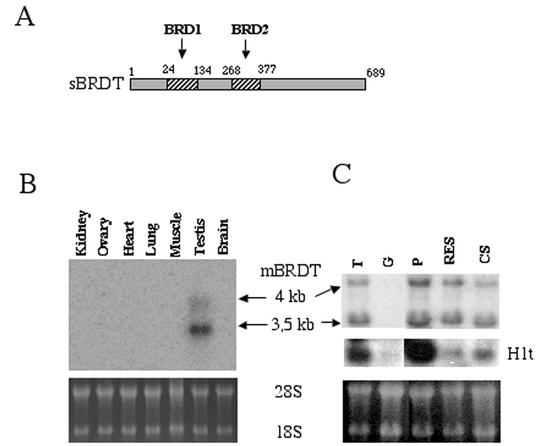
We considered testis-specific bromodomain-containing proteins as candidate factors capable of acting on acetylated chromatin during spermiogenesis. To identify testis-specific bromodomain-containing proteins, we conducted a search of dbEST and dbEST Cumulative Updates using the GCN5 bromodomain as a query. Of all the positive clones, only those present only in a testis cDNA library were finally considered. One of these ESTs corresponded to an already cloned cDNA of unknown function (the protein encoded by this cDNA named BRDT for bromodomain testis-specific protein) (23). We then cloned the murine BRDT cDNA fragments, using oligonucleotides designed on the basis of the human BRDT sequence. Several cDNAs were amplified from a mouse testis library and sequenced, leading to the identification of two splicing products. This was confirmed by Northern blots of different mouse tissues, which showed two BRDT transcripts with sizes of approximately 4 and 3.5 kb, specifically expressed in the adult mouse testis (Fig. 1B). Interestingly, we have noticed that the alternative splicing removes a PEST sequence known to confer protein instability and gives rise to a shorter protein, which should therefore be more stable (not shown). Using RNA purified from germinal cells enriched at successive stages of spermatogenesis by gravity sedimentation, the expression of BRDT was monitored during spermatogenesis. Figure 1C shows that both BRDT transcripts were expressed in germinal cells from the early meiotic (pachytene) spermatocytes (P lanes) and during spermiogenesis in the round and elongating spermatids (RES lanes) until the condensed late spermatids (CS lanes). These transcripts were absent from 6-day-old mouse testis, containing Sertoli cells and spermatogonia only (G lanes), indicating that BRDT was not expressed in spermatogonia. As shown previously, the H1t transcript, encoding a testis-specific linker histone variant associated with postmeiotic cell chromatin (24), was found essentially in the pachytene cell population, providing a good indication of the efficiency of our fractionation procedure. Figure 1C also shows that during the postmeiotic maturation of cells, the smaller BRDT-encoding transcript (3.5 kb) remains the most abundant one, especially in condensing spermatids (CS lanes) where histone hyperacetylation occurs.
FIG. 1.
Murine BRDT is specifically expressed during spermatogenesis. (A) The cDNA corresponding to the most abundant BRDT mRNA was cloned and sequenced. The schematic representation shows the domain organization of the cDNA-encoded protein (sBRDT) and positions of the two bromodomains (BRD1 [amino acids 24 to 134] and BRD2 [amino acids 268 and 377]) are indicated. (B and C) BRDT transcripts were detected by Northern blots of several mouse tissues (B) and of mouse germinal cell populations (C), including 6-day-old mouse testis containing spermatogonia and Sertoli cells (G), pachytene spermatocytes (P), round and elongating spermatids (RES), and condensed spermatids (CS). RNA from adult mouse testis (T) was also used. The blot was reprobed with a H1t probe. Ethidium bromide staining of 18S and 28S RNA in the gels prior to transfer is shown at the bottom.
TSA-dependent chromatin remodelling in BRDT-expressing cells.
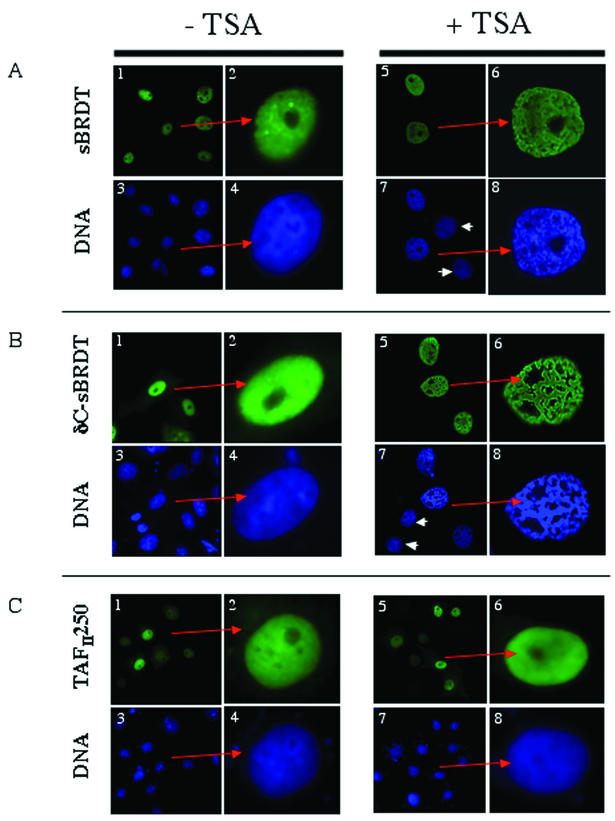
We then investigated whether BRDT could modify the global organization of chromatin in an acetylation-dependent manner. A HA-tagged short version of the protein (sBRDT), corresponding to the mRNA most abundantly expressed in spermatogenic cells, was expressed in somatic cells, which were then treated with TSA in order to induce histone hyperacetylation. In this experiment, we also expressed a HA-tagged truncated BRDT fragment (called δC-sBRDT here) (corresponding to amino acids 1 to 443), obtained during the cloning of BRDT from the testis cDNA library. This fragment lacked the 246 C-terminal amino acids of sBRDT.
In the absence of TSA treatment, either sBRDT or δC-sBRDT was expressed in the nucleus of almost all transfected cells, and no particular modification of the chromatin organization was observed compared with nontransfected cells (Fig. 2A and B, panels 1 to 4). Very interestingly, when cells were treated with TSA, a reorganization of chromatin was observed only in BRDT-expressing cells (Fig. 2A and B, panels 5 to 8). This phenomenon was strongly enhanced in cells expressing δC-sBRDT. Indeed, a dramatic remodelling of chromatin was visualized with DAPI staining in most of the TSA-treated transfected cells expressing δC-sBRDT (Fig. 2B, panels 5 to 8). Such chromatin remodelling was not observed either in TSA-treated nontransfected cells (Fig. 2A and B, panels 7, arrowheads) or in transfected but untreated cells (Fig. 2A and B, panels 1 to 4).
FIG. 2.
TSA-dependent chromatin reorganization after ectopic expression of sBRDT. Cos7 cells were transfected by vectors expressing the indicated proteins, and cells were either treated with TSA (100 ng/ml) (+ TSA) or not treated with TSA (− TSA). A higher magnification of a transfected cell is shown in the right panels (indicated by a red arrow) to better visualize the chromatin reorganization. Arrowheads show TSA-treated nontransfected cells.
These data strongly suggested that sBRDT is able to induce global remodelling of acetylated chromatin and that the removal of a fragment from its C-terminal region considerably enhances this ability. The TSA-dependent chromatin remodelling induced by sBRDT or δC-sBRDT was specific, since the expression of TAFII250, which also contains two bromodomains, did not induce any observable TSA-dependent reorganization of the chromatin (Fig. 2C, panels 1 to 8).
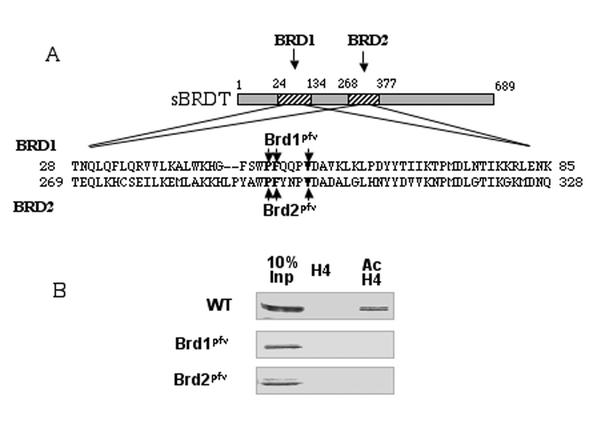
The rationale behind the cloning of BRDT was the assumption that its bromodomains were involved in the reorganization of acetylated chromatin. To confirm this hypothesis, three amino acids known to play an essential role in the structure of P/CAF (9), Gcn5p (35), and TAFII250 (21) bromodomains and conserved in most of the known bromodomains were replaced by alanines in each of the two BRDT bromodomains individually (Fig. 3A, mutants Brd1pfv and Brd2pfv). In order to facilitate our studies, these mutations were introduced in δC-sBRDT, because of its enhanced capacity to induce a TSA-dependent chromatin remodelling. First, a peptide pull-down assay was set up to monitor the ability of the protein to specifically bind a fully acetylated histone H4 tail. δC-sBRDT or proteins bearing a mutation in bromodomain 1 (Brd1pfv) or 2 (Brd2pfv) were expressed in cells. Extracts from these cells were used to monitor the binding of the expressed proteins to beads bound to a peptide corresponding to the histone H4 N-terminal tail or to the same peptide bearing four acetylated lysines. Figure 3B shows that, while wild-type δC-sBRDT efficiently interacted with the acetylated histone H4 tail, no interaction was observed between this protein and the nonmodified histone H4 tail. Mutations of each of the bromodomains abolished the interaction of the protein with the acetylated peptides, thereby confirming the important role of both bromodomains in the recognition of the acetylated histone tail.
FIG. 3.
The two bromodomains of sBRDT are involved in the specific recognition of the acetylated histone H4 tail. (A) Schematic representation of sBRDT proteins bearing mutations in the first bromodomain (Brd1pfv) or in the second bromodomain (Brd2pfv) were expressed in Cos7 cells. In the Brd1pfv mutant, P50, F51 and V55 were replaced by A. In the Brd2pfv mutant, P293, F294, and V298 were replaced by A (the mutations are shown in boldface type). (B) Wild-type (WT) or mutant δC-sBRDT proteins bearing mutations in the first bromodomain (Brd1pfv) or in the second bromodomain (Brd2pfv) were expressed in Cos7 cells. Twenty-four hours posttransfection, an extract was prepared and subjected to a peptide pull-down assay using beads bound to either nonmodified or hyperacetylated (Ac) histone H4 N-terminal tails. Proteins retained on the beads were then analyzed by Western blotting using an anti-HA antibody. The Inp lane contains 10% of the input material.
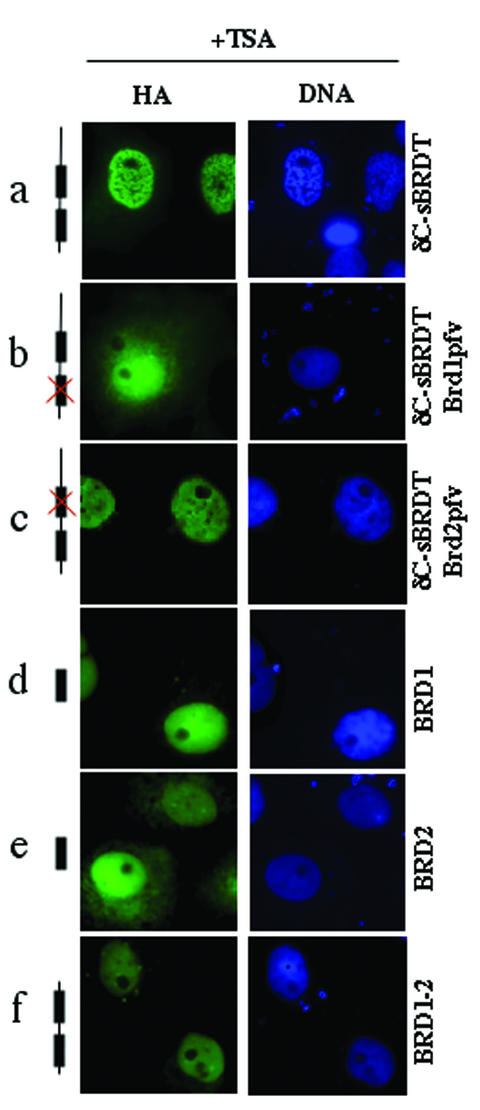
We then analyzed the capacity of δC-sBRDT and its bromodomain mutants to induce a TSA-dependent chromatin remodelling. Figure 4 shows that the inactivation of the first bromodomain of δC-sBRDT leads to an almost homogenous distribution of the protein in both nuclear and cytoplasmic compartments of the cells and that the mutated protein could not induce chromatin remodelling upon TSA treatment (Fig. 4b). A δC-sBRDT protein containing a mutation in the second bromodomain retained a weak TSA-dependent chromatin remodelling activity (Fig. 4c). We also investigated whether the expression of bromodomains isolated from BRDT, individually or together, could induce a reorganization of acetylated chromatin. Figure 4d to f show that none of the isolated bromodomains (BRD1 and BRD2 in Fig. 4d and e, respectively) or linked together (BRD1-2 in Fig. 4f) could modify the organization of chromatin in TSA-treated cells.
FIG. 4.
The integrity of the two bromodomains is necessary but not sufficient to induce TSA-dependent reorganization of the chromatin. Cos7 cells were transfected with vectors expressing the indicated proteins. Twenty-four hours posttransfection, protein expression and chromatin reorganization were visualized in control cells (not shown) or cells treated with TSA.
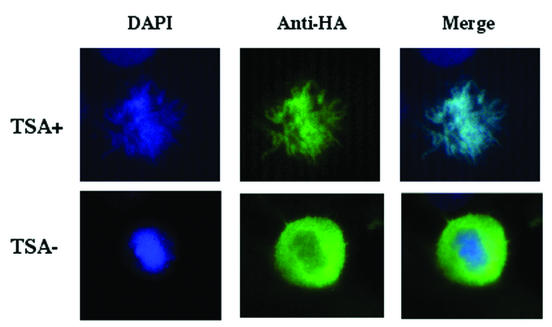
Furthermore, we also investigated whether TSA treatment would allow the association of the protein with the chromatin of condensed mitotic chromosomes. Cells were transfected with δC-sBRDT and treated with TSA or not treated with TSA, and transfected cells in mitosis were examined. Figure 5 shows that, after TSA treatment, δC-sBRDT remained tightly associated with mitotic chromosomes (Fig. 5, TSA+ anti-HA panel), while in nontreated cells, as occurs for many chromatin-associated factors, BRDT was excluded from condensing chromosomes (Fig. 5, TSA− anti-HA panel). It is noteworthy that it was very difficult to find BRDT-expressing cells in mitosis, as if BRDT expression were interfering with the progression of cells in the cell cycle.
FIG. 5.
BRDT remains associated with mitotic chromosomes in TSA-treated cells. Cos7 cells expressing δC-sBRDT were left untreated (TSA−) or treated for 6 h with TSA (TSA+), and the localization of the protein was recorded in mitotic cells. The TSA+ panels show a tight association of BRDT with the mitotic chromosomes, whereas in the absence of TSA, BRDT appears to be excluded from the condensing chromosomes.
Altogether, these data show that sBRDT is a regulatory protein efficiently recruited into chromatin upon histone hyperacetylation and capable of controlling a TSA-dependent chromatin remodelling through a direct interaction of its bromodomains with the acetylated histone tails and that the integrity of both bromodomains is necessary, but not sufficient, for its activity.
Domains flanking the two bromodomains of BRDT are also involved in the ability of the protein to induce acetylation-dependent chromatin reorganization.
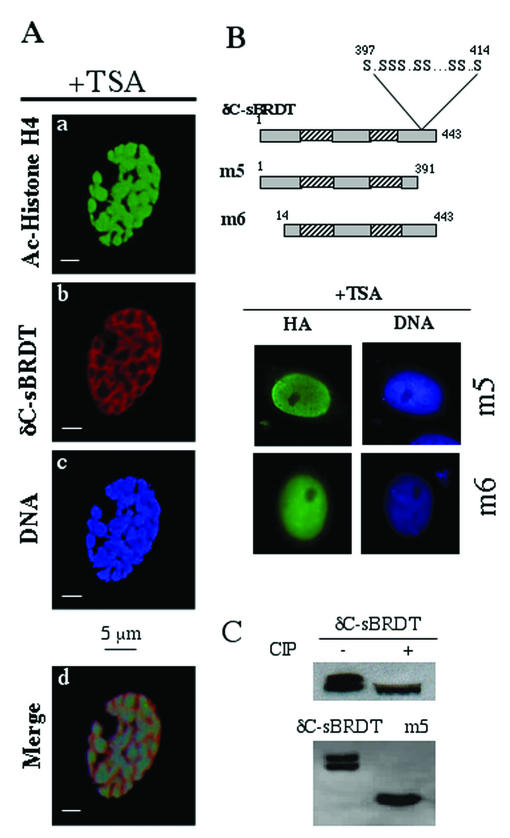
In order to obtain insight into the molecular basis of the activity of BRDT on acetylated chromatin, we first used a scanning confocal microscope to examine the nature of the reorganized chromatin in δC-sBRDT-expressing cells after TSA treatment. Figure 6A shows that the expression of the protein induced the appearance of highly compact chromatin regions containing hyperacetylated histones (Fig. 6A, panels a and c) which were, interestingly, covered and surrounded by δC-sBRDT (panels b and d).
FIG. 6.
Chromatin remodelling by BRDT depends on bromodomains and flanking regions. (A) Confocal analysis of the chromatin in BRDT-expressing and TSA-treated cells shows that δC-sBRDT (shown in red in panel b) is distributed around the condensed hyperacetylated chromatin regions (detected by an anti-acetylated H4 antibody [shown in green in panel a] and also visible after DAPI staining [panel c]). The images of panels a, b, and c were merged (panel d). (B) Deletions of the 52 amino acids in the C-terminal serine-rich region of δC-sBRDT (m5 mutant) or the 13 N-terminal amino acids of δC-sBRDT (m6 mutant) both inhibit the δC-sBRDT's remodelling activity of acetylated chromatin in overexpressing TSA-treated Cos7 cells. (C) Western blots show that δC-sBRDT appears as a double band and as a single band after CIP treatment, indicating a phosphorylation of the protein. The serine-rich region of δC-sBRDT removed in the m5 mutant appears to be the site of phosphorylation, since in contrast to δC-sBRDT, this mutant migrates as a single band (this band was not sensitive to CIP treatment [not shown]).
This observation suggests that acetylated histones may first be recognized by BRDT, which would then undergo a process of protein-protein interaction leading to the compaction of the acetylated chromatin regions. One prediction of this hypothesis is that the deletion of regions surrounding the bromodomains would impair the capacity of the protein to induce this TSA-dependent chromatin compaction. In order to test this hypothesis, new mutants of δC-BRDT were used to analyze the role of the bromodomain-flanking regions. Figure 6B shows that the deletion of the 13 amino acids in the region N-terminal to the first bromodomain (m6 mutant) did indeed abolish the remodelling activity of the protein. Interestingly, the remodelling activity of δC-sBRDT was also dependent on a limited region in the C-terminal part of the protein. Indeed, the removal of 52 amino acids in the C-terminal part of δC-sBRDT (m5 mutant) completely abolished the TSA-dependent reorganization of the chromatin. All the transfection experiments described above were associated with a Western blot analysis of the expression of δC-sBRDT and its mutants (not shown). Interestingly, δC-sBRDT appeared as a doublet after SDS-polyacrylamide gel electrophoresis, whereas after the removal of the serine-rich C-terminal region (mutant m5), the protein appeared as a single band (Fig. 6C), suggesting that this serine-rich region could contain one or several sites of phosphorylation (Fig. 6B). This hypothesis was confirmed after treatment with the calf intestinal alkaline phosphatase (CIP) of an extract containing δC-sBRDT. Figure 6C shows that, after CIP treatment, the protein did indeed migrate as a single band. These data suggest that both C- and N-terminal regions flanking the bromodomains are essential for the TSA-dependent remodelling activity of the protein and that this activity could also be linked to a phosphorylation signaling pathway. However, the role of this phosphorylation is not clear, since we show (below) that a bacterially expressed recombinant δC-sBRDT can induce the remodelling of acetylated chromatin in the absence of ATP.
Acetylation-dependent reorganization of the chromatin in vitro.
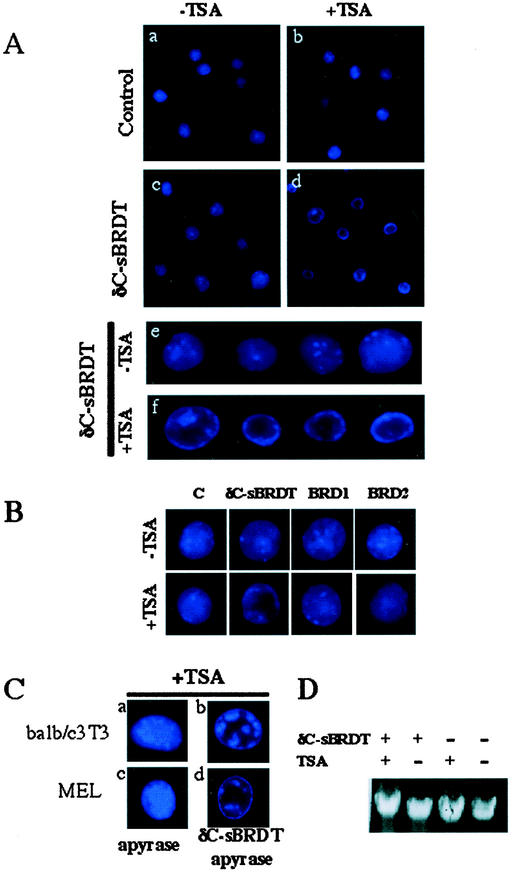
In pursuing our investigations of the molecular basis of the reorganization of acetylated chromatin by BRDT, we wondered whether the recombinant protein would also be able to induce acetylation-dependent remodelling of nuclei in vitro. Purified nuclei, isolated from untreated or TSA-treated MEL cells, were incubated with recombinant δC-sBRDT for 1 h, and the nuclei were examined under a microscope after DAPI staining. Figure 7A shows that δC-sBRDT induces condensation of the chromatin in almost all the nuclei from TSA-treated cells (Fig. 7A, panel d) but not in nuclei from untreated cells (Fig. 7A, panel c), which remain unchanged (higher magnifications are shown in Fig. 7A, panel e [nuclei from untreated cells] and panel f [nuclei from TSA-treated cells]). However, the aspect of this chromatin condensation was different from that observed in δC-sBRDT-expressing cells in vivo. Indeed, the regions of chromatin condensation were observed mostly at the periphery of the nuclei. Neither of the isolated recombinant bromodomains of BRDT (added at the same concentration) could induce chromatin condensation in the nuclei isolated from TSA-treated MEL cells under the same conditions (Fig. 7B, BRD1 and BRD2 panels). Nuclei from BALB/c 3T3 cells, treated with TSA or not treated with TSA, were also used to monitor the in vitro chromatin remodelling activity of recombinant δC-sBRDT. Indeed, we were particularly interested in knowing whether the distinct and visible heterochromatin regions present in these cells could also be modified by BRDT. Figure 7C shows that the incubation of nuclei from TSA-treated 3T3 cells with recombinant δC-sBRDT induced very clear chromatin condensation around the heterochromatin regions (compare panels a and b). As expected, BRDT had no effect on the nuclei isolated from TSA-untreated cells (not shown).
FIG.7.
Recombinant δC-sBRDT induced in vitro remodelling of hyperacetylated chromatin. (A) A total of 105 purified nuclei from untreated (−TSA) or TSA-treated (+TSA) MEL cells were incubated alone (control [panels a and b]) or with 5 μg of recombinant δC-sBRDT (panels c and d) for 1 h, DAPI stained, and examined under a microscope. Panels e and f show higher-magnification views of the structures of several nuclei present in panels c and d, respectively. (B) δC-sBRDT induces an in vitro remodelling of hyperacetylated MEL nuclei, which is absent from control (C) nuclei, and not detected in hyperacetylated nuclei (+TSA) treated with the same amount of recombinant proteins containing isolated bromodomains (BRD1 or BRD2). (C) In vitro remodelling of nuclei from BALB/c 3T3 or MEL cells containing hyperacetylated chromatin in the absence of ATP. Nuclei were incubated either with apyrase alone or with apyrase and recombinant δC-sBRDT. (D) δC-sBRDT treatment is not associated with the degradation of DNA. Nuclei from MEL cells were either treated with TSA (+) or not treated with TSA (−) and then incubated with (+) or without (−) δC-sBRDT as described above. DNA was then purified and analyzed on an agarose gel.
These assays were also used to determine whether this acetylation-dependent chromatin reorganization induced by BRDT required ATP. Nuclei isolated from TSA-treated MEL cells and BALB/c 3T3 cells were incubated with δC-sBRDT in the absence of ATP and in the presence of apyrase in order to deplete the extracts of residual amounts of ATP. Surprisingly, the chromatin condensation property of δC-sBRDT was independent of ATP, since the same reorganization of acetylated chromatin was observed when nuclei from TSA-treated cells were incubated with the recombinant protein in the presence of apyrase (Fig. 7C).
When nuclei from TSA-treated MEL or BALB/c cells were incubated with δC-sBRDT, besides the condensed regions of chromatin, vast regions devoid of DNA appeared in the nuclei. This phenomenon could have been caused by the presence of nucleases in the δC-sBRDT preparations. Although this hypothesis was unlikely (because the phenomenon was not observed in nuclei from untreated cells incubated with δC-sBRDT), we analyzed the genomic DNA from the nuclei isolated from either untreated or TSA-treated MEL cells, incubated or not with δC-sBRDT. Figure 7D shows no sign of DNA degradation by δC-sBRDT. We therefore conclude that the appearance of regions devoid of chromatin observed in the nuclei from TSA-treated cells incubated with δC-sBRDT is due to the extreme compaction of the chromatin by the protein.
The results of these experiments strongly suggest a structural role for BRDT in the reorganization of acetylated chromatin, as opposed to an enzymatic activity.
Altogether, our data present the protein BRDT as the first identified factor capable of inducing condensation of acetylated chromatin, in vivo and in vitro.
DISCUSSION
The most dramatic chromatin remodelling occurs during the postmeiotic maturation of male germinal cells (or spermiogenesis). Indeed, in vertebrates, this remodelling corresponds to the replacement of most of the core histones by transition proteins and protamines. Interestingly, this histone replacement is associated with their hyperacetylation, suggesting a link between the two events. We therefore considered the testis as a source of factors able to recognize acetylated chromatin and participate in the remodelling of acetylated chromatin. An approach based on the analysis of EST databases led us to consider a double-bromodomain-containing protein of unknown function, already identified and named BRDT (23). This protein is a member of the bromodomain and extraterminal protein family, all containing one or two N-terminal bromodomain(s). This family includes several somatic members, and some have been functionally characterized (12). One of these somatic members is human Ring3, which interacts with several transcription factors, including E2F, depending on the presence of acetylated histone H3 and H4 tails (7). The mouse homologue of Ring3, Fsrg1, was found to be associated with euchromatin, reinforcing the idea of the association of this protein with the control of transcription (7). Moreover, another member of this family, Brd4 was found to interact with replication factor C and to inhibit DNA replication in vivo, after overexpression, as well as in vitro (29). In line with these findings, mice bearing null alleles of the Brd4 gene displayed pre- and postnatal growth defects associated with a reduced proliferation rate (18). Another characterized member of the family is MCAP. This mouse protein is particularly tightly bound to chromatin, since it remains associated with mitotic chromosomes when most regulatory proteins are released from chromatin (8).
The yeast homologue of MCAP, BDF1, is also associated with meiotic and mitotic chromosomes (6), also interacts with general transcription factors (30), and regulates transcription (28). Interestingly, very recently, BDF1 has been shown to bind H4 with a preference for multiply acetylated forms, whereas the closely related protein BDF2 could bind H4 with no preference for its acetylated forms, suggesting that bromodomains with different specificities could add further complexity to the histone code (31). Moreover, other recent data have shown that a deletion of BDF1 or a BDF1 mutation that abolishes histone binding leads to the transcriptional down-regulation of genes located at heterochromatin-euchromatin boundaries and that BDF1 competes with the Sir2 deacetylase for binding to acetylated H4, suggesting an active role for BDF1 in euchromatin maintenance and antisilencing (26).
BRDT is also a double-bromodomain-containing member of this family, which is expressed only in the testis (23). However, interestingly, BRDT was also expressed in 12 of 47 cases of non-small cell lung cancer, and its mRNA was found in one case of squamous cell carcinoma of the head and neck and esophagus (38). Moreover, in a highly lethal form of epithelial tumor, the poorly differentiated carcinoma, a chromosome translocation t(15,19)(q13, p13.1) causes the fusion of the BRDT homologue, BRD4, to the novel testis-specific protein NUT (nuclear protein in testis), resulting in a fusion oncogene and deregulation of the activities of both proteins (13). All these observations strongly suggest that the aberrant expression of double-bromodomain BRDT-like proteins may be tightly associated with the appearance of specific type of cancers.
Our analysis shows that BRDT is capable of specifically and efficiently interacting with acetylated histone H4 tail. This property of BRDT is dependent on the integrity of both bromodomains. It is therefore very probable that the two bromodomains of BRDT, like those of TAFII250, recognize at least two acetylated lysines, thereby contributing to its high-affinity binding (21). However, unlike TAFII250, BRDT possesses the extraordinary ability to induce condensation of acetylated chromatin in vivo and in vitro, suggesting that the binding of acetylated nucleosome is not sufficient for this activity and that other regions of BRDT participate to the phenomenon. Accordingly, our investigations show that, besides the two bromodomains, the reorganization of acetylated chromatin requires specific regions of BRDT and that this activity is also ATP independent. These observations strongly argue in favor of a structural role for BRDT in inducing the condensation of acetylated chromatin.
The most likely scenario would be a two-step mechanism based first on the recognition of the acetylated nucleosomes and then on a subsequent protein-protein interaction, bridging acetylated nucleosomes together and condensing the chromatin. Very interestingly, the ability of BRDT to condense acetylated chromatin could be observed in vitro using isolated nuclei containing hyperacetylated histones. Several proteins have been shown to be capable of reorganizing chromatin in vitro. One of these protein, DFF, is a caspase-activated nuclease involved in the internucleosomal DNA cleavage and chromatin condensation during apoptosis. It has been shown that the recombinant activated form of the protein triggers chromatin condensation when added to nuclei isolated from nonapoptotic cells in an ATP-independent manner (27, 42). The other example comes from a protein involved in the formation of heterochromatin in chicken granulocytes, MENT, capable of enhancing the compaction and reversible Mg2+-dependent self association of nucleosome arrays (14). However, BRDT is the only known protein capable of inducing chromatin compaction, which is specifically acetylation dependent. Data available on the interaction of the two bromodomains of TAFII250 with acetylated histone H4 show the ability of the bromodomains to contact two different lysines. Accordingly, BRDT would also need at least two acetylated lysines to efficiently bind to acetylated histones. A detailed analysis of histone acetylation during mouse spermatogenesis shows that the acetylation status of histone H3 does not change significantly during spermiogenesis. In contrast, histone H4 becomes maximally acetylated in elongating spermatids (our unpublished data). These observations suggest that BRDT would have only a limited period of time in stage 9 to 11 spermatids (17) to efficiently bind the nucleosomes. It is therefore possible that, upon the binding to hyperacetylated chromatin in elongating spermatids, BRDT recruits machinery to remove the acetylated histones. The characterization of BRDT is the first step towards the elucidation of the mechanism underlying the replacement of acetylated histones in spermatids, and further identification of BRDT partners will certainly help to better understand the phenomenon. Moreover, these findings may also help to investigate the potential role of BRDT in somatic cell carcinogenesis associated with its aberrant expression.
Acknowledgments
This work was supported by the INSERM Assistance Médicale à la Procréation (AMP) and region Rhône-Alpes emergence programs. C.C. is supported by the INSERM delegation program.
We are grateful to Sandrine Curtet-Benitski for technical assistance.
REFERENCES
- 1.Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111:381-392. [DOI] [PubMed] [Google Scholar]
- 2.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 3.Bellvé, A. R. 1993. Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol. 225:84-113. [DOI] [PubMed] [Google Scholar]
- 4.Christensen, M. E., and G. H. Dixon. 1982. Hyperacetylation of histone H4 correlates with the terminal, transcriptionally inactive stages of spermatogenesis in rainbow trout. Dev. Biol. 93:404-415. [DOI] [PubMed] [Google Scholar]
- 5.Christensen, M. E., J. B. Rattner, and G. H. Dixon. 1984. Hyperacetylation of histone H4 promotes chromatin decondensation prior to histone replacement by protamines during spermatogenesis in rainbow trout. Nucleic Acids Res. 12:4575-4592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chua, P., and G. S. Roeder. 1995. Bdf1, a yeast chromosomal protein required for sporulation. Mol. Cell. Biol. 15:3685-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crowley, T. E., E. M. Kaine, M. Yoshida, A. Nandi, and D. J. Wolgemuth. 2002. Reproductive cycle regulation of nuclear import, euchromatic localization, and association with components of Pol II mediator of a mammalian double-bromodomain protein. Mol. Endocrinol. 16:1727-1737. [DOI] [PubMed] [Google Scholar]
- 8.Dey, A., J. Ellenberg, A. Farina, A. E. Coleman, T. Maruyama, S. Sciortino, J. Lippincott-Schwartz, and K. Ozato. 2000. A bromodomain protein, MCAP, associates with mitotic chromosomes and affects G2-to-M transition. Mol. Cell. Biol. 20:6537-6549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dhalluin, C., J. E. Carlson, L. Zeng, C. He, A. K. Aggarwal, and M. M. Zhou. 1999. Structure and ligand of a histone acetyltransferase bromodomain. Nature 399:491-496. [DOI] [PubMed] [Google Scholar]
- 10.Dorr, A., V. Kiermer, A. Pedal, H. R. Rackwitz, P. Henklein, U. Schubert, M. M. Zhou, E. Verdin, and M. Ott. 2002. Transcriptional synergy between Tat and PCAF is dependent on the binding of acetylated Tat to the PCAF bromodomain. EMBO J. 21:2715-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dyson, M. H., S. Rose, and L. C. Mahadevan. 2001. Acetyllysine-binding and function of bromodomain-containing proteins in chromatin. Front. Biosci. 6:D853-D865. [DOI] [PubMed]
- 12.Florence, B., and D. V. Faller. 2001. You bet-cha: a novel family of transcriptional regulators. Front. Biosci. 6:D1008-D1018. [DOI] [PubMed]
- 13.French, C. A., I. Miyoshi, I. Kubonishi, H. E. Grier, A. R. Perez-Atayde, and J. A. Fletcher. 2003. BRD4-NUT fusion oncogene: a novel mechanism in aggressive carcinoma. Cancer Res. 63:304-307. [PubMed] [Google Scholar]
- 14.Grigoryev, S. A., and C. L. Woodcock. 1998. Chromatin structure in granulocytes. A link between tight compaction and accumulation of a heterochromatin-associated protein (MENT). J. Biol. Chem. 273:3082-3089. [DOI] [PubMed] [Google Scholar]
- 15.Grimes, S. R., Jr., and N. Henderson. 1984. Hyperacetylation of histone H4 in rat testis spermatids. Exp. Cell Res. 152:91-97. [DOI] [PubMed] [Google Scholar]
- 16.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111:369-379. [DOI] [PubMed] [Google Scholar]
- 17.Hazzouri, M., C. Pivot-Pajot, A. K. Faure, Y. Usson, R. Pelletier, B. Sele, S. Khochbin, and S. Rousseaux. 2000. Regulated hyperacetylation of core histones during mouse spermatogenesis: involvement of histone deacetylases. Eur. J. Cell Biol. 79:950-960. [DOI] [PubMed] [Google Scholar]
- 18.Houzelstein, D., S. L. Bullock, D. E. Lynch, E. F. Grigorieva, V. A. Wilson, and R. S. Beddington. 2002. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol. Cell. Biol. 22:3794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs, S. A., and S. Khorasanizadeh. 2002. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295:2080-2083. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs, S. A., S. D. Taverna, Y. Zhang, S. D. Briggs, J. Li, J. C. Eissenberg, C. D. Allis, and S. Khorasanizadeh. 2001. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 20:5232-5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288:1422-1425. [DOI] [PubMed] [Google Scholar]
- 22.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 23.Jones, M. H., M. Numata, and M. Shimane. 1997. Identification and characterization of BRDT: a testis-specific gene related to the bromodomain genes RING3 and Drosophila fsh. Genomics 45:529-534. [DOI] [PubMed] [Google Scholar]
- 24.Khochbin, S. 2001. Histone H1 diversity: bridging regulatory signals to linker histone function. Gene 271:1-12. [DOI] [PubMed] [Google Scholar]
- 25.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 26.Ladurner, A. G., C. Inouye, R. Jain, and R. Tjian. 2003. Bromodomains mediate an acetyl-histone encoded antisilencing function at heterochromatin boundaries. Mol. Cell 11:365-376. [DOI] [PubMed] [Google Scholar]
- 27.Liu, X., P. Li, P. Widlak, H. Zou, X. Luo, W. T. Garrard, and X. Wang. 1998. The 40-kDa subunit of DNA fragmentation factor induces DNA fragmentation and chromatin condensation during apoptosis. Proc. Natl. Acad. Sci. USA 95:8461-8466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lygerou, Z., C. Conesa, P. Lesage, R. N. Swanson, A. Ruet, M. Carlson, A. Sentenac, and B. Seraphin. 1994. The yeast BDF1 gene encodes a transcription factor involved in the expression of a broad class of genes including snRNAs. Nucleic Acids Res. 22:5332-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maruyama, T., A. Farina, A. Dey, J. Cheong, V. P. Bermudez, T. Tamura, S. Sciortino, J. Shuman, J. Hurwitz, and K. Ozato. 2002. A mammalian bromodomain protein, Brd4, interacts with replication factor C and inhibits progression to S phase. Mol. Cell. Biol. 22:6509-6520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matangkasombut, O., R. M. Buratowski, N. W. Swilling, and S. Buratowski. 2000. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 14:951-962. [PMC free article] [PubMed] [Google Scholar]
- 31.Matangkasombut, O., and S. Buratowski. 2003. Different sensitivities of bromodomain factors 1 and 2 to histone H4 acetylation. Mol. Cell 11:353-363. [DOI] [PubMed] [Google Scholar]
- 32.Meistrich, M. L., P. K. Trostle-Weige, R. Lin, Y. M. Bhatnagar, and C. D. Allis. 1992. Highly acetylated H4 is associated with histone displacement in rat spermatids. Mol. Reprod. Dev. 31:170-181. [DOI] [PubMed] [Google Scholar]
- 33.Mujtaba, S., Y. He, L. Zeng, A. Farooq, J. E. Carlson, M. Ott, E. Verdin, and M. M. Zhou. 2002. Structural basis of lysine-acetylated HIV-1 Tat recognition by PCAF bromodomain. Mol. Cell 9:575-586. [DOI] [PubMed] [Google Scholar]
- 34.Oliva, R., and C. Mezquita. 1982. Histone H4 hyperacetylation and rapid turnover of its acetyl groups in transcriptionally inactive rooster testis spermatids. Nucleic Acids Res. 10:8049-8059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Owen, D. J., P. Ornaghi, J.-C. Yang, N. Lowe, P. R. Evans, P. Ballario, D. Neuhaus, P. Filetici, and A. A. Travers. 2000. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase Gcn5p. EMBO J. 19:6141-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parazza, F., C. Humbert, and Y. Usson. 1993. Method for 3D volumetric analysis in intranuclear fluorescence distribution in confocal microscopy. Comput. Med. Imaging Graph. 17:189-200. [DOI] [PubMed] [Google Scholar]
- 37.Polesskaya, A., I. Naguibneva, A. Duquet, E. Bengal, P. Robin, and A. Harel-Bellan. 2001. Interaction between acetylated MyoD and the bromodomain of CBP and/or p300. Mol. Cell. Biol. 21:5312-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Scanlan, M. J., N. K. Altorki, A. O. Gure, B. Williamson, A. Jungbluth, Y. T. Chen, and L. J. Old. 2000. Expression of cancer-testis antigens in lung cancer: definition of bromodomain testis-specific gene (BRDT) as a new CT gene, CT9. Cancer Lett. 150:155-164. [DOI] [PubMed] [Google Scholar]
- 39.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 40.Taverna, S. D., R. S. Coyne, and C. D. Allis. 2002. Methylation of histone H3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell 110:701-711. [DOI] [PubMed] [Google Scholar]
- 41.Turner, B. M. 2002. Cellular memory and the histone code. Cell 111:285-291. [DOI] [PubMed] [Google Scholar]
- 42.Widlak, P., O. Palyvoda, S. Kumala, and W. T. Garrard. 2002. Modeling apoptotic chromatin condensation in normal cell nuclei. Requirement for intranuclear mobility and actin involvement. J. Biol. Chem. 277:21683-21690. [DOI] [PubMed] [Google Scholar]
- 43.Zeng, L., and M. M. Zhou. 2002. Bromodomain: an acetyl-lysine binding domain. FEBS Lett. 513:124-128. [DOI] [PubMed] [Google Scholar]